Introduction
Hepatocellular carcinoma (HCC) is the sixth most
common type of cancer and the third leading cause of cancer-related
mortality worldwide (1). Recently,
the incidence of HCC has rapidly increased and thus, the disease
has received considerable attention. Patients diagnosed with HCC
often exhibit an adverse outcome due to the aggressive nature of
the disease, and surgical resection is usually only effective at
the early stages of the disease (2). However, ~70% of these patients develop
recurrent tumors within five years (3,4). Even
with the advent of the multikinase inhibitor, sorafenib, prolonged
survival is limited (5,6). Thus, the development of novel
molecular targets for the diagnosis and therapy of HCC are urgently
required.
Cirrhosis is the underlying liver disease in 80% of
patients with HCC, which distinguishes these tumors from other
solid neoplasms (7). Although the
high prevalence of hepatitis C virus infection is the main cause of
the increasing incidence of HCC, as observed in Western countries
(3,8), other etiologies may lead to liver
damage and a subsequent increase in HCC incidence, including
chronic viral hepatitis B infection, alcohol consumption and
exposure to aflatoxin (2).
Therefore, clinical approaches for treating HCC are complex and
must contend with high molecular variability. Previous studies
investigating the molecular mechanisms of carcinogenesis have
revealed that the development and progression of HCC is caused by
an accumulation of genetic changes, which alter the expression of
genes that promote malignant transformation (9–11). The
development of novel genomic technologies, such as microarrays and
next-generation sequencing, led to the identification of numerous
genetic alterations in HCC; however, their clinical significance
and the functions of the mutated genes remain largely unclear
(2,12).
Current studies have focused on the expression of
genes encoding tumor-specific antigens and their association with
tumorigenesis and progression (13). Melanoma-associated antigens (MAGEs)
represent tumor-specific antigens, which have been increasingly
utilized as therapeutic targets for immunotherapy (14). MAGE proteins are classified into
types I and II (13,15,16).
Type I MAGE genes are located on the X-chromosome and
include MAGE-A, B and C, which are expressed during germ
cell development, but not by mature somatic cells. By contrast, the
localization, expression and oncological functions of type II MAGE
proteins, which include MAGE-D, E, F, G and H, are less
clear (13,17). Our previous study analyzed the
expression of MAGE-D4 in HCC and esophageal cancer and found
that the overexpression of MAGE-D4 was significantly
associated with the malignant phenotypes of these cancers (18,19).
However, little is known with regard to the oncological functions
of other MAGE-D genes. Since melanoma-associated antigen-D2
(MAGE-D2) is involved in cell adhesion (17), we hypothesized that MAGE-D2
and MAGE-D4 contribute to the progression of HCC. The aim of
the present study was to evaluate the clinical significance of
MAGE-D2 expression in HCC.
Materials and methods
Ethics
This study complied with the ethical guidelines of
the World Medical Association Declaration of Helsinki Ethical
Principles for Medical Research Involving Human Subjects 3(Seoul,
Korea; 2008). Written informed consent was obtained from all
patients and the study was approved by the Institutional Review
Board of Nagoya University (Nagoya, Japan; approval no.
2013-0295-2).
Sample collection
A total of nine HCC cell lines (Hep3B, HepG2, HLE,
HLF, HuH1, HuH2, HuH7, PLC/PRF/5 and SK-Hep1), which were obtained
from the American Type Culture Collection (Manassas, VA, USA), were
stored at −80°C in Cell Banker® preservative solution
(Mitsubishi Chemical Medience Corporation, Tokyo, Japan) and
cultured in RPMI-1640 medium (Sigma-Aldrich, St. Louis, MO, USA)
supplemented with 10% fetal bovine serum at 37°C in an atmosphere
containing 5% CO2. Primary HCC tissues and corresponding
non-cancerous tissues were collected consecutively from 151
patients undergoing liver resection for the treatment of HCC at
Nagoya University Hospital (Nagoya, Japan) between January 1998 and
January 2012. Specimens were classified histologically according to
the Union for International Cancer Control tumor-node-metastasis
classification (seventh edition) (20). Furthermore, background liver status,
Child-Pugh classification, hepatitis virus infection status,
pre-operative serum tumor markers, tumor multiplicity and maximum
size, and pathological observations, including tumor
differentiation and vascular invasion, were analyzed.
Post-operative follow-up included physical examination, measurement
of serum tumor markers every three months, and enhanced chest and
abdominal computed tomography examinations every six months.
Treatment following recurrence included surgery, radiofrequency
ablation, transcatheter arterial chemoembolization and
chemotherapy, which was selected according to tumor status and
liver function. Tissue samples were immediately flash-frozen in
liquid nitrogen and stored at −80°C until RNA was extracted (mean,
28 days). RNA was extracted from tumor samples, which were
~5-mm2, without necrotic components and were confirmed
to contain >80% tumor cells. Corresponding non-cancerous liver
tissue samples from the respective patients were collected >2 cm
from the tumor edge, and did not contain any regenerative or
dysplastic nodules (12).
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR)
The expression of MAGE-D2 mRNA was analyzed
using RT-qPCR. Total RNA (10 μg) was isolated from each of the nine
aforementioned HCC cell lines, the 151 primary HCC tissues and the
corresponding non-cancerous tissues, and used as templates to
obtain cDNA. The PCR primer sequences for MAGE-D2 were as
follows: Sense, 5′-TAGAGAAGGCAGACGCATCC-3′ in exon 1 and antisense,
5′-AAGCGAGTTAGACCTGCACC-3′ in exon 2, which amplify a 110-bp
sequence. RT-qPCR was performed using nine HCC cell lines and 151
pairs of clinical samples, as well as samples without templates,
which served as negative controls, with the SYBR-Green PCR core
reagents kit (Perkin-Elmer, Applied Biosystems, Foster City, CA,
USA). The SYBR-Green emission intensity was detected using an ABI
StepOnePlus Real-Time PCR System (Perkin-Elmer, Applied Biosystems)
under the following conditions: One cycle at 95°C for 10 min,
followed by 40 cycles at 95°C for 5 sec and 6°C for 30 sec. The
expression of glyceraldehyde-3-phosphate dehydrogenase
(GAPDH) mRNA was quantified in each sample for
standardization. mRNA quantification was calculated using the
2−ΔΔCT method. Biological and technical replicates of
the cell lines and HCC tissues were performed in triplicate. The
expression level of each sample is presented as the value of
MAGE-D2 divided by that of GAPDH. In the tumor
tissues, MAGE-D2 mRNA expression was considered to be
increased when mRNA levels were higher than those of the
corresponding non-cancerous tissues (21).
Immunohistochemistry (IHC)
IHC was conducted to investigate the localization of
MAGE-D2 in 40 representative sections of well-preserved HCC
tissue. Formalin-fixed, paraffin-embedded tissues were dewaxed in
xylene twice for 5 min, rehydrated in a graded alcohol series (100,
90 and 70%) followed by H2O for 2 min each, then treated
with 3% H2O2 to inhibit endogenous peroxidase
activity. Next, epitope retrieval was performed by subjecting
samples to five incubations with 10 mM citrate buffer at 95°C for 5
min each. The samples were incubated with Histofine®
SAB-PO (Nichirei Biosciences. Inc., Tokyo, Japan) for 5 min to
limit non-specific reactivity, then incubated for 1 h at room
temperature with a rabbit polyclonal antibody against MAGE-D2 (cat.
no. HPA031573; Atlas Antibodies, Stockholm, Sweden), which was
diluted (1:500) in antibody diluent (Dako, Glostrup, Denmark).
Sections were developed for 2 min using liquid
3,3′-diaminobenzidine substrate (Nichirei Biosciences, Inc.). The
staining patterns of the HCC and corresponding non-cancerous
tissues were compared. Specimens were randomized and coded prior to
analysis by two independent observers who were blinded to the
status of the samples. Each observer evaluated all specimens at
least twice within a specific time interval to decrease
intra-observer variation (21).
Statistical analysis
The relative mRNA expression levels
(MAGE-D2/GAPDH) between two groups were compared
using the Mann-Whitney U test. The χ2 test was used to
analyze the association between the expression and methylation
status of MAGE-D2 and the clinicopathological parameters.
Overall and disease-free survival rates were calculated using the
Kaplan-Meier method, and the difference in survival curves was
analyzed using the log-rank test. Multivariable regression analysis
was performed to detect prognostic factors using the Cox
proportional hazards model, and variables with P<0.05 were
entered into the final model. All statistical analyses were
performed using JMP® 10 software (SAS Institute Inc.,
Cary, NC, USA). P<0.05 was considered to indicate a
statistically significant difference.
Results
MAGE-D2 mRNA expression in HCC
cell lines and clinical tissues
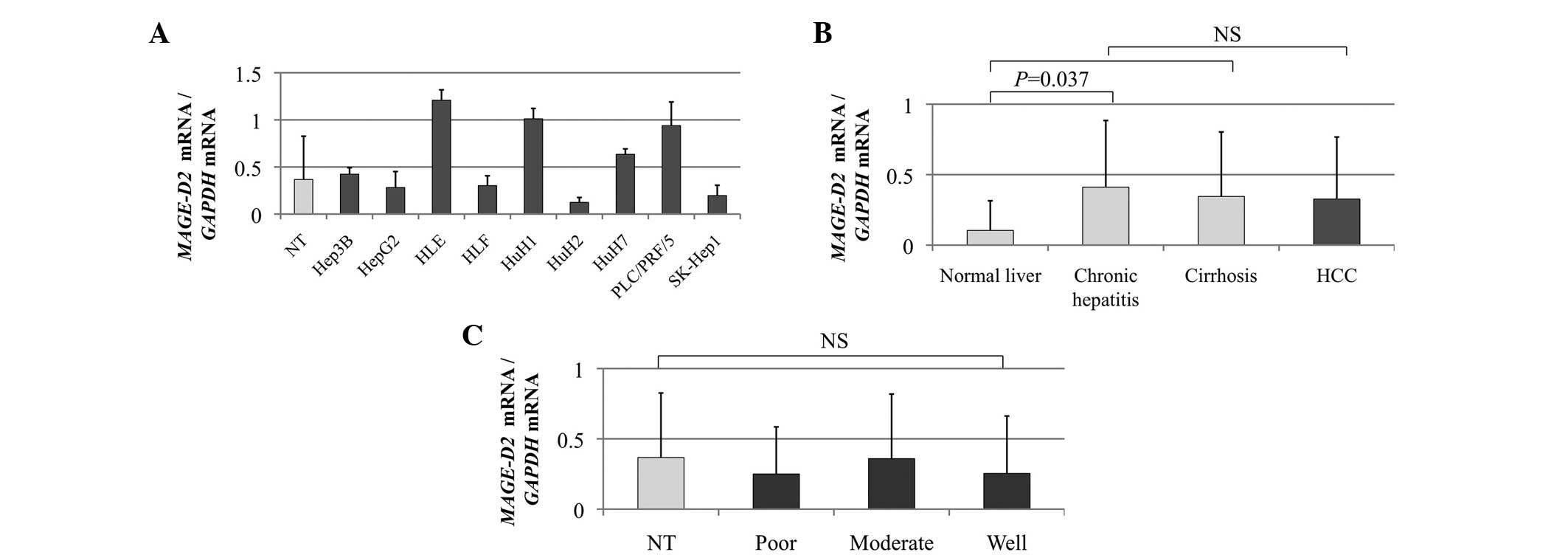
The heterogeneity of MAGE-D2 expression in
the HCC cell lines was determined using qPCR analysis (Fig. 1A). The MAGE-D2 mRNA
expression levels were compared in the non-cancerous tissues
categorized by the background liver status as follows: Normal liver
(n=10), chronic hepatitis (n=87), cirrhosis (n=54) and HCC tissues.
A significant difference was observed between normal liver and
chronic hepatitis tissues (P=0.037), whereas chronic hepatitis and
cirrhosis were comparable, indicating that MAGE-D2
expression was stimulated by chronic inflammation, but not fibrosis
(Fig. 1B). The expression level of
MAGE-D2 mRNA in 66 (44%) of the 151 patients was higher in
the HCC tissues compared with the corresponding non-cancerous
tissues. However, no significant difference in the mean expression
level of MAGE-D2 mRNA was identified between the
non-cancerous and HCC tissues (Fig.
1B), indicating that the upregulation of MAGE-D2
expression is not involved in hepatocarcinogenesis. Furthermore,
MAGE-D2 mRNA expression levels were independent of tumor
differentiation (Fig. 1C).
IHC
The expression of MAGE-D2 protein was determined
using IHC in 30 cases exhibiting relative overexpression,
underexpression or equivalent MAGE-D2 mRNA expression in the
HCC tissues compared with the corresponding non-cancerous tissues.
Two representative cases with high expression levels of
MAGE-D2 mRNA in HCC tissues showed increased expression of
MAGE-D2 in the cytoplasm and the nuclei of tumor cells compared
with the adjacent non-cancerous tissues (Fig. 2A and B). The results of
immunohistochemical staining were consistent with the RT-qPCR
data.
Prognostic value of MAGE-D2 expression in
151 HCC patients
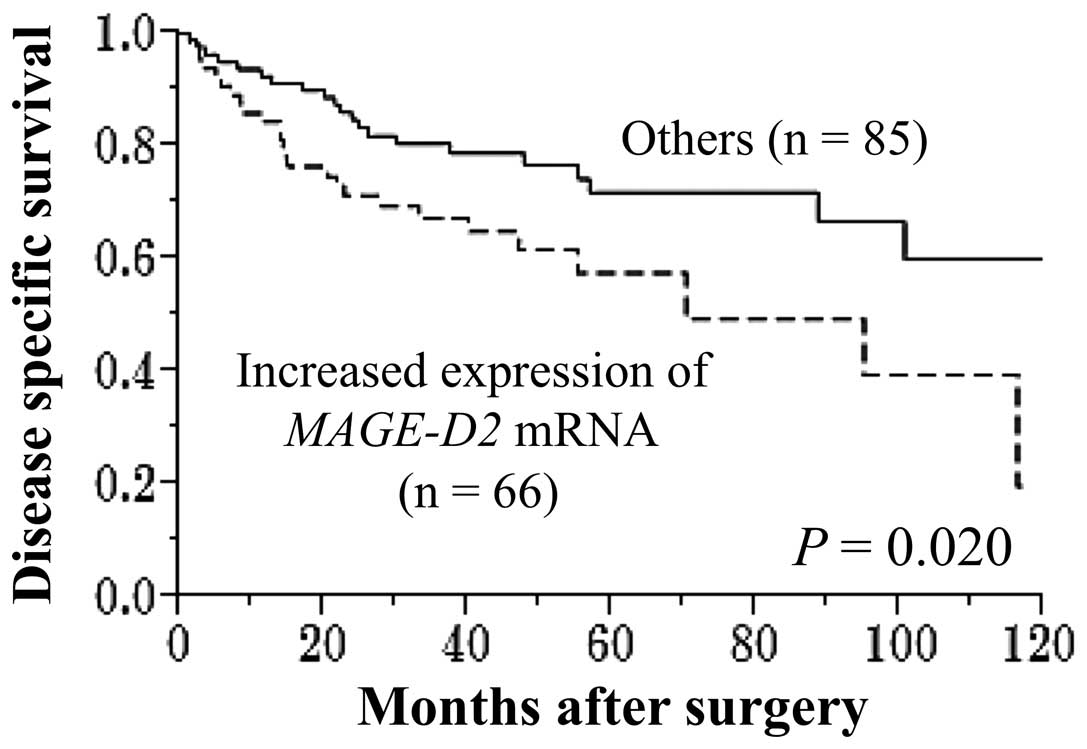
Increased expression of MAGE-D2 mRNA was
detected in the tumor samples from 66 of the 151 (44%) patients
with HCC. The disease-specific survival rate was significantly
reduced in the patients with increased expression of MAGE-D2
mRNA (five-year survival rate, 58 vs. 72%; P=0.020; Fig. 3). The MAGE-D2 expression
level was not associated with recurrence-free survival. Univariate
analysis for disease-specific survival showed that advanced age,
α-fetoprotein levels of >20 ng/ml, protein induced by vitamin K
antagonists II levels of >40 mAU/ml, multiple tumors, a tumor
size of ≥3.0 cm, serosal infiltration, vascular invasion, positive
margin status and increased expression of MAGE-D2 mRNA were
all significant prognostic indicators of adverse outcomes.
Multivariate analysis identified increased expression of
MAGE-D2 mRNA as an independent prognostic factor for
disease-specific survival (hazard ratio, 2.65; 95% confidence
interval, 1.43–4.98; P=0.002; Table
I). Increased expression of MAGE-D2 mRNA was not
significantly associated with other clinicopathological parameters,
including extrahepatic recurrence.
 | Table IPrognostic factors of
disease-specific survival in 151 hepatocellular carcinoma
patients. |
Table I
Prognostic factors of
disease-specific survival in 151 hepatocellular carcinoma
patients.
| | Univariate | Multivariate |
|---|
| |
|
|
|---|
| Variables | n | Hazard ratio | 95% CI | P-value | Hazard ratio | 95% CI | P-value |
|---|
| Age (≥65
years) | 84 | 1.92 | 1.07–3.57 | 0.030a | 1.60 | 0.87–3.05 | 0.133 |
| Gender (male) | 126 | 1.27 | 0.60–3.13 | 0.553 | | | |
| Background liver
(cirrhosis) | 54 | 1.58 | 0.88–2.81 | 0.123 | | | |
| Pugh-Child’s
classification (B) | 11 | 0.93 | 0.28–2.32 | 0.889 | | | |
| α-FP (>20
ng/ml) | 70 | 1.90 | 1.07–3.42 | 0.029a | 1.32 | 0.69–2.50 | 0.395 |
| PIVKA II (>40
mAU/ml) | 93 | 2.10 | 1.14–4.07 | 0.016a | 1.20 | 0.60–2.53 | 0.610 |
| Tumor multiplicity
(multiple) | 34 | 2.09 | 1.11–3.76 | 0.023a | 1.31 | 0.67–2.48 | 0.418 |
| Tumor size (≥3.0
cm) | 104 | 2.20 | 1.13–4.71 | 0.020a | 1.37 | 0.61–3.36 | 0.453 |
| Tumor
differentiation (well) | 35 | 0.55 | 0.25–1.10 | 0.095 | | | |
| Growth type
(invasive growth) | 24 | 1.44 | 0.69–2.76 | 0.318 | | | |
| Serosal
infiltration | 37 | 2.51 | 1.32–4.61 | 0.006a | 1.47 | 0.70–3.02 | 0.304 |
| Formation of
capsule | 104 | 1.05 | 0.57–2.02 | 0.884 | | | |
| Infiltration to
capsule | 83 | 1.20 | 0.67–2.18 | 0.537 | | | |
| Septum
formation | 98 | 0.87 | 0.49–1.60 | 0.651 | | | |
| Vascular
invasion | 37 | 3.40 | 1.87–6.07 | <0.001a | 2.42 | 1.17–4.97 | 0.017a |
| Margin status
(positive) | 28 | 2.64 | 1.42–4.73 | 0.003a | 2.84 | 1.48–5.36 | 0.002a |
| Increased
expression of MAGE-D2 mRNA | 66 | 1.96 | 1.10–3.54 | 0.022a | 2.65 | 1.43–4.98 | 0.002a |
Discussion
Extensive studies and the development of novel
genomic technologies may improve our understanding of the molecular
pathogenesis of HCC (21–23). Gene signatures derived from tumors
and corresponding non-cancerous tissues may identify patients who
are at high risk of developing HCC and would benefit from potential
chemopreventive strategies (24,25).
MAGE-D2 is encoded by one of the cancer testis
family of genes and is located on chromosome Xp11.21 (26,27).
In contrast to the testis- and tumor-specific expression of
numerous MAGE type I genes, MAGE-D2 mRNA is expressed in
healthy human tissues and the majority of cell types that have been
examined (28,29). In previous studies, MAGE-D2
expression has been analyzed in a clinical setting using
high-density oligonucleotide DNA arrays and served as a marker to
predict the occurrence of liver metastases from colorectal tumors
(30,31). The function of MAGE-D2 is unclear,
however, its increased expression may promote the cancer cell
adhesion to the vascular epithelium (17). Since MAGE-D2 protects melanoma cells
from tumor necrosis factor-related apoptosis-inducing ligand
(TRAIL)-induced apoptosis, this observation is of particular note,
as TRAIL is involved in the killing of melanoma cells by the immune
system and is expressed by a number of immune cells, including
activated CD4 and CD8 T lymphocytes, natural killer cells and
dendritic cells (32). To the best
of our knowledge, the present study is the first to evaluate
MAGE-D2 expression in HCC.
In the present study, once the direct correlation
between the expression patterns of MAGE-D2 mRNA and MAGE-D2
had been identified, the clinical significance of MAGE-D2
expression was evaluated using RT-qPCR. Increased expression of
MAGE-D2 mRNA in HCC tissues was found to significantly
correlate with an adverse outcome and was identified as one of the
independent prognostic factors after curative hepatectomy. However,
future studies using a larger patient cohort are required to
confirm these observations. These results question whether the
aberrant expression of MAGE-D2 contributes to the
carcinogenesis and progression of HCC. Although the present study
indicated that MAGE-D2 expression is modulated by chronic
inflammation, the expression levels of MAGE-D2 in HCC and
the corresponding non-cancerous tissues were equivalent, indicating
that the upregulation of MAGE-D2 was incidental in hepatic
carcinogenesis. By contrast, increased expression levels of
MAGE-D2 were significantly associated with earlier mortality
following curative resection, indicating that the upregulation of
MAGE-D2 contributed to the progression of HCC rather than to
carcinogenesis.
The use of cDNA microarrays has identified
MAGE-D2 expression as a predictor of the metastatic
potential of colorectal cancer (31). By contrast, in the present study, no
significant correlation was identified between the expression
pattern of MAGE-D2 mRNA and extrahepatic recurrence.
Therefore, the biological functions of MAGE-D2 in HCC and
colorectal cancer may differ. Notably, the increased expression of
MAGE-D2 demonstrated high prognostic value despite the
absence of a significant association with other important
prognostic factors, including tumor size, multiplicity, vascular
invasion, and advanced stage. This reveals the unique prognostic
value of MAGE-D2 for HCC and indicates that HCC patients
with increased expression of MAGE-D2 must be categorized
into a high-risk group with an adverse prognosis even during the
early stage of HCC.
The present study was limited by the lack of a
functional analysis of MAGE-D2. Future studies, which include
pathway analysis in hepatocarcinogenesis and functional analysis,
are required to analyze the molecular mechanisms that underlie the
biological function of MAGE-D2 in HCC.
In conclusion, the results of this study indicated
that MAGE-D2 mRNA overexpression contributes to tumor
progression and thus, may serve as a prognostic indicator following
curative resection, as well as a potential therapeutic target in
HCC.
References
|
1
|
Jemal A, Bray F, Center MM, et al: Global
cancer statistics. CA Cancer J Clin. 61:69–90. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Shiraha H, Yamamoto K and Namba M: Human
hepatocyte carcinogenesis (review). Int J Oncol. 42:1133–1138.
2013.PubMed/NCBI
|
|
3
|
Llovet JM, Burroughs A and Bruix J:
Hepatocellular carcinoma. Lancet. 362:1907–1917. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Mínguez B and Lachenmayer A: Diagnostic
and prognostic molecular markers in hepatocellular carcinoma. Dis
Markers. 31:181–190. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Cheng AL, Kang YK, Chen Z, et al: Efficacy
and safety of sorafenib in patients in the Asia-Pacific region with
advanced hepatocellular carcinoma: a phase III randomised,
double-blind, placebo-controlled trial. Lancet Oncol. 10:25–34.
2009. View Article : Google Scholar
|
|
6
|
Llovet JM, Ricci S, Mazzaferro V, et al:
SHARP Investigators Study Group: Sorafenib in advanced
hepatocellular carcinoma. N Engl J Med. 359:378–390. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
El-Serag HB: Hepatocellular carcinoma:
recent trends in the United States. Gastroenterology. 127(5 Suppl
1): S27–S34. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Herath NI, Leggett BA and MacDonald GA:
Review of genetic and epigenetic alterations in
hepatocarcinogenesis. J Gastroenterol Hepatol. 21:15–21. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Kanda M, Nomoto S, Nishikawa Y, et al:
Correlations of the expression of vascular endothelial growth
factor B and its isoforms in hepatocellular carcinoma with
clinico-pathological parameters. J Surg Oncol. 98:190–196. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Miki D, Ochi H, Hayes CN, Aikata H and
Chayama K: Hepatocellular carcinoma: towards personalized medicine.
Cancer Sci. 103:846–850. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Villanueva A, Newell P, Chiang DY,
Friedman SL and Llovet JM: Genomics and signaling pathways in
hepatocellular carcinoma. Semin Liver Dis. 27:55–76. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Kanda M, Nomoto S, Okamura Y, et al:
Detection of metallothionein 1G as a methylated tumor suppressor
gene in human hepatocellular carcinoma using a novel method of
double combination array analysis. Int J Oncol. 35:477–483.
2009.PubMed/NCBI
|
|
13
|
Sang M, Wang L, Ding C, et al:
Melanoma-associated antigen genes-an update. Cancer Lett.
302:85–90. 2011. View Article : Google Scholar
|
|
14
|
Chang CC, Campoli M, Luo W, et al:
Immunotherapy of melanoma targeting human high molecular weight
melanoma-associated antigen: potential role of nonimmunological
mechanisms. Ann NY Acad Sci. 1028:340–350. 2004. View Article : Google Scholar
|
|
15
|
Barker PA and Salehi A: The MAGE proteins:
emerging roles in cell cycle progression, apoptosis, and
neurogenetic disease. J Neurosci Res. 67:705–712. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Xiao J and Chen HS: Biological functions
of melanoma-associated antigens. World J Gastroenterol.
10:1849–1853. 2004.PubMed/NCBI
|
|
17
|
Langnaese K, Kloos DU, Wehnert M, Seidel B
and Wieacker P: Expression pattern and further characterization of
human MAGED2 and identification of rodent orthologues. Cytogenet
Cell Genet. 94:233–240. 2001. View Article : Google Scholar
|
|
18
|
Oya H, Kanda M, Takami H, et al:
Overexpression of melanoma-associated antigen D4 is an independent
prognostic factor in squamous cell carcinoma of the esophagus. Dis
Esophagus. Oct 21–2013.(Epub ahead of print). View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Takami H, Kanda M, Oya H, et al:
Evaluation of MAGE-D4 expression in hepatocellular carcinoma in
Japanese patients. J Surg Oncol. 108:557–562. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Sobin LH, Gospodarowicz MK and Wittekind
C: International Union Against Cancer. TNM classification of
malignant tumors. 7th edition. Wiley-Blackwell; New York, NY:
2009
|
|
21
|
Kanda M, Nomoto S, Okamura Y, et al:
Promoter hypermethylation of fibulin 1 gene is associated with
tumor progression in hepatocellular carcinoma. Mol Carcinog.
50:571–579. 2011. View
Article : Google Scholar : PubMed/NCBI
|
|
22
|
Arzumanyan A, Reis HM and Feitelson MA:
Pathogenic mechanisms in HBV- and HCV-associated hepatocellular
carcinoma. Nat Rev Cancer. 13:123–135. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Khare S, Zhang Q and Ibdah JA: Epigenetics
of hepatocellular carcinoma: role of microRNA. World J
Gastroenterol. 19:5439–5445. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Hernandez-Gea V, Toffanin S, Friedman SL
and Llovet JM: Role of the microenvironment in the pathogenesis and
treatment of hepatocellular carcinoma. Gastroenterology.
144:512–527. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Kanda M, Nomoto S, Oya H, et al:
Downregulation of DENND2D by promoter hypermethylation is
associated with early recurrence of hepatocellular carcinoma. Int J
Oncol. 44:44–52. 2014.
|
|
26
|
Bertrand M, Huijbers I, Chomez P and De
Backer O: Comparative expression analysis of the MAGED genes during
embryogenesis and brain development. Dev Dyn. 230:325–334. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Harper R, Xu C, Di P, et al:
Identification of a novel MAGE D2 antisense RNA transcript in human
tissues. Biochem Biophys Res Commun. 324:199–204. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Kidd M, Modlin IM, Mane SM, et al: The
role of genetic markers - NAP1L1, MAGE-D2, and MTA1 - in defining
small-intestinal carcinoid neoplasia. Ann Surg Oncol. 13:253–262.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Papageorgio C, Brachmann R, Zeng J, et al:
MAGED2: a novel p53-dissociator. Int J Oncol. 31:1205–1211.
2007.PubMed/NCBI
|
|
30
|
Kidd M, Modlin IM, Mane SM, et al: Utility
of molecular genetic signatures in the delineation of gastric
neoplasia. Cancer. 106:1480–1488. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Li M, Lin YM, Hasegawa S, et al: Genes
associated with liver metastasis of colon cancer, identified by
genome-wide cDNA microarray. Int J Oncol. 24:305–312.
2004.PubMed/NCBI
|
|
32
|
Tseng HY, Chen LH, Ye Y, et al: The
melanoma-associated antigen MAGE-D2 suppresses TRAIL receptor 2 and
protects against TRAIL-induced apoptosis in human melanoma cells.
Carcinogenesis. 33:1871–1881. 2012. View Article : Google Scholar : PubMed/NCBI
|