Introduction
Ovarian cancer is the leading cause of mortality
from gynecological cancers and the fifth most common cause of
cancer-related mortality in females in the USA (1). Despite significant advances in surgical
management and chemotherapy over the past two decades, the survival
rate for ovarian cancer has not improved significantly.
Paclitaxel-based chemotherapy is important in the therapy of newly
diagnosed and recurrent ovarian cancer, however drug resistance may
seriously impede the clinical effect (2,3). Therefore
researchers are eager to understand how drug resistance develops in
ovarian carcinoma.
MicroRNAs (miRNAs) are RNAs of ~19–23 nucleotides in
length, and consist of a cluster of short non-protein-coding RNAs
(4). Increasingly, studies have
demonstrated that miRNAs are crucial in tumor cell response to
chemotherapeutic drugs by acting as oncogenes and tumor suppressors
(5–7).
More and more novel miRNAs, including miR-200c, miR-125b, miR-214,
have been reported to be associated with paclitaxel resistance in
ovarian cancer (8–10).
MiR-134, which is located in the 14q32.31, was
initially identified in cloning research of rat in 2002 (11); since this identification, reports have
shown that abnormal expression of miR-134 is associated with tumor
formation, cell proliferation and even chemoresistance. In 2012,
Hirota et al (12)
demonstrated that miR-134 expression was significantly decreased in
lung cancer tissues, and that miR-134 affects the fluorouracil
sensitivity of lung cancer by decreasing the expression of
dihydropyrimidine dehydrogenase. The study by Guo et al
(13) in 2010 revealed that in
multi-resistant small cell lung cancer cell line H69AR, miR-134
expression was decreased significantly, and that increasing the
expression of miR-134 in drug-resistant cells can significantly
increase therapeutic sensitivity to cisplatin, etoposide and
doxorubicin. However, there is currently no study with regard to
miR-134 in ovarian carcinoma chemoresistance.
A conventional way to identify miRNA targets is by
using bioinformatics, however different algorithms always produce
divergent results (14–16). Additionally, miRNAs often exhibit a
temporal or tissue-specific expression pattern, and their target
mRNAs share this characteristic, therefore, prediction of targets
by bioinformatic methods such as TargetScan, cannot be modulated by
these factors when researchers seek tissue and stage specific mRNA
of a certain miRNA. For these reasons, in the current study,
hybrid-polymerase chain reaction (PCR) (17), a rapid experimental approach for
screening putative target mRNAs of any known miRNA, was conducted
to identify target genes of miR-134 in SKOV3-TR30 cells. This study
is the first study aiming to target mRNA for a certain miRNA in the
study of ovarian cancer chemoresistance, as well as aiming to
identify novel targets of miR-134 to elucidate how miR-134
participates in the formation of ovarian cancer paclitaxel
resistance.
Materials and methods
Cell culture
The ovarian carcinoma cell line SKOV3 was provided
by the Tumor Cell Bank of the Chinese Academy of Medical Sciences
(Beijing, China). The paclitaxel-resistant ovarian carcinoma cell
line, SKOV3-TR30 with a resistant ability 27-fold greater than its
parental cell line, was derived from SKOV3 cell line and provided
by The Obstetrics and Gynecology Hospital Affiliated to Zhejiang
University (Hangzhou, China). SKOV3 cells were maintained in
McCoy's 5A medium (Gibco Life Technologies, Grand Island, NY, USA)
containing 10% fetal bovine serum (Gibco), 100 µg/ml penicillin
(Hyclone, Logan, UT, USA) and 100 µg/ml streptomycin (Hyclone).
SKOV3-TR30 cells were maintained in McCoy's 5A medium supplemented
with 10% fetal bovine serum, 100 µg/ml penicillin, 100 µg/ml
streptomycin and 30 nmol/l paclitaxel; paclitaxel was withdrawn 1
week prior to the experiment. All cells were maintained in a
humidified atmosphere of 5% CO2 at 37°C. HEK293 cells
were maintained in a Dulbecco's modified Eagle's medium (Gibco)
containing 10% fetal bovine serum. Cells in the logarithmic phase
of growth were used for all studies described, cultured in a
humidified atmosphere of 5% CO2 at 37°C.
RNA extraction and mRNA
purification
RNA was obtained from the SKOV-3 and SKOV-3TR30
cells. Total RNA was isolated using Trizol agent (Qiagen, Shanghai,
China) according to the manufacturer's instructions. RNA was
dissolved in 40 µl RNase free H2O and treated with the
TURBO DNA-free™ kit (Ambion Life Technologies, Carlsbad, CA, USA).
Additionally, the RNA quality was determined via ethidium bromide
staining following agarose/formaldehyde gel electrophoresis.
Quantitative reverse transcription
(qRT)-PCR for miR-134 expression
Total RNA from treated cells was extracted using
TRIzol reagent (Invitrogen Life Technologies, Carlsbad, CA, USA)
and quantified using an ultraviolet spectrophotometer (UVP Inc.,
Upland, CA, USA) at a wavelength of 260 nm. For miR-134 qRT-PCR,
cDNA was synthesized from 10 ng of total RNA using TaqManTM miRNA
hsa-miR-134-specific primers (Applied Biosystems Life Technologies,
Beijing, China) and a TaqManTM MicroRNA Reverse Transcription Kit
(Applied Biosystems Life Technologies). qPCR was performed on the
ABI PRISM 7900 Sequence Detection System (Applied Biosystems Life
Technologies). U6 snRNA was used as an endogenous control. All
reactions were performed in triplicate. The relative expression of
miR-134 was normalized to U6 RNA using the 2−ΔΔCT
method.
Hybrid-PCR
An miR-134 specific primer was designed for
hybrid-PCR. The sequence of miR-134 is UGUGACUGGUUGACCAGAGGGG. A
reverse and complementary sequences of miR-134 were generated for
the miR-134 hybrid primer and the primer sequence was
5′CCCCTCTGGTCRRCCRGTCRC3′. The seed region of miR-134 was
correspondingly located in the 3′-terminal of hybrid-primer. [As
G:U pairs are allowed for the miRNA:mRNA duplexes, the primer was
synthesized as a compatible primer. R in the primer represents
adenine (A) or guanine (G)]. After reverse transcription using
3′-Full RACE Core Set kit (Takara Bio, Inc., Otsu, Japan),
sequences between miR-134 binding sites and polyA signal were
amplified with the hybrid-primer, the outer primer
(5′-TACCGTCGTTCCACTAGTGATTT-3′) and the inner primer
(5′-CGCGGATCCTCCACTAGTGATTTCACTATAGG-3′) provided in the kit. To
acquire the actual sequences of miR-134 putative target mRNAs, all
hybrid-PCR products were harvested using the Qiaex® || gel
extraction kit (Qiagen), cloned into pMD 18-T vectors (TakaraBio,
Inc.,) and transformed into E. coli to produce a pool which
should contain partial sequences of putative mRNAs that miR-134
would bind to. Insertions were identified by PCR using M13 primers,
and confirmed by electrophoresis on a 2% agarose gel to determine
the size of inserted fragments in the pool. Clones observed in
different sizes were selected and the corresponding plasmids were
sequenced on an ABI 3730 automated sequencer (Applied Biosystems
Life Technologies).
Sequences blast and analysis
In order to identify the putative target genes of
miR-134, mRNA specific sequences located between the corresponding
sequence of miR-134 hybrid primer and polyA structure were
intercepted and analyzed using the online Basic Local Alignment
Search Tool (BLAST) (http://www.ncbi.nlm.nih.gov/blast).
Plasmid construction
MiR-134 putative binding sites within 200–300 bases
of flanking sequences of each target gene were amplified from
mRNA-derived cDNA and subsequently cloned into BamHI and
MluI sites of the multiple cloning regions of the luciferase
reporter constructor pMIR (Ambion Life Technologies). The sequence
predicted to express miR-134 was cloned into miRNA expression
vector pSilencer4.1 (Ambion Life Technologies) at the
BamHI-HindⅢ sites. All primer sequences used in
plasmid construction are listed in Table
I. All constructs were confirmed by DNA sequencing.
 | Table I.Primers designed for constructing the
plasmids used for luciferase reporter assays. |
Table I.
Primers designed for constructing the
plasmids used for luciferase reporter assays.
| Genes inserted | Primer sepuences |
|---|
| C16orf72 | F:
5′CCCAAGCTTTTGATAAAACGTGCCATT |
|
| R:
5′GGACTAGTTTACGCTTCTACTGCTGA |
| SRM | F:
5′CCCAAGCTTATGAATAATAGCAGTTCT |
|
| R:
5′GGACTAGTGGGAGATAGGTAGGAGTAGC |
| PNAS-105 | F:
5′CCCAAGCTTGCGCCGCCCGCCCGCCCG |
|
| R:
5′GGACTAGTTGAGGGGCAACAGAAGGCAG |
| VIM | F:
5′CCCAAGCTTGGAGCCCGCTGAGACTTGAA |
|
| R:
5′GGACTAGTAAAGATTTATTGAAGCAGAACC |
| F-box protein
22 | F:
5′CCCAAGCTTCCTGGCGGAGGCCGGCCACC |
|
| R:
5′GGACTAGTCTCTTCCTATGCAGGAAGAC |
| GAPDH | F:
5′CCCAAGCTTTGGTAAAGTGGATATTGTTG |
|
| R:
5′GGACTAGTGTTGAGCACAGGGTACTT |
| PRPF6 | F:
5′CCCAAGCTTGACGCGACGACGGCGACACT |
|
| R:
5′GGACTAGTGCCTGTTCTGACACGAGACA |
| RPL41 | F:
5′CCCAAGCTTGTGGAGGAAGAAGCGAATG |
|
| R:
5′GGACTAGTTTTATGAGCAAGGTGGGT |
| miR-134 | F:
5′CGCGGATCCTGTGAGGTGACGCTGGTG |
|
| R:
5′CCCAAGCTTTCGTGGTGGATTCGCTTT |
Luciferase Reporter Assays
HEK293 cells were plated at 2×105 cells
per well in a 24-well plate 24 h before transfection. pMIR
construct (200 ng) carrying the putative target sequence was
co-transfected with miR-134 expression plasmid, pSilencer-miR-134
(pSilencer was used as normal control) and 200 ng control renilla
plasmid, pRL-TK (Promega Corporation, Madison, WI, USA) into HEK293
cells using lipofectamine 2000 (Invitrogen Life Technologies).
Cells were collected 24 h post transfection and luciferase activity
levels were measured using the Dual Luciferase Assay System
(Promega Corporation). Firefly Luciferase activity was normalized
to renilla luciferase activity for each reaction. Transfected wells
were analyzed in triplicate for each group.
MicroRNA transfection
MiR-134 mimics and miRNA mimic negative control were
chemically synthesized by RiboBio (Guangzhou, China). RNA
oligonucleotides were transfected into cells at a final
concentration of 50 nM using Lipofectamine 2000 according to the
manufacturer's protocol.
Protein expression/western
blotting
Proteins were harvested using RIPA lysis buffer
(ThermoFisher Scientific, Waltham, MA, USA), diluted with buffer
containing SDS (Life Technologies, Grand Island, NY, USA) and
denatured at 95°C for 10 min. Protein lysates were subjected to 10%
Precise Tris-Glycine gel (Thermo-Fisher Scientific) for 75 min at
100 V and transferred onto a nitrocellulose membrane for 65 min at
100 V. Membranes were washed with Tris buffered saline (TBS;
Sigma-Aldrich, St. Louis, MO, USA), blocked with 5% dried nonfat
milk (Bio-Rad Laboratories, Hercules, CA, USA) in 1% tween-TBS and
probed using a rabbit polyclonal antibody against vimentin (VIM;
catalog no. ab45939; Abcam, Cambridge, UK; dilution, 1:1,000), with
a rabbit anti-GAPDH antibody (catalog no. ab181603; Abcam;
dilution, 1:2,000) as a visual loading control. Bands were
visualized using a horseradish peroxidase-conjugated polyclonal
goat anti-rabbit IgG secondary antibody (catalog no. ab136817;
Abcam; dilution, 1:2,000) and enhanced chemiluminescence, and
quantified using Chemi-Doc XRS imaging software version 2.0
(Bio-Rad Laboratories).
Statistical analysis
Statistical comparison between two groups was
performed using Student's t-test. P<0.05 was considered
to indicate a statistically significant difference. All data are
presented as the mean and standard deviation from at least three
separate experiments.
Results
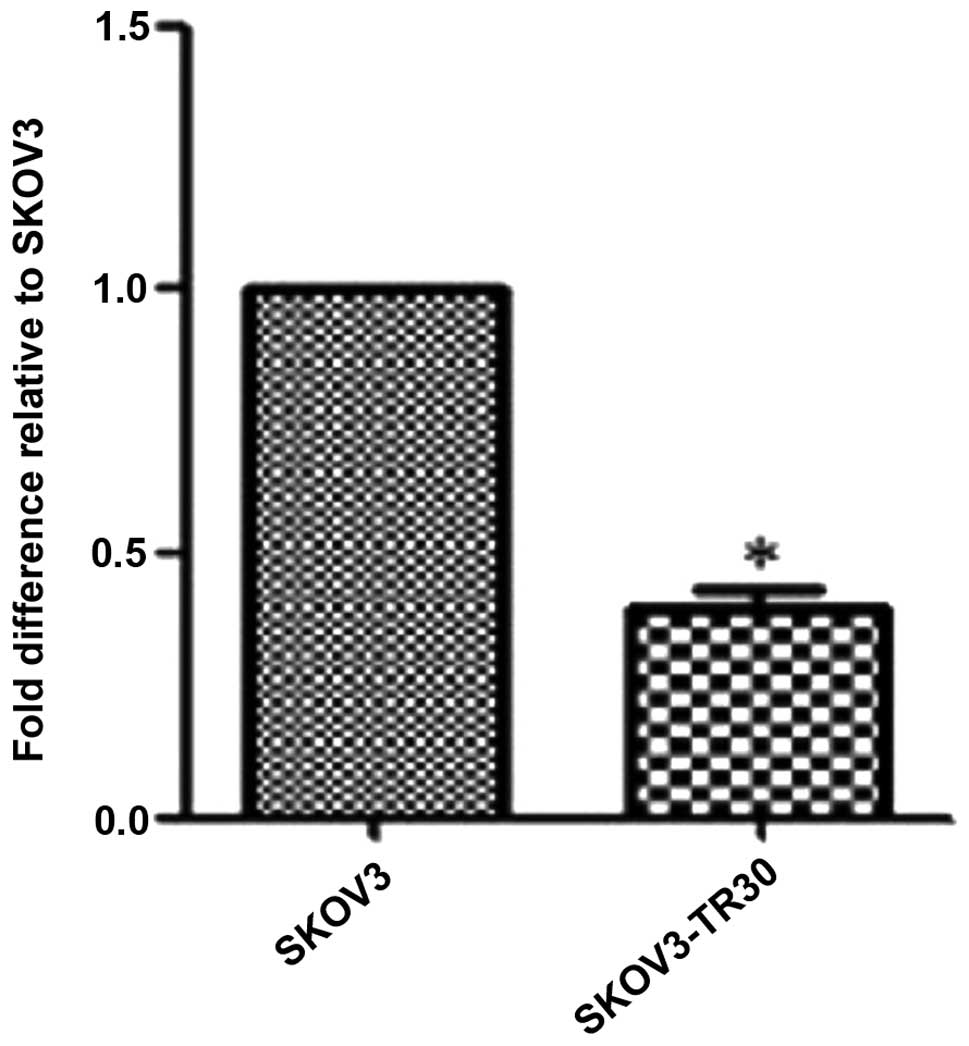
MiR-134 is down-regulated in
SKOV3-TR30 cells compared with in SKOV3
In our previous study we applied MicroRNA Gene Chip
analysis and found that miR-134 had a significantly decreased
expression in paclitaxel resistant SKOV3-TR30 cell line compared
with in its parental human ovarian carcinoma SKOV3 cell line
(18). In the current study, this
result is further confirmed by using qRT-PCR. SKOV3 and SKOV3-TR30
cells were analyzed to determine the miR-134 expression level, as
shown in Fig. 1. miR-134 expression
was significantly decreased in SKOV3-TR30 cells compared with SKOV3
cells, suggesting that the decreased expression of miR-134 may be
associated with the paclitaxel resistance of SKOV3-TR30 cells
(Fig. 1).
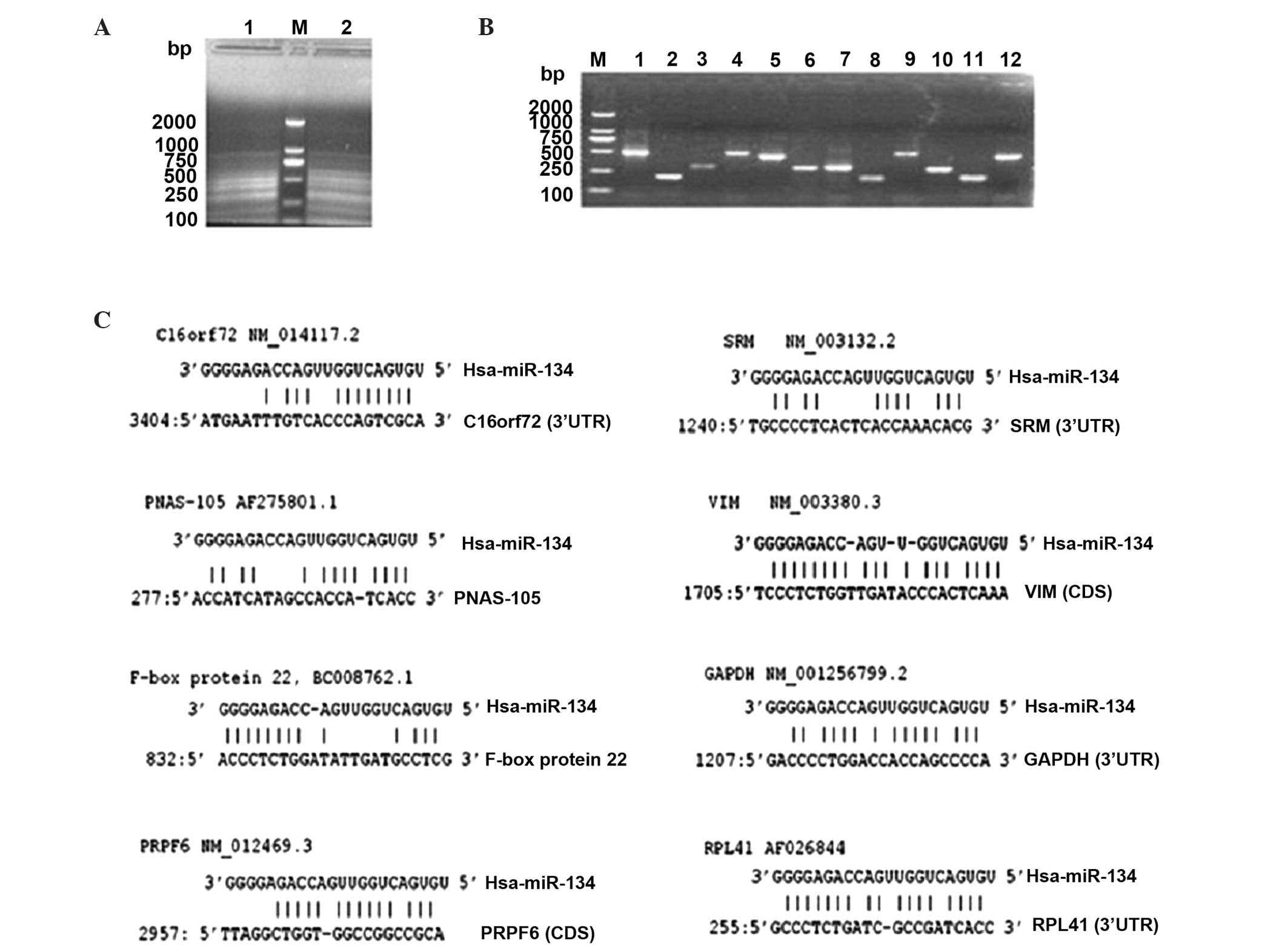
Hybrid-PCR may amplify and aid in the
identification of the putative target mRNA of miR-134 in SKOV3-TR30
cells
To explore the mechanism of how miR-134 may cause
paclitaxel resistance in SKOV3-TR30 cells, a newly established
method hybrid-PCR, was applied (17)
to identify the target mRNAs of miR-134 in SKOV3-TR30 cells.
Hybrid-PCR was projected as semi-nested PCR using the hybrid-primer
and the outer/inner primers homologous to the oligo dT-3 site
adaptor primer. The products of amplification were variable in
length (Fig. 2A). To acquire the
actual sequences of miR-134 putative target mRNAs, all hybrid-PCR
products (with different lengths shown in Fig. 2A and B) were harvested by gel
extraction, cloned into T-vectors and then transformed into E.
coli for further selection and amplification. Positive colony
forming units were picked for sequencing (Fig. 2B shows partial of the positive colony
units). In total, 28 sequences were obtained successfully in the
current study. Hybrid-primer sequences and polyA structure were
confirmed for a complete extremity of mRNA. These 28 mRNA sequences
located between hybrid-primer and polyA were intercepted and the
BLAST tool was used to identify their host genes. Of the 28
sequences, 8 matched sequences in GenBank and their host mRNAs were
identified successfully. The remaining 20 were not identified.
Detailed information is reported in Table II and Fig.
2C.
 | Table II.8 putative target mRNAs of miR-134
identified by Hybrid-PCR in SKOV3-TR30 cells. |
Table II.
8 putative target mRNAs of miR-134
identified by Hybrid-PCR in SKOV3-TR30 cells.
| Putative target
mRNA | Accession
number |
|---|
| Homo sapiens
chromosome 16 open reading frame 72 (C16orf72) | NM_014117.2 |
| Homo sapiens
PNAS-105 (PNAS-105) | AF275801.1 |
| Homo sapiens
spermidine synthase (SRM) | NM_003132.2 |
| Homo sapiens
vimentin (VIM) | NM_003380.3 |
| Homo sapiens F-box
protein 22(F-box protein 22) | BC008762.1 |
| Homo sapiens
glyceraldehyde-3-phosphate dehydrogenase (GAPDH) | NM_001256799.2 |
| Homo sapiens PRP6
pre-mRNA processing factor 6 homolog (PRPF6) | NM_012469.3 |
| Homo sapiens
ribosomal protein L41 mRNA(RPL41) | AF026844 |
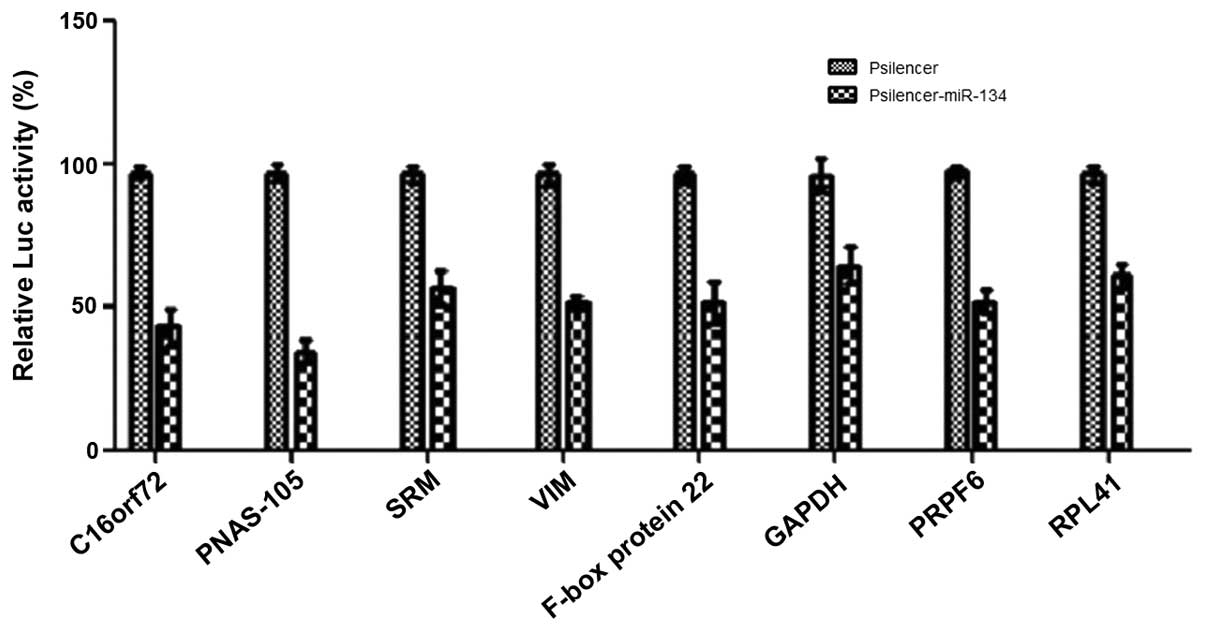
Validation of predicted targets of
miR-134 by luciferase reporter assays
The current study aimed to validate the interaction
between miR-134 and the predicted target genes obtained from
hybrid-PCR. The majority of studies have shown that miRNAs regulate
target gene expression by binding to the sites located in the
3′untranslated regions (UTR) of mRNAs. In recent years, increasing
evidence has demonstrated that miRNAs also bind in the coding
region (CDS). As we previously found that miR-134 binds to the CDS
of VIM and PRPF6, in the current study, we also
determined whether or not the putative binding sequences located in
the CDS represent functional target sites for miR-134. Putative
binding sequences of each target gene were successfully amplified
and cloned into luciferase reporter constructor pMIR (data not
shown). Luciferase activity was measured in the presence of
pSilencer-miR-134 or pSilencer negative control and normalized
using Renilla activity. Compared with the pSilencer negative
control group, co-transfection of Psilencer-miR-134 with pMIR
containing candidate target sequences all led to a decrease in
luciferase activity. These data demonstrate that the putative
binding sites that have been validated in the current study may be
recognized by miR-134. Using this approach, we were able to
validate all of the target genes predicted by hybrid-PCR (Fig. 3).
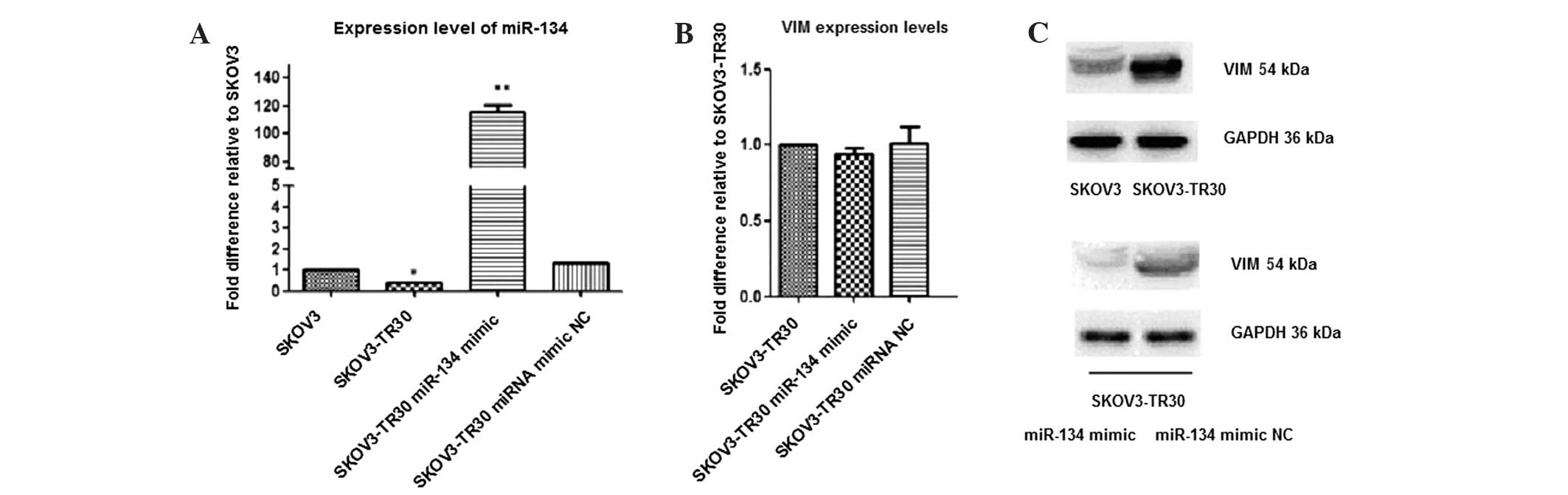
VIM is a direct target of miR-134 in
SKOV3-TR30 cells
To further clarify whether the target mRNA protein
levels were mediated through miR-134, VIM was selected for
further investigation. Recently, it has been demonstrated that
miRNAs can regulate target mRNAs by binding to the CDS (19–22).
Notably, in the current study, 8 putative target mRNAs of
has-miR-134 were obtained overall in SKOV3-TR30 cells, and among
these, putative binding sites of VIM and PRPF6 were
not located in the 3′UTR, but rather in the CDS region.
Additionally, studies have shown that VIM functions in cell
adhesion, migration, survival, and cell signaling processes via
dynamic assembly/disassembly in cancer cells (23,24).
Therefore, in the current study VIM was selected to
investigate whether miR-134 may target VIM in SKOV3-TR30
cells.
SKOV3-TR30 cells were transfected with miR-134 mimic
(while using transfected miRNA mimc negative control as the control
group) and subsequently RT-PCR and western blot analysis were
performed to measure the VIM mRNA and protein levels. The results
demonstrated that miR-134 had only a limited effect on the mRNA of
the endogenous target VIM, however, the corresponding protein was
affected more substantially. Combined with luciferase reporter
assay (Fig. 4), the results indicate
that VIM is likely to be direct target of miR-134 and that
VIM protein was regulated at the post-transcriptional level in
SKOV-TR30 cells.
Discussion
Among gynecological malignancies, ovarian carcinoma
is the leading cause of cancer death worldwide (25) and the majority of cases are associated
with recurrence and chemoresistance (26). Studies have shown that abnormal
expression of certain miRNAs such as miR-200c, miR-125b,
miRNA-31,miR-182 in ovarian cancer were associated with activated
tumor cell proliferation, enhanced migration ability and may
increase chemoresistance (8,9,27,28).
Increasing evidence has shown that miR-134 is
involved in the promotion of tumorigenesis, chemoresistance and its
expression is downregulated in a number of cancers. In our previous
study following analysis by miRNA microarray, we reported for the
first time that miR-134 is downregulated in ovarian cancer
paclitaxel-resistant SKOV3-TR30 cells compared with the parental
SKOV3 cells (18); in the current
study we confirm this result by applying real-time PCR, as shown in
Fig. 1.
miRNAs often show temporal and tissue-specific
expression, which means that the differential expression of certain
miRNA only occurs in a certain cell or at a particular stage; their
target mRNA also exhibits this characteristic. However, the
prediction of targets by bioinformatics methods such as TargetScan
cannot accommodate these factors when users require tissue- and
stage-specific mRNA of a certain miRNA. Hybrid-PCR, a recently
reported tissue- and stage-specific method (17) was used in order to identify specific
mRNA targets of miR-134 in ovarian cancer SKOV3-TR30 cells to gain
an improved understanding of how miR-134 functions in ovarian
carcinoma paclitaxel resistance.
By applying this method, 8 putative target genes of
miR-134 were identified in SKOV3-TR30 cells, including
C16orf72, PNAS-105, SRM, VIM, F-box
protein 2, GAPDH, PRPF6 and RPL41.
Furthermore, by successfully constructing PMIR luciferase reporter
plasmids of putative binding sites in these 8 mRNAs and applying
luciferase reporter assays, the current results demonstrated that,
compared with the pSilencer negative control group, co-transfection
of Psilencer-miR-134 with pMIR containing the candidate target
sequences all led to a decrease in luciferase activity. These data
demonstrated that the putative binding sites that have been
identified by hybrid-PCR may be recognized by miR-134.
Subsequently, VIM was selected out of the 8 putative target
genes to further investigate whether miR-134 regulates the protein
expression level of the target genes. VIM was selected for
the following reasons: Firstly, it is well known that VIM is an
essential constituent of cytoskeletal proteins of mesenchymal cells
and VIM is a marker of epithelial-mesenchymal transition (EMT)
(29). Many studies have shown that
EMT is critical for chemoresistance, particularly in ovarian cancer
(30,31). Secondly, VIM has a putative
miR-134 target site in its CDS region. A number of recent reports
have shown that miRNAs also target the CDS of mammalian genes
(32,33) and have a critical function in certain
aspects of cancer cell biology, including in apoptosis. The results
in the current study demonstrate that miR-134 mimic (compared with
miRNA mimic negative control) was successfully transfected into
SKOV3-TR30 cells and the protein level of VIM was significantly
decreased in SKOV3-TR30 cells transfected with miR-134 mimic.
In summary, in this study demonstrated that miR-134
is downregulated in the ovarian carcinoma paclitaxel-resistant cell
line SKOV3-TR30, compared with the parental SKOV3 cell line. By
conducting hybrid-PCR, 8 putative targets of miR-134 were
identified in SKOV3-TR30 cells, and by applying a luciferase
reporter assay we further confirmed that hybrid-PCR may be used as
a simple and effective method to screen putative target mRNAs of
miR-134 in the study of ovarian cancer chemoresistance. By
transfecting SKOV3-TR30 cells with miR-134 mimic and subsequently
conducting RT-PCR and western blot experiments, this study presents
the first evidence that VIM is regulated at the
post-transcriptional level by miR-134, which directly targets
VIM within its coding region; this finding provides an
important and novel perspective for determining the complex
mechanism of miRNA/mRNA interactions. Follow-up studies will
investigate the mechanism of miR-134 in paclitaxel resistance in
SKOV3-TR30 cells by regulating the target genes identified in this
study, including VIM.
Acknowledgements
This study is supported by the Natural Science
Foundation of China (Grant No. 30973189).
References
|
1
|
Siegel R, Naishadham D and Jemal A: Cancer
statistics, 2013. CA Cancer J Clin. 63:11–30. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
See HT, Kavanagh JJ, Hu W and Bast RC:
Targeted therapy for epithelial ovarian cancer: current status and
future prospects. Int J Gynecol Cancer. 13:701–734. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Yusuf RZ, Duan Z, Lamendola DE, et al:
Paclitaxel resistance: molecular mechanisms and pharmacologic
manipulation. Curr Cancer Drug Targets. 3:1–19. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Barbarotto E, Schmittgen TD and Calin GA:
MicroRNAs and cancer: profile. Int J Cancer. 122:969–977. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Chen CZ: MicroRNAs as oncogenes and tumor
suppressors. N Engl J Med. 353:1768–1771. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Cho WC: OncomiRs: the discovery and
progress of microRNAs in cancers. Mol Cancer. 6:602007. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Hu X, Macdonald DM, Huettner PC, et al: A
miR-200 microRNA cluster as prognostic marker in advanced ovarian
cancer. Gynecol Oncol. 114:457–464. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Cittelly DM, Dimitrova I, Howe EN, et al:
Restoration of miR-200c to ovarian cancer reduces tumor burden and
increases sensitivity to paclitaxel. Mol Cancer Ther. 11:2556–2565.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Kong F, Sun C, Wang Z, et al: miR-125b
confers resistance of ovarian cancer cells to cisplatin by
targeting pro-apoptotic Bcl-2 antagonist killer 1. J Huazhong Univ
Sci Technolog Med Sci. 31:543–549. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Yang H, Kong W, He L, et al: MicroRNA
expression profiling in human ovarian cancer: miR-214 induces cell
survival and cisplatin resistance by targeting PTEN. Cancer Res.
68:425–433. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Lagos-Quintana M, Rauhut R, Yalcin A,
Meyer J, Lendeckel W and Tuschl T: Identification of
tissue-specific microRNAs from mouse. Curr Biol. 12:735–739. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Hirota T, Date Y, Nishibatake Y, et al:
Dihydropyrimidine dehydrogenase (DPD) expression is negatively
regulated by certain microRNAs in human lung tissues. Lung Cancer.
77:16–23. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Guo L, Liu Y, Bai Y, Sun Y, Xiao F and Guo
Y: Gene expression profiling of drug-resistant small cell lung
cancer cells by combining microRNA and cDNA expression analysis.
Eur J Cancer. 46:1692–1702. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Baek D, Villen J, Shin C, Camargo FD, Gygi
SP and Bartel DP: The impact of microRNAs on protein output.
Nature. 455:64–71. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Bentwich I: Prediction and validation of
microRNAs and their targets. FEBS Lett. 579:5904–5910. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Rajewsky N: microRNA target predictions in
animals. Nat Genet. 38:Suppl. 8–13. 2006. View Article : Google Scholar
|
|
17
|
Huang Y, Qi Y, Ruan Q, et al: A rapid
method to screen putative mRNA targets of any known microRNA. Virol
J. 8:82011. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Shuang T, Shi C, Chang S, Wang M and Bai
CH: Downregulation of miR-17~92 expression increase
paclitaxel sensitivity in human ovarian carcinoma SKOV3-TR30 cells
via BIM instead of PTEN. Int J Mol Sci. 14:3802–3816. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Hausser J, Syed AP, Bilen B and Zavolan M:
Analysis of CDS-located miRNA target sites suggests that they can
effectively inhibit translation. Genome Res. 23:604–615. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Duursma AM, Kedde M, Schrier M, le Sage C
and Agami R: miR-148 targets human DNMT3b protein coding region.
RNA. 14:872–877. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Forman JJ, Legesse-Miller A and Coller HA:
A search for conserved sequences in coding regions reveals that the
let-7 microRNA targets Dicer within its coding sequence. Proc Natl
Acad Sci USA. 105:14879–14884. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Tay Y, Zhang J, Thomson AM, Lim B and
Rigoutsos I: MicroRNAs to Nanog, Oct4 and Sox2 coding regions
modulate embryonic stem cell differentiation. Nature.
455:1124–1128. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
McInroy L and Mӓӓttӓ A: Down-regulation of
vimentin expression inhibits carcinoma cell migration and adhesion.
Biochem Biophys Res Commun. 360:109–114. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Satelli A and Li S: Vimentin in cancer and
its potential as a molecular target for cancer therapy. Cell Mol
Life Sci. 68:3033–3046. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Jemal A, Siegel R, Xu J and Ward E: Cancer
statistics, 2010. CA Cancer J Clin. 60:277–300. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Matulonis U and Abrahm JL: Cancer of the
ovary. N Engl J Med. 352:1268–1269. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Mitamura T, Watari H, Wang L, et al:
Downregulation of miRNA-31 induces taxane resistance in ovarian
cancer cells through increase of receptor tyrosine kinase MET.
Oncogenesis. 2:e402013. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Wang YQ, Guo RD, Guo RM, Sheng W and Yin
LR: MicroRNA-182 promotes cell growth, invasion and chemoresistance
by targeting programmed cell death 4 (PDCD4) in human ovarian
carcinomas. J Cell Biochem. 114:1464–1473. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Satelli A and Li S: Vimentin in cancer and
its potential as a molecular target for cancer therapy. Cell Mol
Life Sci. 68:3033–3046. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Ahmed N, Abubaker K, Findlay J and Quinn
M: Epithelial mesenchymal transition and cancer stem cell-like
phenotypes facilitate chemoresistance in recurrent ovarian cancer.
Curr Cancer Drug Targets. 10:268–278. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Marchini S, Fruscio R, Clivio L, et al:
Resistance to platinum-based chemotherapy is associated with
epithelial to mesenchymal transition in epithelial ovarian cancer.
Eur J Cancer. 49:520–530. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Qin W, Shi Y, Zhao B, Yao C, Jin L, Ma J
and Jin Y: miR-24 regulates apoptosis by targeting the open reading
frame (ORF) region of FAF1 in cancer cells. PLoS One. 5:e94292010.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Zhang J, Guo H, Qian G, Ge S, Ji H, Hu X
and Chen W: MiR-145, a new regulator of the DNA fragmentation
factor-45 (DFF45)-mediated apoptotic network. Mol Cancer.
9:2112010. View Article : Google Scholar : PubMed/NCBI
|