Introduction
Mesenchymal stem cells (MSCs) are pluripotent adult
stromal cells that possess the ability to differentiate into bone,
cartilage and adipose tissues, and the cells exhibit promising
potential for regenerative medicine (1). An increasing number of studies have
demonstrated that MSCs demonstrate potential therapeutic effects
for multiple diseases, including liver injury,
ischemia/reperfusion-induced acute kidney injury and acute
myocardial infarction (2–4). However, transplantation therapy using
MSCs continues to be subject to investigation, particularly into
the safety of MSCs in clinical application. In a previous study
that aimed to determine the induction of MSC differentiation and
immune function, tumorigenesis was observed unexpectedly and a
cloned a novel tumor cell line, the F6 cell line, transformed from
human fetal bone marrow MSCs (5).
These findings initiated interest into investigating the mechanism
of mutation in fetal MSCs (FMSCs).
Previous studies have demonstrated that epigenetic
and genetic alterations markedly contribute to tumorigenesis. The
hypermethylation of tumor suppressor genes has been detected in
various types of tumors (6). Ohta
et al first reported the tumor suppressor gene fragile
histidine triad (FHIT), providing the molecular evidence for
the association between fragile sites and cancer (7). The loss or reduction of FHIT
expression has been revealed to be important in the initiation of
tumorigenesis in a variety of tumors (8,9). It has
also been reported that aberrant methylation of the FHIT
gene occurs in certain types of cancer (10,11).
Therefore, the methylation of the FHIT gene within F6 tumor
cells may be a novel direction for the identification of the
mechanism of transformation of FMSCs.
DNA methylation plays an important role in cancer
development, and has also become a therapeutic target due to the
reversibility of DNA methylation. The DNA methyltransferase (DNMT)
inhibitor, 5-Aza-2′-deoxycytidine (5-Aza-CdR) is able to restore
the expression of genes that induce growth arrest and apoptosis in
tumor cells (12,13). In the F6 tumor cell line, the effect
of 5-Aza-CdR on the cell cycle and cell apoptosis requires
additional investigation.
The aim of the present study is to investigate the
methylation status of the FHIT gene promoter in FMSC and F6
cells, and explore the effect of 5-Aza-CdR on the growth and
apoptosis of FMSC and F6 cells. The present results revealed that
methylation of the promoter of the FHIT gene occurred in the
F6 cell line. The expression of the FHIT gene was restored
in F6 cells, resulting in growth arrest and apoptosis, subsequent
to treatment with 5-Aza-CdR. The present results indicated the
significant role of FHIT in the malignant transformation of
FMSCs and provided a potential target for tumor therapy.
Materials and methods
Cell culture
Human FMSCs and F6 cells were from our laboratory
and cultured as previous described (5). FMSCs were derived from fetal bone marrow
and the F6 cells were the mutated cell line. FMSCs and F6 cells
were maintained in Dulbecco's modified Eagle's medium with low
glucose (LG-DMEM; Gibco Life Technologies, Carlsbad, CA, USA) that
was supplemented with 10% fetal bovine serum (FBS; Gibco Life
Technologies), 100 units/ml penicillin and 100 µg/ml streptomycin.
The cells were incubated at 37°C in a 5% CO2
atmosphere.
Reverse transcription polymerase chain
reaction (RT-PCR)
The total RNA of the MSCs and F6 cells was extracted
using TRIzol reagent (Invitrogen, Carlsbad, CA, USA). The cDNA was
synthesized using 4 mg of total RNA as the template and Oligo(dT)
as the primer, with the SuperScript II RT kit (Invitrogen),
according to the manufacturer's instructions. The cDNA was stored
at −20°C until RT-PCR was performed. The primer sequences were
designed as described in Table I.
 | Table I.Primer sequences for the amplification
of target genes. |
Table I.
Primer sequences for the amplification
of target genes.
| Genes | Primer sequence | Amplicon size,
bp | Annealing
temperature, °C |
|---|
| FHIT-M | F,
5′-AGGTACGGGGTTACGTTAGC-3′ | 133 | 60 |
|
| R,
5′-CAAACCTATTAAAACGAATTTTGC-3′ |
|
|
| FHIT-U | F,
5′-AATTGAGGTATGGGGTTATGTTAGT-3′ | 136 | 56 |
|
| R,
5′-AAACCTATTAAAACAAATTTTCACT-3′ |
|
|
| FHIT | F,
5′-CACGAAACTACCTTCAACTCC-3′ | 244 | 60 |
|
| R,
5′-AACTGTCCTTCGCTCTTGTG-3′ |
|
|
| β-actin | F,
5′-CACGAAACTACCTTCAACTCC-3′ | 265 | 56 |
|
| R,
5′-CATACTCCTGCTTGCTGATC-3′ |
|
|
DNA isolation and bisulfite
modification
DNA was isolated from F6 cells and FMSC using
Universal Genomic DNA Extraction kit (Takara Bio, Inc., Otsu,
Japan). Subsequently, 1 µg of genomic DNA was modified using the
CpGenome DNA Modification kit (Chemicon, Temecula, CA, USA)
according to the manufacturer's instructions. The modified DNA was
resuspended in water and used immediately or stored at −70°C until
use. Bisulfite treatment converted unmethylated cytosines to
uracils, while leaving the methylated cytosines unaffected.
Methylation-specific PCR and
sequencing
Methylation-specific PCR was performed as described
by Lin et al (14). DNA
methylation patterns in the CpG island of FHIT were
determined by chemical treatment with sodium bisulfite and
subsequent use of the aforementioned PCR procedure. Primer
sequences for the methylated FHIT reaction (M) and primer
sequences for the unmethylated FHIT reaction (U) are
described in Table I. The reaction
mixture (25 µl) contained 10X PCR buffer, 2.5 mmol/l
MgCl2, 2.5 mmol/l deoxyribonucleotide triphosphates,
0.25 units Hot Start DNA polymerase (Takara Bio, Inc.) and 2 µl of
modified DNA. Subsequent to the initial denaturation at 95°C for 10
min, 40 cycles of 45 sec at 94°C, 45 sec at 65°C (M) or 63°C (U)
and 45 sec at 72°C were performed. These cycles were followed by a
final 7 min elongation step at 72°C. DNA was treated in
vitro with DNA methylase from Spiroplasma sp. strain
MQ1, and untreated DNA was used as the positive control for the
methylated DNA. Negative control samples that lacked DNA were
included for each set of PCR. The PCR products were analyzed on 2%
agarose gels and visualized under the Gene Genius imaging system
(Synoptics Inc., Santa Clara, CA, USA) subsequent to ethidium
bromide staining. Products of the methylation reaction of the
FHIT gene DNA in F6 cells were cloned and sequenced
(Shanghai Genecore Biotechnologies Co., Ltd., Shanghai, China).
Treatment with 5-Aza-CdR
The F6 cells or FMSCs were plated into six-well
plates. After 24 h, 5-Aza-CdR was administered to each well at a
concentration of 1, 5, 25, 50 or 100 µM, and the morphological
changes were observed at 4, 8, 12 and 24 h. Subsequent to treatment
with 5-Aza-CdR, the expression of the FHIT gene was detected
by RT-PCR. Flow cytometry and Hoechst 33342 staining were used to
detect DNA content and apoptosis. All images were obtained using a
fluorescent microscope (Eclipse Ti-S; Nikon, Tokyo, Japan).
Statistical analysis. Differences between the groups
were examined for statistical significance using Student's
t-test and P<0.05 was considered to indicate a
statistically significant difference. Statistical analysis was
performed using GraphPad Prism 5 software (Graph Pad Software,
Inc., La Jolla, CA, USA). All results were expressed as the mean ±
standard deviation and all experiments were repeated at least three
times.
Results
FHIT gene expression level in FMSCs
and F6 cells
The expression of FHIT was initially detected
in FMSCs and F6 cells by RT-PCR. The results revealed that
FHIT mRNA was detectable in FMSCs, but not in F6 cells
(Fig. 1).
Methylation status of the FHIT gene in
F6 cells
The methylation status of the FHIT gene in F6
cells was detected by methylation-specific PCR. The promoter region
of FHIT contains numerous CpG islands. The results from
methylation-specific PCR revealed that FMSCs were amplified by
un-methylated and methylated primers, while F6 cells were amplified
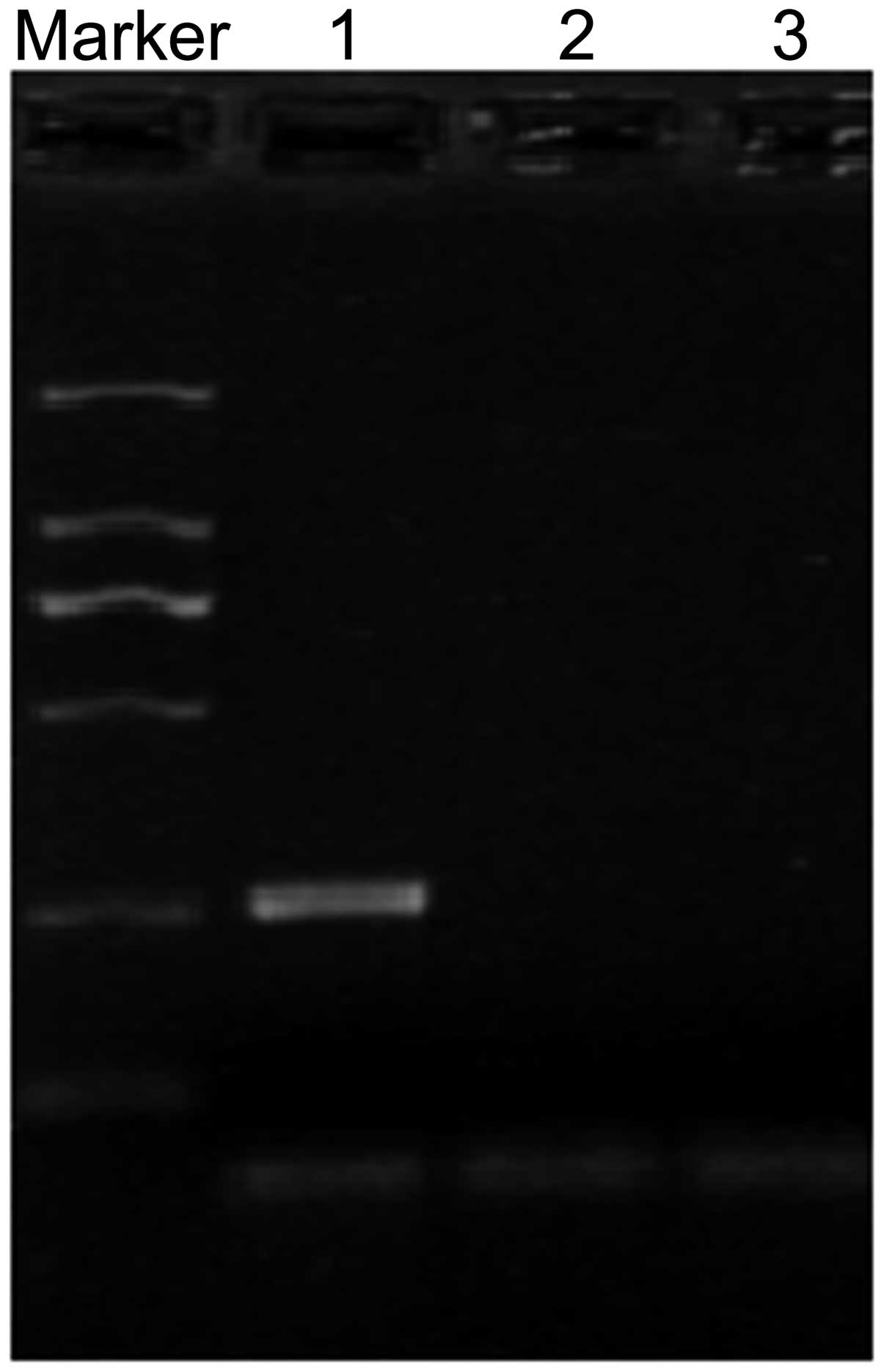
by methylated primers (Fig. 2A).
Sequencing of bisulfite-treated DNA was performed to investigate
the methylation status of the FHIT gene. The results of
sequencing demonstrated that FHIT gene promoter methylation
occurred in F6 cells (Fig. 2B).
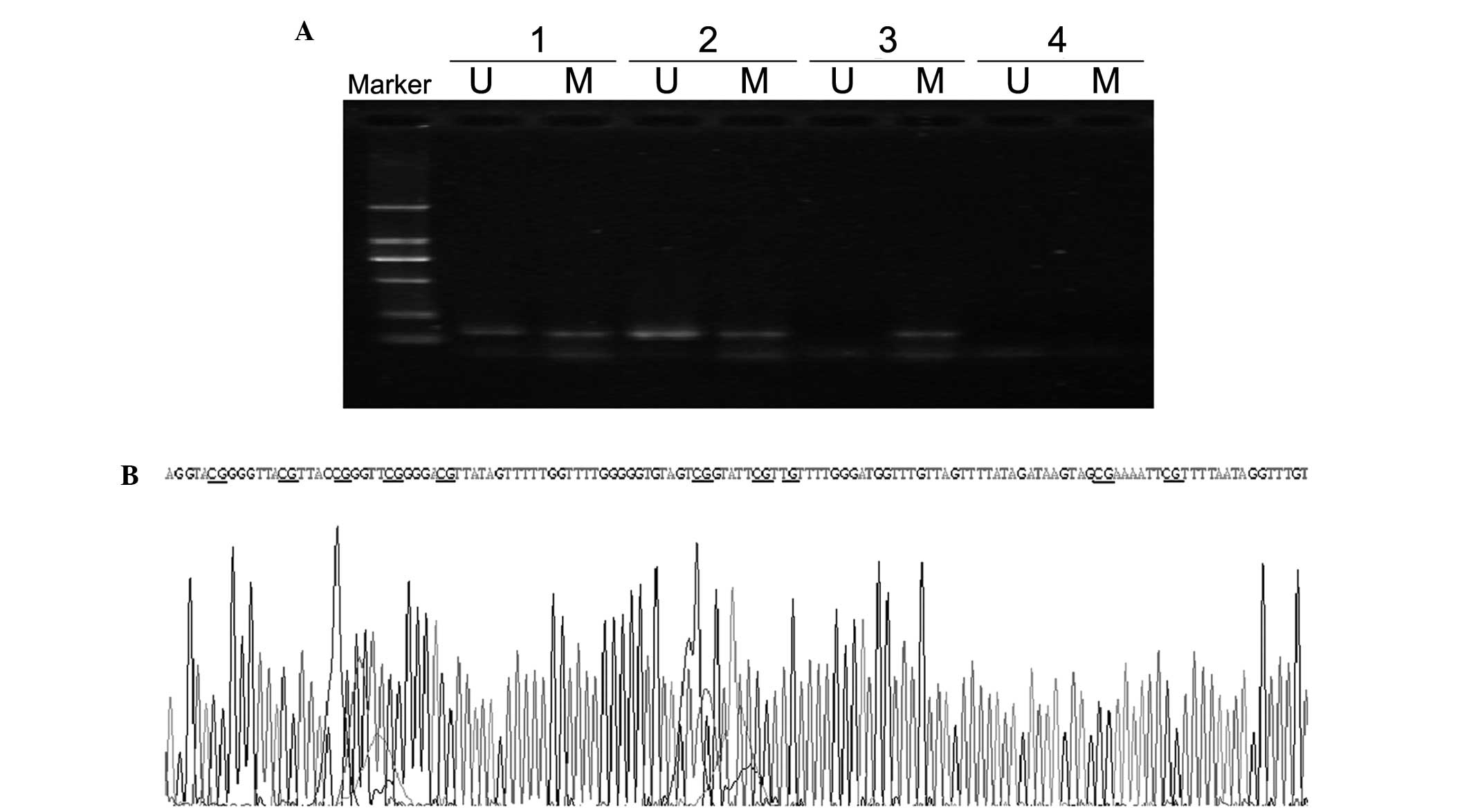 | Figure 2.Methylation status of the FHIT
gene in F6 cells and FMSCs. (A) Methylation-specific PCR of the
FHIT gene. U and M represent PCR results by using primer
sets for methylated and unmethylated FHIT gene,
respectively. Lane 1, positive controls for methylation and
unmethylation, comprising genomic DNA from normal bone marrow that
was modified with or without DNA methylase from Spiroplasma
sp. strain MQ1. Lane 2, FMSCs. Lane 3, F6 cells. Lane 4,
double-distilled H2O control. Marker, DL2000 DNA ladder.
(B) Sequencing of the methylated FHIT gene promoter in F6
cells. The CG sequences were not changed subsequent to bisulfite
treatment. FHIT, fragile histidine triad; PCR, polymerase
chain reaction; U, unmethylated; M, methylated; FMSCs, fetal
mesenchymal stem cells. |
Change in FHIT gene expression
subsequent to treatment with 5-Aza-CdR
Subsequent to treatment with 50 µM 5-Aza-CdR for 8
h, the expression of the FHIT gene was detected in F6 cells
by RT-PCR. The results demonstrated that the FHIT gene was
expressed in F6 cells (Fig. 3).
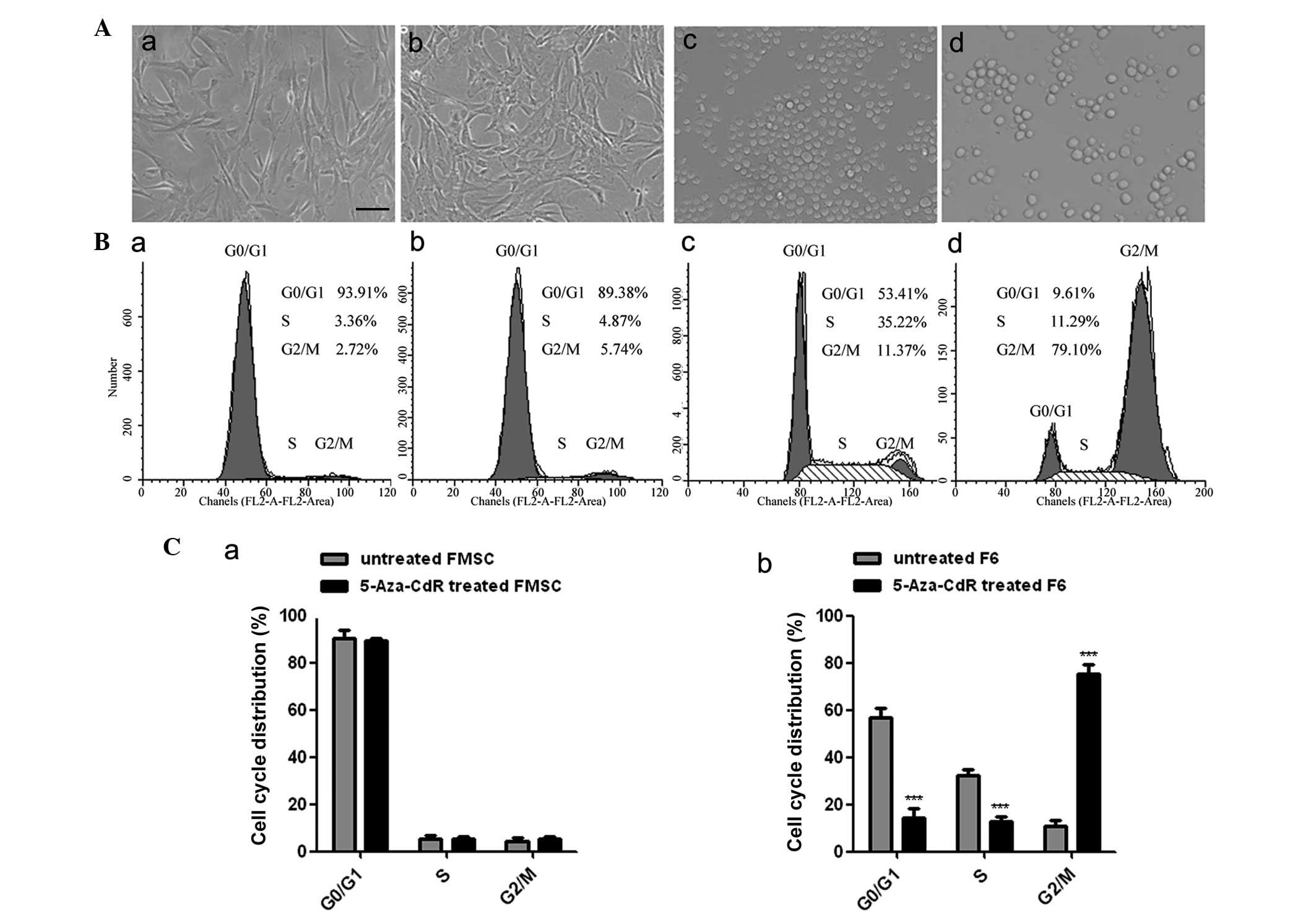
Morphological characteristics and cell
cycle
The cell morphology of F6 and FMSCs was assessed
subsequent to treatment with 5-Aza-CdR for 8 h. It was found that
treatment with 5-Aza-CdR inhibited the growth of F6 cells in
vitro compared with the control. The F6 cells became enlarged,
and the frequency of cell granulations increased. However, the
morphology of FMSCs remained unchanged at the same concentration of
5-Aza-CdR treatment (Fig. 4A). Flow
cytometry analysis revealed that the proportion of F6 cells in the
G2 phase was markedly increased following treatment with
5-Aza-CdR, while the cell cycle of the FMSCs was not changed
(Fig. 4B and C).
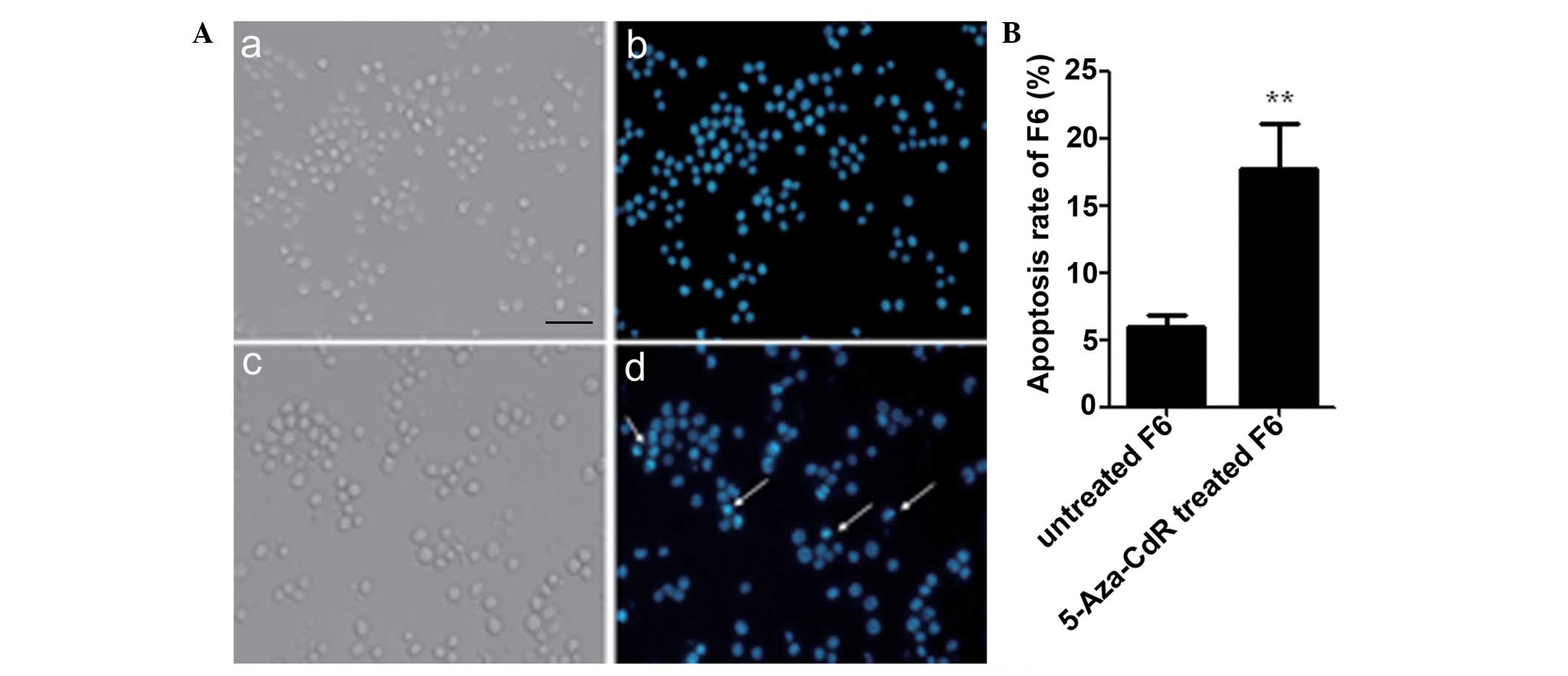
Apoptosis in F6 cells treated with
5-Aza-CdR
As demonstrated by the Hoechst 33342 staining
results in Fig. 5, the proportion of
apoptotic F6 cells was increased by 5-Aza-CdR. The nuclei of the
apoptotic F6 cells were pyknotic with fragmentation that was
stained bright blue (Fig. 5A). The
apoptotic ratio of F6 cells was increased following 5-Aza-CdR
treatment compared with the untreated cells (P<0.01; Fig. 5B).
Discussion
Previously, the methylation of CpG islands in tumor
suppressor genes during the initiation and development of tumors
initiated general interest as a subject for studies. Methylation of
the FHIT gene promoter plays an important role in several
types of solid tumor and hematological malignancy (15,16). In
the present study, the F6 cell line, a tumor cell line derived from
mutant human FMSCs, was focused on. The methylation status of the
FHIT gene was investigated, and the growth and apoptosis of
F6 cells was observed following demethylation by treatment with
5-Aza-CdR. It was found that the promoter region of FHIT was
methylated in the F6 cells. Treatment with 5-Aza-CdR induced an
increase in the expression of FHIT and also promoted
apoptosis in F6 cells.
Previous studies have indicated that the methylation
status of FHIT may lead to the silencing of gene expression,
promoting the occurrence and development of acute lymphoblastic
leukemia (17). In the present study,
the FHIT gene was found to be expressed in FMSCs, but not in
F6 cells. To confirm that the silencing of gene expression was due
to the methylation of FHIT during the transformation of
FMSCs into F6 cells, methylation-specific PCR was used. The
methylated FHIT gene was amplified using primers which only
anneal to sequences that are methylated. To verify DNA integrity
subsequent to bisulfite modification and to determine the
specificity of PCR amplification, the products of PCR were cloned
and sequenced. The results revealed that the CG sequence in the CpG
island was not sulfurized, and the FHIT gene demonstrated
complete methylation in F6 cells and partial methylation in FMSCs.
These findings indicated that the FHIT gene in FMSCs may
methylate progressively until the FMSCs undergo transformation into
F6 tumor cells.
Clinically, 5-Aza-CdR is used for the treatment of
myelodysplastic syndrome and demonstrates anti-leukemic activity
against acute myeloid leukemia (AML). The clinical activity of
5-Aza-CdR against solid tumors is under investigation (18). To confirm the possibility that the
silenced expression of the FHIT gene was caused by
methylation, the present study examined whether 5-Aza-CdR was able
to restore the expression of the FHIT gene. Subsequent to
treatment with 5-Aza-CdR, the promoter region of the FHIT
gene was identified as demethylated, and the expression of
FHIT was increased at the mRNA levels. Following exposure to
5-Aza-CdR for 8 h, the cell cycle and apoptosis were analyzed by
flow cytometry. In contrast to the results reported by Sard et
al (19), the cell cycle of F6
cells was arrested in the G2 phase, but not in the
G0 phase, and apoptosis in F6 cells occurred after 24 h.
By contrast, the morphology and cell cycle of FMSCs was not changed
throughout this process.
In the present study, 50 µM 5-Aza-CdR induced the
expression of the FHIT gene, but when the concentration of
5-Aza-CdR was increased to 100 µM, FHIT gene expression was
not restored. This result suggests that a low dose of 5-Aza-CdR
affects the demethylation and growth of cells, but a high dose may
result in severe side-effects, although a high dose may also
increase the rate of apoptosis. Therefore, the concentration and
exposure time of 5-Aza-CdR requires optimization in future studies
and a reasonable dose schedule is extremely important for clinical
application. A previous study revealed that the administration of
5-Aza-CdR in combination with tetrahydrouridine was effective in
inhibiting the growth of murine melanoma tumors (20). Thus, 5-Aza-CdR in combination with an
additional DNMT may decrease the side-effects of 5-Aza-CdR and
provide a novel approach for tumor therapy.
The novel tumor cell line F6 was mutated from human
FMSCs, and a previous study confirmed that F6 cells contain a
population of cancer stem cells (CSCs) that contribute to the
heterogeneity and tumorigenic potential of F6 cells (21). The present study revealed that
FHIT gene promoter methylation occurred in F6 cells
transformed from FMSCs. Additional studies are required to examine
the biological characteristics of F6 cells and the associated
pathways in this process. However, it is not clear whether other
tumor suppressor genes have epigenetic alterations. The association
between DNA methylation and F6 cells or CSCs requires additional
investigation.
In summary, the present study demonstrates that the
FHIT gene was methylated in F6 cells transformed from FMSCs,
and that treatment with 5-Aza-CdR restored the expression of
FHIT, altered the biological characteristics of F6 cells and
induced apoptosis in F6 cells. The present results not only
indicate that the silencing of the tumor suppressor gene
FHIT was due to the DNA methylation in the transformed human
mesenchymal stem cell F6 cell line, but also clarified the
epigenetic mechanism of tumorigenesis and safety of MSCs in
clinical application. Methylation of tumor suppressor genes may
provide potential molecular targets for tumor therapy and also act
as markers in the early detection of cancer.
Acknowledgements
The authors thank Qiaolin Chen for excellent
assistance with flow cytometry analysis. This study was supported
by the Key Project Supported by Medical Science and Technology
Development Foundation, Nanjing Department of Health (grant no.,
QYK11163).
References
|
1
|
Caplan AI: Adult mesenchymal stem cells
for tissue engineering versus regenerative medicine. J Cell
Physiol. 213:341–347. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Xu H, Qian H, Zhu W, Zhang X, Yan Y, Mao
F, Wang M, Xu H and Xu W: Mesenchymal stem cells relieve fibrosis
of Schistosoma japonicum-induced mouse liver injury. Exp Biol Med
(Maywood). 237:585–592. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Chen Y, Qian H, Zhu W, Zhang X, Yan Y, Ye
S, Peng X, Li W and Xu W: Hepatocyte growth factor modification
promotes the amelioration effects of human umbilical cord
mesenchymal stem cells on rat acute kidney injury. Stem Cells Dev.
20:103–113. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Dayan V, Yannarelli G, Billia F, Filomeno
P, Wang XH, Davies JE and Keating A: Mesenchymal stromal cells
mediate a switch to alternatively activated monocytes/macrophages
after acute myocardial infarction. Basic Res Cardiol.
106:1299–1310. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Xu W, Qian H, Zhu W, Chen Y, Shao Q, Sun
X, Hu J, Han C and Zhang X: A novel tumor cell line cloned from
mutated human embryonic bone marrow mesenchymal stem cells. Oncol
Rep. 12:501–508. 2004.PubMed/NCBI
|
|
6
|
Esteller M: CpG island hypermethylation
and tumor suppressor genes: A booming present, a brighter future.
Oncogene. 21:5427–5440. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Ohta M, Inoue H, Cotticelli MG, Kastury K,
Baffa R, Palazzo J, Siprashvili Z, Mori M, McCue P, Druck T, et al:
The FHIT gene, spanning the chromosome 3p14.2 fragile site and
renal carcinoma-associated t(3;8) breakpoint, is abnormal in
digestive tract cancers. Cell. 84:587–597. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Connolly DC, Greenspan DL, Wu R, Ren X,
Dunn RL, Shah KV, Jones RW, Bosch FX, Muñoz N and Cho KR: Loss of
fhit expression in invasive cervical carcinomas and intraepithelial
lesions associated with invasive disease. Clin Cancer Res.
6:3505–3510. 2000.PubMed/NCBI
|
|
9
|
Guler G, Uner A, Guler N, Han SY,
Iliopoulos D, McCue P and Huebner K: Concordant loss of fragile
gene expression early in breast cancer development. Pathol Int.
55:471–478. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Jeong YJ, Jeong HY, Lee SM, Bong JG, Park
SH and Oh HK: Promoter methylation status of the FHIT gene and Fhit
expression: Association with HER2/neu status in breast cancer
patients. Oncol Rep. 30:2270–2278. 2013.PubMed/NCBI
|
|
11
|
Sinha R, Hussain S, Mehrotra R, Kumar RS,
Kumar K, Pande P, Doval DC, Basir SF and Bharadwaj M: Kras gene
mutation and RASSF1A, FHIT and MGMT gene promoter hypermethylation:
Indicators of tumor staging and metastasis in adenocarcinomatous
sporadic colorectal cancer in Indian population. PLoS One.
8:e601422013. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Zhang JF, Zhang JG, Kuai XL, Zhang H,
Jiang W, Ding WF, Li ZL, Zhu HJ and Mao ZB: Reactivation of the
homeotic tumor suppressor gene CDX2 by
5-aza-2′-deoxycytidine-induced demethylation inhibits cell
proliferation and induces caspase-independent apoptosis in gastric
cancer cells. Exp Ther Med. 5:735–741. 2013.PubMed/NCBI
|
|
13
|
Kang HF, Dai ZJ, Bai HP, Lu WF, Ma XB, Bao
X, Lin S and Wang XJ: RUNX3 gene promoter demethylation by
5-Aza-CdR induces apoptosis in breast cancer MCF-7 cell line. Onco
Targets Ther. 6:411–417. 2013.PubMed/NCBI
|
|
14
|
Lin J, Yao DM, Qian J, Wang YL, Han LX,
Jiang YW, Fei X, Cen JN and Chen ZX: Methylation status of fragile
histidine triad (FHIT) gene and its clinical impact on prognosis of
patients with myelodysplastic syndrome. Leuk Res. 32:1541–1545.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Cecener G, Tunca B, Egeli U, Bekar A,
Tezcan G, Erturk E, Bayram N and Tolunay S: The promoter
hypermethylation status of GATA6, MGMT, and FHIT in glioblastoma.
Cell Mol Neurobiol. 32:237–244. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Griffiths EA, Gore SD, Hooker CM, Mohammad
HP, McDevitt MA, Smith BD, Karp JE, Herman JG and Carraway HE:
Epigenetic differences in cytogenetically normal versus abnormal
acute myeloid leukemia. Epigenetics. 5:590–600. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Chen X, Zhang H, Li P, Yang Z, Qin L and
Mo W: Gene expression of WWOX, FHIT and p73 in acute lymphoblastic
leukemia. Oncol Lett. 6:963–969. 2013.PubMed/NCBI
|
|
18
|
Karahoca M and Momparler RL:
Pharmacokinetic and pharmacodynamic analysis of
5-aza-2′-deoxycytidine (decitabine) in the design of its
dose-schedule for cancer therapy. Clin Epigenetics. 5:32013.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Sard L, Accornero P, Tornielli S, Delia D,
Bunone G, Campiglio M, Colombo MP, Gramegna M, Croce CM, et al: The
tumor-suppressor gene FHIT is involved in the regulation of
apoptosis and in cell cycle control. Proc Natl Acad Sci USA.
96:8489–8492. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Alcazar O, Achberger S, Aldrich W, Hu Z,
Negrotto S, Saunthararajah Y and Triozzi P: Epigenetic regulation
by decitabine of melanoma differentiation in vitro and in vivo. Int
J Cancer. 131:18–29. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Xu X, Qian H, Zhu W, Zhang X, Yan Y, Wang
M and Xu W: Isolation of cancer stem cells from transformed human
mesenchymal stem cell line F6. J Mol Med Berl. 88:1181–1190. 2010.
View Article : Google Scholar : PubMed/NCBI
|