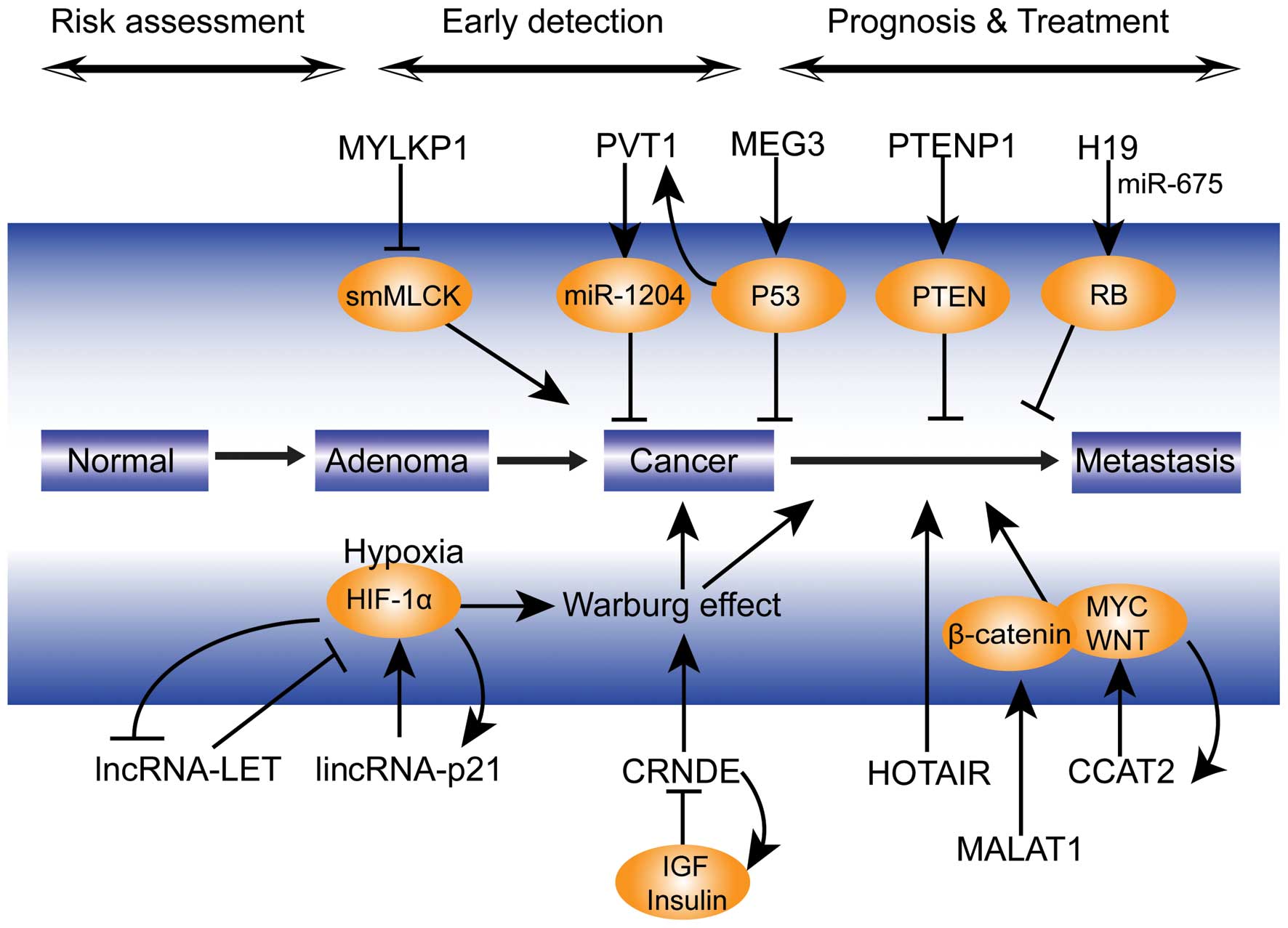
|
1
|
Jemal A, Bray F, Center MM, Ferlay J, Ward
E and Forman D: Global cancer statistics. CA Cancer J Clin.
61:69–90. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Esteller M: Non-coding RNAs in human
disease. Nat Rev Genet. 12:861–874. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Ponting CP, Oliver PL and Reik W:
Evolution and functions of long noncoding RNAs. Cell. 136:629–641.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Kapranov P, Cheng J, Dike S, et al: RNA
maps reveal new RNA classes and a possible function for pervasive
transcription. Science. 316:1484–1488. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Bartel DP: MicroRNAs: genomics,
biogenesis, mechanism, and function. Cell. 116:281–297. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Gutschner T and Diederichs S: The
hallmarks of cancer: a long non-coding RNA point of view. RNA Biol.
9:703–719. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Tsang WP, Ng EK, Ng SS, et al: Oncofetal
H19-derived miR-675 regulates tumor suppressor RB in human
colorectal cancer. Carcinogenesis. 31:350–358. 2010. View Article : Google Scholar
|
|
8
|
Yoshimizu T, Miroglio A, Ripoche MA, et
al: The H19 locus acts in vivo as a tumor suppressor. Proc Natl
Acad Sci USA. 105:12417–12422. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Moulton T, Crenshaw T, Hao Y, et al:
Epigenetic lesions at the H19 locus in Wilms’ tumour patients. Nat
Genet. 7:440–447. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Cui H, Onyango P, Brandenburg S, Wu Y,
Hsieh CL and Feinberg AP: Loss of imprinting in colorectal cancer
linked to hypomethylation of H19 and IGF2. Cancer Res.
62:6442–6446. 2002.PubMed/NCBI
|
|
11
|
Fellig Y, Ariel I, Ohana P, et al: H19
expression in hepatic metastases from a range of human carcinomas.
J Clin Pathol. 58:1064–1068. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Ohana P, Schachter P, Ayesh B, et al:
Regulatory sequences of H19 and IGF2 genes in DNA-based therapy of
colorectal rat liver metastases. J Gene Med. 7:366–374. 2005.
View Article : Google Scholar
|
|
13
|
Sorin V, Ohana P, Mizrahi A, et al:
Regional therapy with DTA-H19 vector suppresses growth of colon
adenocarcinoma metastases in the rat liver. Int J Oncol.
39:1407–1412. 2011.PubMed/NCBI
|
|
14
|
Gupta RA, Shah N, Wang KC, et al: Long
non-coding RNA HOTAIR reprograms chromatin state to promote cancer
metastasis. Nature. 464:1071–1076. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Pádua Alves C, Fonseca AS, Muys BR, et al:
Brief report: The lincRNA Hotair is required for
epithelial-to-mesenchymal transition and stemness maintenance of
cancer cell lines. Stem cells. 31:2827–2832. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Kogo R, Shimamura T, Mimori K, et al: Long
noncoding RNA HOTAIR regulates polycomb-dependent chromatin
modification and is associated with poor prognosis in colorectal
cancers. Cancer Res. 71:6320–6326. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Gutschner T, Hämmerle M, Eissmann M, et
al: The noncoding RNA MALAT1 is a critical regulator of the
metastasis phenotype of lung cancer cells. Cancer Res.
73:1180–1189. 2013. View Article : Google Scholar :
|
|
18
|
Chang JL, Li ZG, Wang XY and Yang MH:
Detection of p53, MALAT1, ki-67 and β-catenin mRNA expression and
its significance in molecular diagnosis of colorectal carcinoma.
World Chinese J Digestol. 16:3849–3854. 2008.
|
|
19
|
Xu C, Yang M, Tian J, Wang X and Li Z:
MALAT-1: a long non-coding RNA and its important 3′ end functional
motif in colorectal cancer metastasis. Int J Oncol. 39:169–175.
2011.PubMed/NCBI
|
|
20
|
Ji Q, Liu X, Fu X, et al: Resveratrol
inhibits invasion and metastasis of colorectal cancer cells via
MALAT1 mediated Wnt/β-catenin signal pathway. PLoS One.
8:e787002013. View Article : Google Scholar
|
|
21
|
Panzitt K, Tschernatsch MM, Guelly C, et
al: Characterization of HULC, a novel gene with striking
up-regulation in hepatocellular carcinoma, as noncoding RNA.
Gastroenterology. 132:330–342. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Matouk IJ, Abbasi I, Hochberg A, Galun E,
Dweik H and Akkawi M: Highly upregulated in liver cancer noncoding
RNA is overexpressed in hepatic colorectal metastasis. Eur J
Gastroenterol Hepatol. 21:688–692. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Xie H, Ma H and Zhou D: Plasma HULC as a
promising novel biomarker for the detection of hepatocellular
carcinoma. Biomed Res Int. 2013:1361062013. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Miyoshi N, Wagatsuma H, Wakana S, et al:
Identification of an imprinted gene, Meg3/Gtl2 and its human
homologue MEG3, first mapped on mouse distal chromosome 12 and
human chromosome 14q. Genes Cells. 5:211–220. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Zhang X, Zhou Y, Mehta KR, et al: A
pituitary-derived MEG3 isoform functions as a growth suppressor in
tumor cells. J Clin Endocrinol Metab. 88:5119–5126. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Zhou Y, Zhang X and Klibanski A: MEG3
noncoding RNA: a tumor suppressor. J Mol Endocrinol. 48:R45–R53.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Nissan A, Stojadinovic A,
Mitrani-Rosenbaum S, et al: Colon cancer associated transcript-1: a
novel RNA expressed in malignant and pre-malignant human tissues.
Int J Cancer. 130:1598–1606. 2012. View Article : Google Scholar
|
|
28
|
Kam Y, Rubinstein A, Naik S, et al:
Detection of a long non-coding RNA (CCAT1) in living cells and
human adenocarcinoma of colon tissues using FIT-PNA molecular
beacons. Cancer Lett. 352:90–96. 2014. View Article : Google Scholar
|
|
29
|
Ling H, Spizzo R, Atlasi Y, et al: CCAT2,
a novel noncoding RNA mapping to 8q24, underlies metastatic
progression and chromosomal instability in colon cancer. Genome
Res. 23:1446–1461. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Graham LD, Pedersen SK, Brown GS, et al:
Colorectal neoplasia differentially expressed (CRNDE), a novel gene
with elevated expression in colorectal adenomas and
adenocarcinomas. Genes Cancer. 2:829–840. 2011. View Article : Google Scholar
|
|
31
|
Ellis BC, Molloy PL and Graham LD: CRNDE:
A long non-coding RNA Involved in CanceR, Neurobiology, and
DEvelopment. Front Genet. 3:2702012. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Ellis BC, Graham LD and Molloy PL: CRNDE,
a long non-coding RNA responsive to insulin/IGF signaling,
regulates genes involved in central metabolism. Biochim Biophys
Acta. 1843:372–386. 2014. View Article : Google Scholar
|
|
33
|
Liu Q, Huang J, Zhou N, et al: LncRNA
loc285194 is a p53-regulated tumor suppressor. Nucleic Acids Res.
41:4976–4987. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Pibouin L, Villaudy J, Ferbus D, et al:
Cloning of the mRNA of overexpression in colon carcinoma-1: a
sequence overexpressed in a subset of colon carcinomas. Cancer
Genet Cytogenet. 133:55–60. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Peng JC, Shen J and Ran ZH: Transcribed
ultraconserved region in human cancers. RNA Biol. 10:1771–1777.
2013. View Article : Google Scholar
|
|
36
|
Scaruffi P: The transcribed-ultraconserved
regions: a novel class of long noncoding RNAs involved in cancer
susceptibility. ScientificWorldJournal. 11:340–352. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Calin GA, Liu CG, Ferracin M, et al:
Ultraconserved regions encoding ncRNAs are altered in human
leukemias and carcinomas. Cancer Cell. 12:215–229. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Sana J, Hankeova S, Svoboda M, Kiss I,
Vyzula R and Slaby O: Expression levels of transcribed
ultraconserved regions uc.73 and uc388 are altered in colorectal
cancer. Oncology. 82:114–118. 2012. View Article : Google Scholar
|
|
39
|
Wojcik SE, Rossi S, Shimizu M, et al:
Non-coding RNA sequence variations in human chronic lymphocytic
leukemia and colorectal cancer. Carcinogenesis. 31:208–215. 2010.
View Article : Google Scholar :
|
|
40
|
Ng SY, Gunning P, Eddy R, et al: Evolution
of the functional human beta-actin gene and its multi-pseudogene
family: conservation of noncoding regions and chromosomal
dispersion of pseudogenes. Mol Cell Biol. 5:2720–2732.
1985.PubMed/NCBI
|
|
41
|
Poliseno L: Pseudogenes: newly discovered
players in human cancer. Sci Signal. 5:re52012.PubMed/NCBI
|
|
42
|
Wezel F, Pearson J, Kirkwood LA and
Southgate J: Differential expression of Oct4 variants and
pseudogenes in normal urothelium and urothelial cancer. Am J
Pathol. 183:1128–1136. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Kastler S, Honold L, Luedeke M, et al:
POU5F1P1, a putative cancer susceptibility gene, is overexpressed
in prostatic carcinoma. Prostate. 70:666–674. 2010.
|
|
44
|
Panagopoulos I, Möller E, Collin A and
Mertens F: The POU5F1P1 pseudogene encodes a putative protein
similar to POU5F1 isoform 1. Oncol Rep. 20:1029–1033.
2008.PubMed/NCBI
|
|
45
|
Ali A, Saluja SS, Hajela K, Mishra PK and
Rizvi MA: Mutational and expressional analyses of PTEN gene in
colorectal cancer from Northern India. Mol Carcinog. 53(Suppl 1):
E45–E52. 2014. View Article : Google Scholar
|
|
46
|
Johnsson P, Ackley A, Vidarsdottir L, et
al: A pseudogene long-noncoding-RNA network regulates PTEN
transcription and translation in human cells. Nat Struct Mol Biol.
20:440–446. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Poliseno L, Salmena L, Zhang J, Carver B,
Haveman WJ and Pandolfi PP: A coding-independent function of gene
and pseudogene mRNAs regulates tumour biology. Nature.
465:1033–1038. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Lazar V and Garcia JG: A single human
myosin light chain kinase gene (MLCK; MYLK). Genomics. 57:256–267.
1999. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Han YJ, Ma SF, Yourek G, Park YD and
Garcia JG: A transcribed pseudogene of MYLK promotes cell
proliferation. FASEB J. 25:2305–2312. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Rack KA, Delabesse E, Radford-Weiss I, et
al: Simultaneous detection of MYC, BVR1, and PVT1 translocations in
lymphoid malignancies by fluorescence in situ hybridization. Genes
Chromosomes Cancer. 23:220–226. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Guan Y, Kuo WL, Stilwell JL, et al:
Amplification of PVT1 contributes to the pathophysiology of ovarian
and breast cancer. Clin Cancer Res. 13:5745–5755. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Takahashi Y, Sawada G, Kurashige J, et al:
Amplification of PVT-1 is involved in poor prognosis via apoptosis
inhibition in colorectal cancers. Br J Cancer. 110:164–171. 2014.
View Article : Google Scholar :
|
|
53
|
Barsotti AM, Beckerman R, Laptenko O,
Huppi K, Caplen NJ and Prives C: p53-Dependent induction of PVT1
and miR-1204. J Biol Chem. 287:2509–2519. 2012. View Article : Google Scholar :
|
|
54
|
Nakagawa H, Chadwick RB, Peltomaki P,
Plass C, Nakamura Y and de La Chapelle A: Loss of imprinting of the
insulin-like growth factor II gene occurs by biallelic methylation
in a core region of H19-associated CTCF-binding sites in colorectal
cancer. Proc Natl Acad Sci USA. 98:591–596. 2001. View Article : Google Scholar :
|
|
55
|
Murakami K, Oshimura M and Kugoh H:
Suggestive evidence for chromosomal localization of non-coding RNA
from imprinted LIT1. J Hum Genet. 52:926–933. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Mitsuya K, Meguro M, Lee MP, et al: LIT1,
an imprinted antisense RNA in the human KvLQT1 locus identified by
screening for differentially expressed transcripts using
monochromosomal hybrids. Hum Mol Genet. 8:1209–1217. 1999.
View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Nakano S, Murakami K, Meguro M, et al:
Expression profile of LIT1/KCNQ1OT1 and epigenetic status at the
KvDMR1 in colorectal cancers. Cancer Sci. 97:1147–1154. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Zhai H, Fesler A, Schee K, Fodstad O,
Flatmark K and Ju J: Clinical significance of long intergenic
noncoding RNA-p21 in colorectal cancer. Clin Colorectal Cancer.
12:261–266. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Wang G, Li Z, Zhao Q, et al: LincRNA-p21
enhances the sensitivity of radiotherapy for human colorectal
cancer by targeting the Wnt/β-catenin signaling pathway. Oncol Rep.
31:1839–1845. 2014.PubMed/NCBI
|
|
60
|
Yang F, Zhang H, Mei Y and Wu M:
Reciprocal regulation of HIF-1α and lincRNA-p21 modulates the
Warburg effect. Mol Cell. 53:88–100. 2014. View Article : Google Scholar
|
|
61
|
Yu M, Ohira M, Li Y, et al: High
expression of ncRAN, a novel non-coding RNA mapped to chromosome
17q25.1, is associated with poor prognosis in neuroblastoma. Int J
Oncol. 34:931–938. 2009.PubMed/NCBI
|
|
62
|
Qi P, Xu MD, Ni SJ, et al: Down-regulation
of ncRAN, a long non-coding RNA, contributes to colorectal cancer
cell migration and invasion and predicts poor overall survival for
colorectal cancer patients. Mol Carcinog. 2014. View Article : Google Scholar
|
|
63
|
Yang L, Lin C, Jin C, et al:
lncRNA-dependent mechanisms of androgen-receptor-regulated gene
activation programs. Nature. 500:598–602. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Ge X, Chen Y, Liao X, et al:
Overexpression of long noncoding RNA PCAT-1 is a novel biomarker of
poor prognosis in patients with colorectal cancer. Med Oncol.
30:5882013. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Meyer LR, Zweig AS, Hinrichs AS, et al:
The UCSC Genome Browser database: extensions and updates 2013.
Nucleic Acids Res. 41:D64–D69. 2013. View Article : Google Scholar :
|
|
66
|
Chung S, Nakagawa H, Uemura M, et al:
Association of a novel long non-coding RNA in 8q24 with prostate
cancer susceptibility. Cancer Sci. 102:245–252. 2011. View Article : Google Scholar
|
|
67
|
Li L, Sun R, Liang Y, et al: Association
between polymorphisms in long non-coding RNA PRNCR1 in 8q24 and
risk of colorectal cancer. J Exp Clin Cancer Res. 32:1042013.
View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Yang F, Huo XS, Yuan SX, et al: Repression
of the long noncoding RNA-LET by histone deacetylase 3 contributes
to hypoxia-mediated metastasis. Mol Cell. 49:1083–1096. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Salmena L, Poliseno L, Tay Y, Kats L and
Pandolfi PP: A ceRNA hypothesis: the Rosetta Stone of a hidden RNA
language? Cell. 146:353–358. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Tay Y, Rinn J and Pandolfi PP: The
multilayered complexity of ceRNA crosstalk and competition. Nature.
505:344–352. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Wang F, Li X, Xie X, Zhao L and Chen W:
UCA1, a non-protein-coding RNA up-regulated in bladder carcinoma
and embryo, influencing cell growth and promoting invasion. FEBS
Lett. 582:1919–1927. 2008. View Article : Google Scholar : PubMed/NCBI
|