Introduction
microRNAs (miRNA/miRs) are a conserved family of
small non-coding RNA molecules that are recognized as key
regulators of gene expression at the post-transcriptional level via
base pair binding to the binding sites of the 3′ untranslated
region (UTR) of target genes, leading to target gene mRNA cleavage
or translational repression (1,2).
Previous studies have shown that miRNAs are involved in diverse
cellular processes, including cell growth, development, apoptosis
and cancer (3). The observation
that ~50% of miRNAs are located in tumor-associated or fragile
regions validates the hypothesis that abnormal miRNA expression is
closely associated with cancer initiation and progression (4). A previous study also showed that ~60%
of protein-coding genes are regulated by miRNAs (5). Depending on the potential functions of
their targets in tumors, miRNAs may function as oncogenes or tumor
suppressors. For example, miR-125b inhibits liver cancer cell
growth and metastasis by targeting LIN28B, functioning as a tumor
suppressor (6). These results
indicate that miRNAs are crucial in cancer processes and may
present novel biomarkers for cancer diagnosis and progression.
Human colon cancer is one of the most common
malignancies and is the third leading cause of cancer-related
mortality worldwide (7). However,
the molecular mechanism underlying colon cancer growth and
progression remains unclear. Therefore, the identification of novel
molecules responsible for colon cancer development is crucial.
Recent studies have demonstrated that the abnormal expression of
certain miRNAs, including miR-145 (8), miR-203 (9) and miR-365 (10), is involved in colon cancer. However,
the function of miR-214 in colon cancer has not yet been
identified.
Materials and methods
Tissue samples, cell culture and
transfection
A total of 24 human colon cancer tissues and paired
adjacent normal tissues were obtained from the Second Xiangya
Hospital (Changsha, China). Written informed consent was obtained
from all colon cancer patients who were diagnosed by
immunohistochemical staining and pathological diagnosis. The
tissues were stored at −80°C.
The human colon cancer SW480 cells were cultured in
Dulbecco’s modified Eagle’s medium (Invitrogen Life Technologies,
Carlsbad, CA, USA) supplemented with 10% fetal bovine serum (FBS)
and 2 mM L-glutamine (Invitrogen). SW620 cells were cultured in
L-15 medium supplemented with 10% FBS. All the cells were
maintained in a humidified incubator with 5% CO2 at
37°C. miR-214 mimics and controls were purchased from Shanghai
GenePharma Co. Ltd (Shanghai, China). The cells were transfected
with Lipofectamine 2000 (Invitrogen) according to the
manufacturer’s instructions. This study was approved by the ethics
committee of the Second Xiangya Hospital.
RNA isolation and quantitative polymerase
chain reaction (qPCR)
Total RNA was isolated using TRIzol reagent
(Invitrogen) according to the manufacturer’s instructions. Next,
500 ng of RNA was used for the reverse transcription (RT) reaction
and specific RT primers were used for cDNA synthesis of miR-214. U6
small nuclear B non-coding RNA (Shanghai GenePharma Co. Ltd) was
used as an internal control for the normalization of miR-214. For
cDNA synthesis of large oligonucleotides, oligo(dT) was used as a
common primer. GAPDH (Shanghai GenePharma Co. Ltd) was used as an
internal control for the normalization of ADP-ribosylation
factor-like protein 2 (ARL2) expression (Shanghai GenePharma Co.
Ltd). qPCR was performed using the SYBR Green PCR Master Mix
(Applied Biosystems, Carlsbad, CA, USA) according to the following
conditions: 95°C for 5 min, followed by 40 cycles of amplification
at 95°C for 30 sec, 57°C for 30 sec and 72°C for 30 sec.
Western blot analysis
The transfected cells were collected at 48 h
post-transfection and lysed using radioimmunoprecipitation assay
(RIPA) buffer [50 mM Tris-HCl (pH 8.8), 150 mM NaCl, 1% NP-40, 1%
sodium deoxycholate and 0.1% SDS] for 30 min at 4°C. The protein
concentration was measured using the bicinchoninic acid method. A
total of 50 μg of protein was used for the analysis of ARL2
expression and GAPDH was used as a loading control. Rabbit
monoclonal anti-ARL2 (1:200) and anti-GAPDH (1;1,000) (Abcam,
Cambridge, MA, USA) were used as the primary antibodies. Goat
anti-rabbit immunoglobulin G conjugated to horseradish peroxidase
(1:1,000) was used as the secondary antibody (Abcam). The bound
antibodies were detected using the Electrochemiluminescence Plus
Western Blotting Detection System (GE Healthcare Bio-Sciences,
Pittsburgh, PA, USA) and the chemiluminiscent signals were detected
using high-performance chemiluminescence film (GE Healthcare
Bio-Sciences).
WST-1 assay
The transfected cells were plated at a density of
4×103 cells/well into 96-well plates. Following
transfection for 12, 24 and 48 h, the cells were incubated with
WST-1 reagent (Beijing Dingguo Biotechnology Co., Ltd., Beijing,
China), which is similar to
3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide, for
~1 h at 37°C. The absorbance at a wavelength of 490 nm was measured
using a spectrophometer (F-4500, Hitachi, Tokyo, Japan).
Colony formation assay
The transfected cells were seeded at a density of
200 cells/well into 12-well plates. The medium was replaced every
three days until the majority of the colonies consisted of >50
cells. The colonies were then washed, fixed and stained using
crystal violet (Sigma-Aldrich, St. Louis, MO, USA). Finally, images
of the stained colonies were captured and the colonies were counted
(G16, Canon Inc., Tokyo, Japan).
Annexin V fluorescein isothiocyanate
(FITC)/propidium iodide (PI) apoptosis assay
Camptothecin (Sigma-Aldrich) was added to the medium
of the transfected cells for the induction of cell apoptosis. At 24
h post-incubation, the cells were collected and detected by an
Annexin V-FITC/PI double staining kit using the BD FACSCalibur
system (Becton Dickinson, Franklin Lakes, NJ, USA) according to the
manufacturer’s instructions, as described previously (11).
Cell cycle analysis
The cells were starved for 24 h post-transfection
and then incubated with normal medium for an additional 24 h. Next,
the cells were washed with cold phosphate-buffered saline (PBS),
digested to form single cells and fixed with 70% ethanol for ≥1 h.
The cells were then washed again and stained with PI
(Sigma-Aldrich) supplemented with RNase A and Triton X-100 for 40
min at 37°C. Finally, the stained cells were washed and resuspended
in PBS for cell cycle analysis using the BD FACSCalibur system
(Becton Dickinson).
Luciferase reporter assay
The 3′UTR of ARL2 was amplified and inserted
downstream of the luciferase reporter gene. The mutant 3′UTR of
ARL2 (GCUG to AAGC) was amplified using wild-type ARL2 3′UTR as the
template. The cells were co-transfected with miRNA mimics and
wild-type or mutant ARL2 3′UTR. Following transfection for 48 h,
the cells were collected and lysed using RIPA buffer. The
luciferase intensity was measured using the Dual Luciferase
Reporter Assay System (Promega Corporation, Madison, WI, USA)
according to the manufacturer’s instructions.
Statistical analysis
All data are presented as the mean ± standard
deviation and represent three independent experiments. The
difference between groups was analyzed using the paired Student’s
t-test and P<0.05 was considered to indicate a statistically
significant difference.
Results
miR-214 downregulation in human colon
cancer
To investigate the function of miR-214 in human
colon cancer development, miRNAMap2.0 (12) was used for the analysis of miR-214
in diverse normal tissues and tumor tissues, including colon cancer
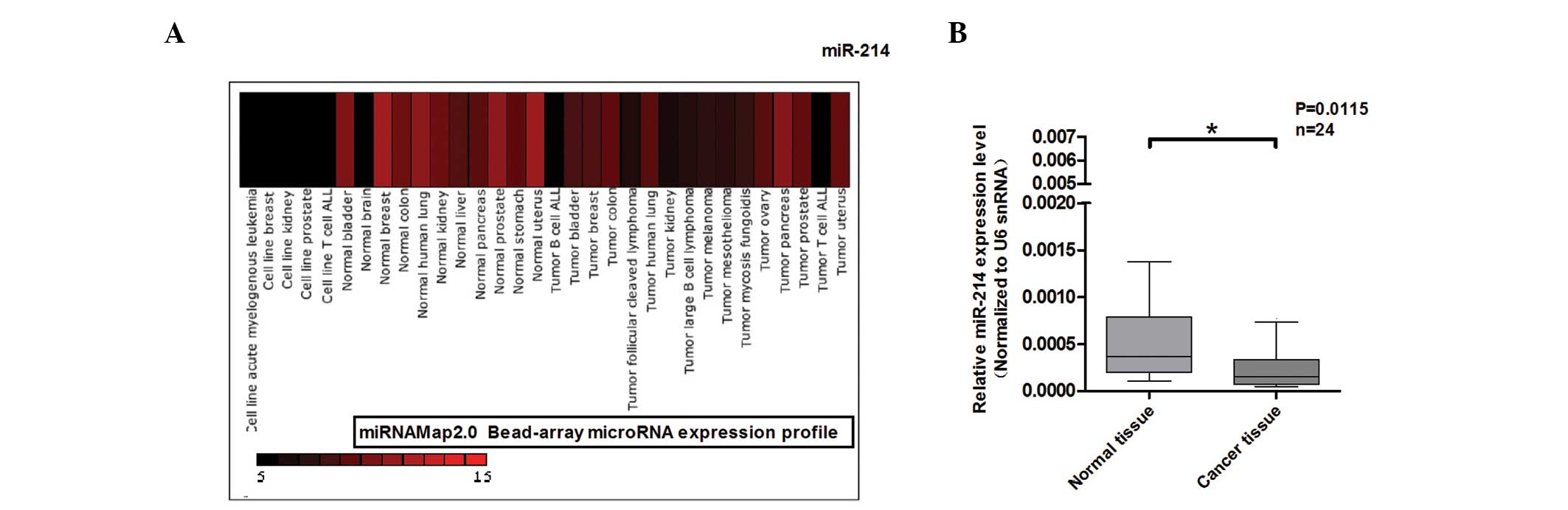
tissues. As shown in Fig. 1A,
miR-214 was found to be downregulated in colon cancer. Based on the
analysis of miRNAmap2.0, qPCR was performed to detect miR-214
expression in 24 paired normal and colon cancer tissues (Fig. 1B). miR-214 expression was found to
be downregulated in colon cancer. These results indicate that the
abnormal expression of miR-214 may be significant in colon
cancer.
miR-214 overexpression inhibits colon
cancer cell viability and colony formation
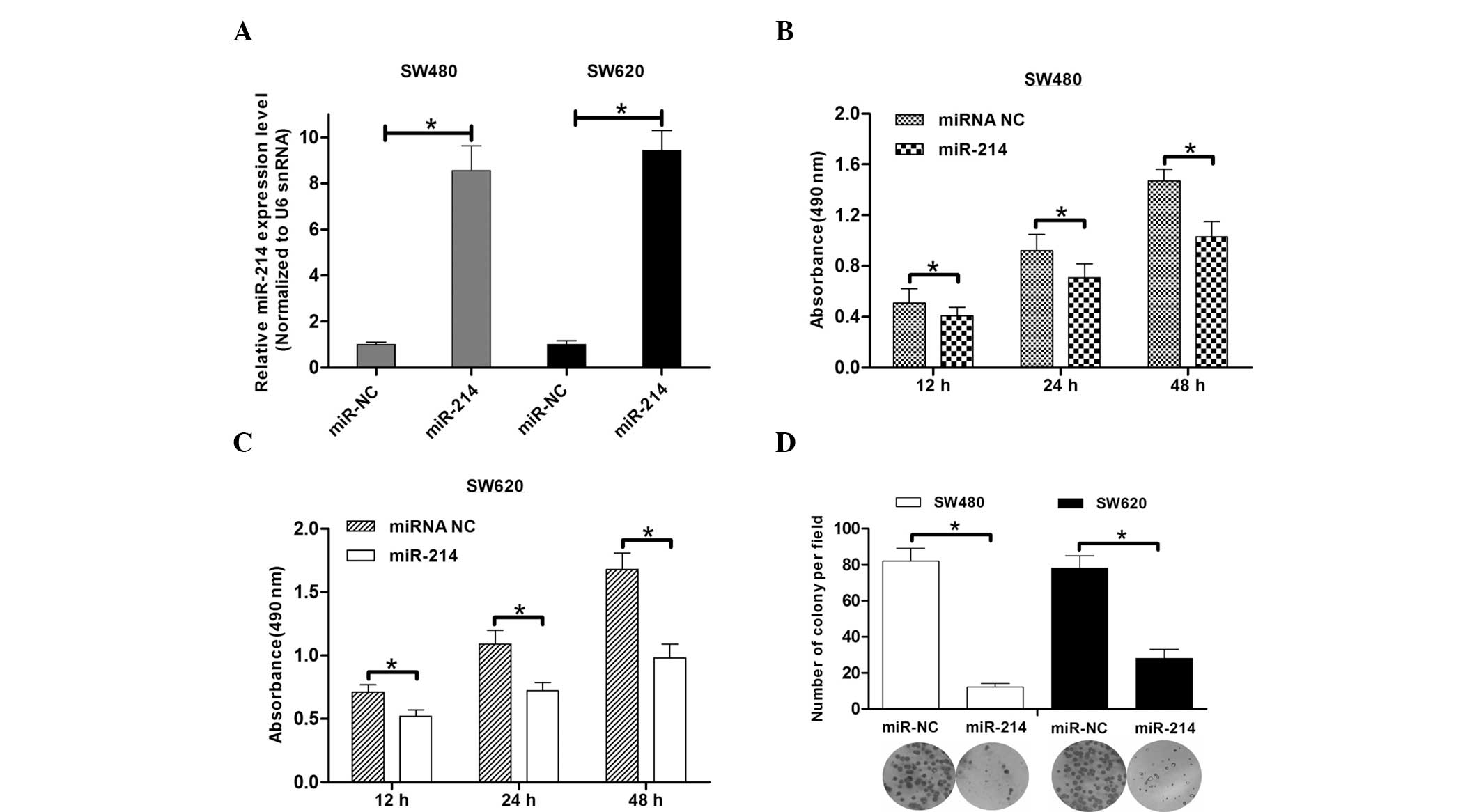
To investigate the functional role of miR-214
downregulation in colon cancer, cell viability and colony formation
assays were performed to analyze cell growth. The overexpression of
miR-214 in the SW480 and SW620 colon cancer cells treated with
miR-214 mimics was confirmed (Fig.
2A). The results from the WST-1 assay showed that miR-214 led
to the inhibition of SW480 cell viability by 20–30% at various
time-points compared with the miR-214 control cells (Fig. 2B). Accordingly, miR-214 inhibited
the cell viability of the SW620 cells (Fig. 2C). Consistent with the effect of
miR-214 on cell viability, miR-214 inhibited the number of SW480
and SW620 cell colonies by ~75 and 60%, respectively (Fig. 2D). These results indicate that
miR-214 may exhibit a key function in colon cancer growth.
miR-214 overexpression promotes colon
cancer cell apoptosis
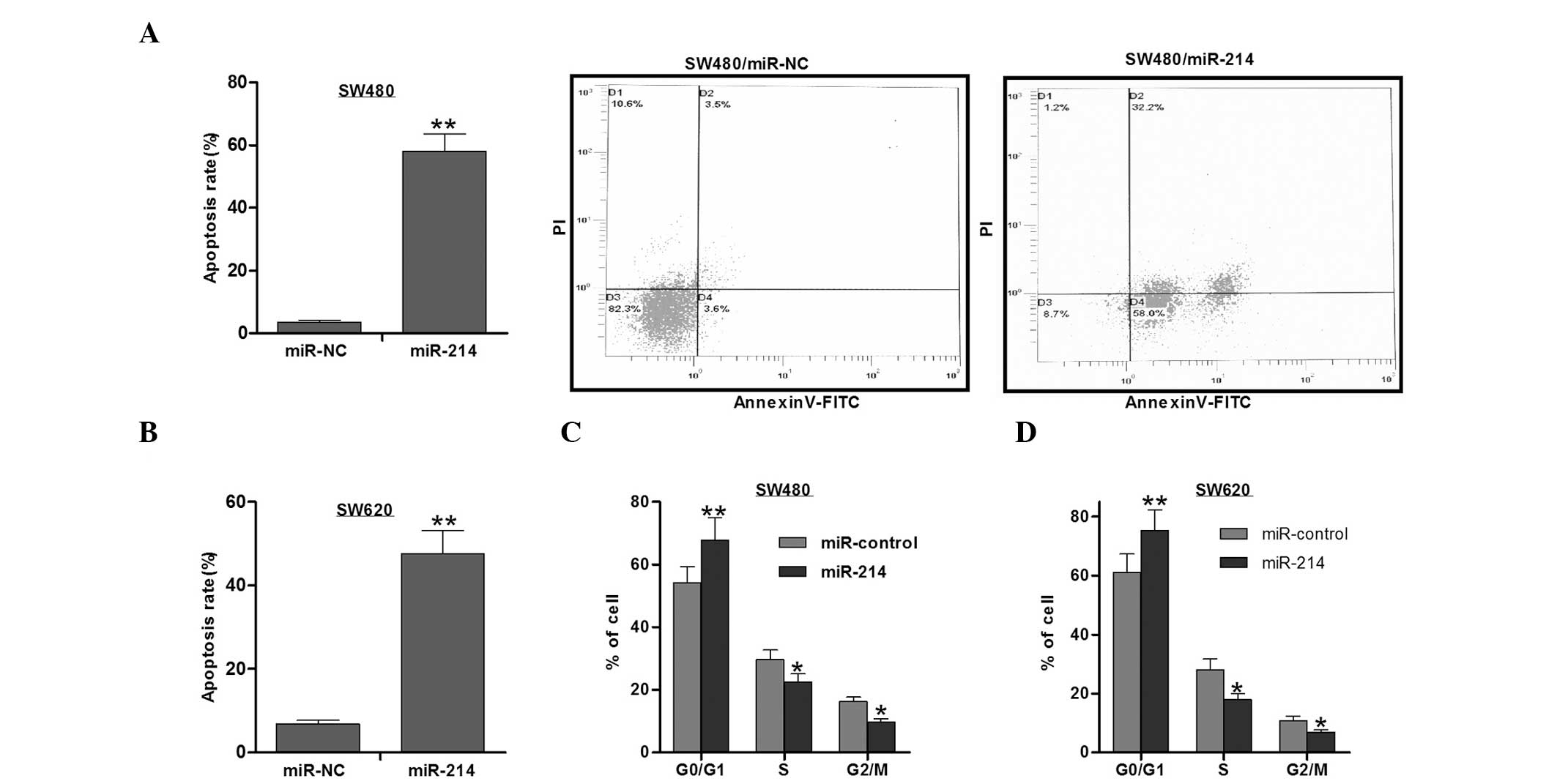
The analysis of the apoptotic rate of the colon
cancer cells was performed using the Annexin V-FITC/PI double
staining method. As shown in Fig. 3A
and B, the cells treated with miR-214 mimics exhibited a higher
apoptotic rate than the cells with the miR-214 control. These
results indicate that miR-214 inhibits cell growth partly through
promoting cell apoptosis.
miR-214 overexpression inhibits colon
cancer cell proliferation
Cell proliferation was analyzed by cell cycle
analysis using PI. It was found that miR-214 increased the number
of cells in the G1 phase, while reducing the number of
cells in the S phase, leading to G1/S arrest in the
SW480 and SW620 cells (Fig. 3D).
Therefore, the inhibition of cell proliferation may be responsible
for the inhibitory role of miRNA-214 in cell growth.
ARL2 is a direct target gene of miR-214
in colon cancer
To analyze the molecular mechanisms underlying the
regulation of miR-214 in colon cancer growth, Targetscan and Pictar
software was used to predict the target of miR-214. From the
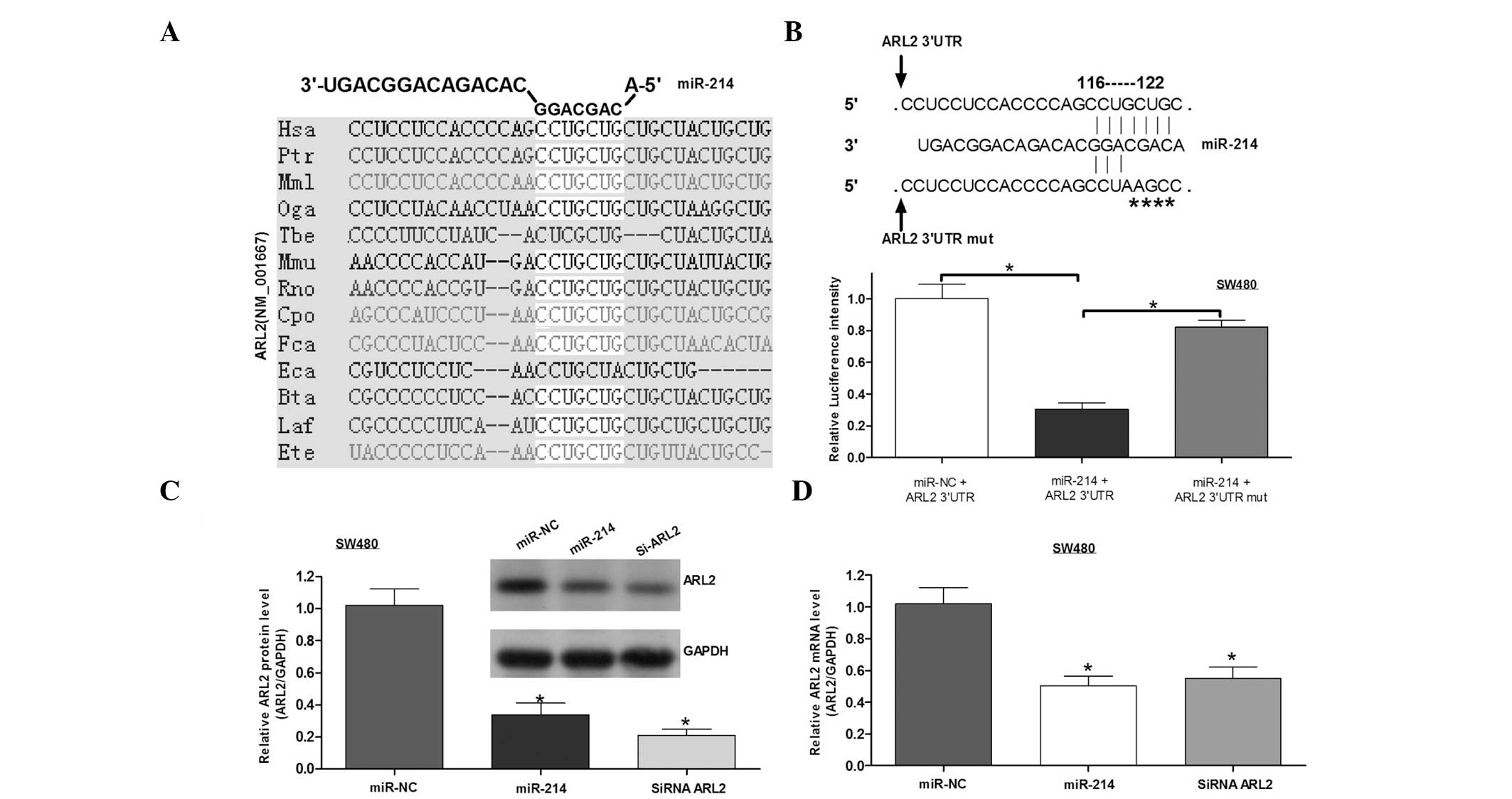
candidates, ARL2 was selected for further study. A binding site for
miR-214 was identified in the 3′UTR of ARL2, and the binding sites
were conserved among species (Fig.
4A). To validate whether ARL2 is a direct target of miR-214, a
point mutation was generated with binding sites and cloned into the
downstream region of the luciferase reporter gene. The cells were
then co-transfected with miR-214 mimics and wild-type or mutant
ARL2 3′UTRs. The results from the luciferase reporter assay
indicated that miR-214 lead to the inhibition of the luciferase
intensity of ARL2 3′UTR, whereby this inhibition was eliminated in
the mutant ARL2 3′UTR (Fig. 4B). To
investigate the function of miR-214 in ARL2 expression, western
blot analysis and qPCR assays were performed. It was identified
that miR-214 inhibited the expression of ARL2 protein and mRNA
(Fig. 4C and D), similar to the
function of ARL2 siRNA. These results indicated that miR-214
negatively regulates ARL2 expression by directly binding to its
3′UTR.
Discussion
miRNAs are considered to be key regulators of
protein-coding gene expression, exhibiting a crucial function in
cellular processes (1).
Accumulating evidence shows that the dysregulation of miRNAs is
associated with cancer initiation and development (3). A previous study showed that miR-214
expression is decreased in human cervical cancer and that it
inhibits cell proliferation, migration and invasion (13). In addition, miR-214 is downregulated
in hepatoma and inhibits tumor angiogenesis by inducing
hepatoma-derived growth factor (14). In agreement with these results, the
present study analyzed the expression of miR-214 in various normal
and cancer tissues, including colon cancer tissues, using
miRNAmap2.0. The analysis showed that miR-214 expression was
downregulated in colon cancer. In addition, miR-124 overexpression
was shown to inhibit cell growth and promote cell apoptosis,
functioning as a tumor suppressor. Controversy remains with regard
to the function of miR-214 in tumor progression. In certain
cancers, miR-214 has been shown to function as a tumor suppressor
(13,14), consistent with the results of the
present study. However, miR-214 has also been shown to be
downregulated in ovarian cancer and to induce cell survival and
cisplatin resistance by targeting the phosphate and tensin homolog
3′UTR (15), indicating that it may
function as an oncogene. In addition, miR-214 has been found to be
upregulated in gastric carcinoma (16), melanoma (17,18)
and hepatocellular carcinoma (19),
playing an important role in the promotion of tumor malignancy.
These results indicate that the different roles of miR-214 may be
tissue or cell-specific, and the main target genes of miR-214 may
be responsible for its diverse functions.
ARL2 is a GTPase belonging to the ADP-ribosylation
factor family (20) and is located
on chromosome 11 (11q13). ARL2 is considered to be involved in the
regulation of tubulin peptide folding and microtubule dynamics in
breast cancer cells (21). Previous
studies have revealed that ARL2-knockdown results in
G1/S phase arrest and the inhibition of cell
proliferation, which is a direct target of miR-16 (22). In addition, ARL2 has been reported
to form a complex with the tumor suppressor, protein phosphatase 2A
(PP2A), leading to changes in the phosphorylation status or the
cellular sublocalization of p53, a specific PP2A target. The
altered localization of p53 induces cell sensitivity to anticancer
compounds in breast cancer cells (23). These results demonstrate that ARL2
exhibits an oncogenic role in disease. Based on the results
reported in previous studies and the binding sites of miR-214 in
the ARL2 3′UTR, in the current study, ARL2 was selected as a
candidate target of miR-214. The luciferase assay indicated that
miR-214 reduced the luciferase intensity controlled by ARL2 3′UTR.
However, the inhibitory function of miR-214 in the mutant 3′UTR of
ARL2 was eliminated. In addition, miR-214 inhibited the ARL2
protein and mRNA levels. The function of ARL2 in cell
proliferation, as shown in previous studies, indicated that
ARL2-knockdown may provide a phenocopy of the effect of miR-214 on
colon cancer cells. These results confirm that ARL2 is a direct
target gene of miR-214 and that miR-214 negatively regulates its
expression.
During the investigation of the function of miR-214
in ARL2 protein and mRNA expression, the siRNA of ARL2 was used as
a positive control. miR-214 was found to inhibit ARL2 expression by
~60%, similar to the inhibitory effect of siRNA on ARL2 expression
(Fig. 3C and D). These results
further indicated that miRNAs and siRNAs exhibit similar functions
in target gene expression (24).
In conclusion, we hypothesize the following
explanation for the regulation mechanism of miR-124 in colon
cancer: miR-214 suppresses cell growth and promotes cell apoptosis
by targeting ARL2. However, additional confirmed target genes of
miR-214 may also mediate its function in colon cancer, although
this requires further investigation. The results of the present
study indicate that miR-214 may present a novel biomarker for the
evaluation of colon cancer progression, and may present a potential
miRNA-based therapeutic target for colon cancer patients.
References
|
1
|
Bartel DP: MicroRNAs: genomics,
biogenesis, mechanism, and function. Cell. 116:281–297. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Zeng Y and Cullen BR: Sequence
requirements for micro RNA processing and function in human cells.
RNA. 9:112–123. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Ambros V: The functions of animal
microRNAs. Nature. 431:350–355. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Calin GA, Sevignani C, Dumitru CD, et al:
Human microRNA genes are frequently located at fragile sites and
genomic regions involved in cancers. Proc Natl Acad Sci USA.
101:2999–3004. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Zhang LY, Liu M, Li X and Tang H:
miR-490-3p modulates cell growth and epithelial to mesenchymal
transition of hepatocellular carcinoma cells by targeting
endoplasmic reticulum-Golgi intermediate compartment protein 3
(ERGIC3). J Biol Chem. 288:4035–4047. 2013. View Article : Google Scholar :
|
|
6
|
Liang L, Wong CM, Ying Q, et al:
MicroRNA-125b suppresses human liver cancer cell proliferation and
metastasis by directly targeting oncogene LIN28B2. Hepatology.
52:1731–1740. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Xu Q, Liu LZ, Qian X, et al: MiR-145
directly targets p70S6K1 in cancer cells to inhibit tumor growth
and angiogenesis. Nucleic Acids Res. 40:761–774. 2012. View Article : Google Scholar :
|
|
8
|
Gregersen LH, Jacobsen AB, Frankel LB, Wen
J, Krogh A and Lund AH: MicroRNA-145 targets YES and STAT1 in colon
cancer cells. PLoS One. 5:e88362010. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Li J, Chen Y, Zhao J, Kong F and Zhang Y:
miR-203 reverses chemoresistance in p53-mutated colon cancer cells
through downregulation of Akt2 expression. Cancer Lett. 304:52–59.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Nie J, Liu L, Zheng W, et al:
microRNA-365, down-regulated in colon cancer, inhibits cell cycle
progression and promotes apoptosis of colon cancer cells by
probably targeting Cyclin D1 and Bcl-2. Carcinogenesis. 33:220–225.
2012. View Article : Google Scholar
|
|
11
|
Wang CM, Wang Y, Fan CG, et al: miR-29c
targets TNFAIP3, inhibits cell proliferation and induces apoptosis
in hepatitis B virus-related hepatocellular carcinoma. Biochem
Biophys Res Commun. 411:586–592. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Hsu SD, Chu CH, Tsou AP, et al: miRNAMap
2.0: genomic maps of microRNAs in metazoan genomes. Nucleic Acids
Res. 36:D165–D169. 2008. View Article : Google Scholar :
|
|
13
|
Peng RQ, Wan HY, Li HF, Liu M, Li X and
Tang H: MicroRNA-214 suppresses growth and invasiveness of cervical
cancer cells by targeting
UDP-N-acetyl-α-D-galactosamine:polypeptide
N-acetylgalactosaminyltransferase 7. J Biol Chem. 287:14301–14309.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Shih TC, Tien YJ, Wen CJ, et al:
MicroRNA-214 downregulation contributes to tumor angiogenesis by
inducing secretion of the hepatoma-derived growth factor in human
hepatoma. J Hepatol. 57:584–591. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Yang H, Kong W, He L, et al: MicroRNA
expression profiling in human ovarian cancer: miR-214 induces cell
survival and cisplatin resistance by targeting PTEN. Cancer Res.
68:425–433. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Li X, Zhang Y, Zhang H, et al: miRNA-223
promotes gastric cancer invasion and metastasis by targeting tumor
suppressor EPB41L3. Mol Cancer Res. 9:824–833. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Penna E, Orso F, Cimino D, et al:
microRNA-214 contributes to melanoma tumour progression through
suppression of TFAP2C. EMBO J. 30:1990–2007. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Bar-Eli M: Searching for the
‘melano-miRs’: miR-214 drives melanoma metastasis. EMBO J.
30:1880–1881. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Duan Q, Wang X, Gong W, et al: ER stress
negatively modulates the expression of the miR-199a/214 cluster to
regulates tumor survival and progression in human hepatocellular
cancer. PLoS One. 7:e315182012. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Kahn RA, Volpicelli-Daley L, Bowzard B, et
al: Arf family GTPases: roles in membrane traffic and microtubule
dynamics. Biochem Soc Trans. 33:1269–1272. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Beghin A, Honore S, Messana C, et al: ADP
ribosylation factor like 2 (Arl2) protein influences microtubule
dynamics in breast cancer cells. Exp Cell Res. 313:473–485. 2007.
View Article : Google Scholar
|
|
22
|
Wang K, Li P, Dong Y, et al: A
microarray-based approach identifies ADP ribosylation factor-like
protein 2 as a target of microRNA-16. J Biol Chem. 286:9468–9476.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Béghin A, Matera EL, Brunet-Manquat S and
Dumontet C: Expression of Arl2 is associated with p53 localization
and chemosensitivity in a breast cancer cell line. Cell Cycle.
7:3074–3082. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Zeng Y, Yi R and Cullen BR: MicroRNAs and
small interfering RNAs can inhibit mRNA expression by similar
mechanisms. Proc Natl Acad Sci USA. 100:9779–9784. 2003. View Article : Google Scholar : PubMed/NCBI
|