Introduction
Pancreatic cancer has the highest mortality rate
among all cancer types, with an overall five-year survival rate of
<5% (1–3). In the United States, pancreatic cancer
has been estimated to cause ≥36,800 mortalities annually (2). Thus, novel treatment and detection
methods are urgently required for pancreatic cancer patients.
Aberrant methylation of CpG-rich sequences is a
common epigenetic alteration in human cancers, including pancreatic
cancer (4,5). In numerous cases, a tumor may arise
following the methylation of the promoter of a tumor suppressor
gene, leading to gene silencing. This is typically an early event
in tumorigenesis, thus these events may be used as a diagnostic
marker to detect early-stage pancreatic cancer (6). Therefore, an understanding of the
altered gene methylation patterns and the underlying molecular
mechanisms in pancreatic cancer may have a significant clinical
impact.
GS homeobox 2 (GSH2), also know as GSX2, is a
homeobox gene involved in the regulation of mammalian organ
development downstream of the sonic hedgehog (Shh) signaling
pathway (7). GSH2 is hypermethylated
in astrocytomas (8), and Shh
signaling is involved in the initiation and progression of
pancreatic cancer (9–11). However, the methylation status of GSH2
in pancreatic cancer patients remains unclear. In the present
study, the methylation status of the GSH2 gene transcriptional
regulation region (TRR) was examined in primary carcinoma and
paired normal tissues derived from 47 patients with pancreatic
cancer, and the association of methylation with the
clinicopathological features of the patients was also assessed.
These findings suggest that GSH2 methylation status may provide a
novel diagnostic tool for pancreatic cancer.
Materials and methods
Cell line and culture
The pancreatic cancer PANC1 cell line was purchased
from the American Type Culture Collection (Manassas, VA, USA), and
PaTu8988 cells were a gift from Dr H.P. Elsasser (Phillips
University, Marburg, Germany). The cells were grown in Dulbecco's
modified Eagle's medium supplemented with 10% fetal bovine serum
(Life Technologies Inc., Rockville, MD, USA) and incubated at 37°C
in a humidified chamber with 95% air and 5% CO2.
Sample collection and DNA
preparation
The tissue and patient data usage protocol was
approved by the Ethics Committee of the General Hospital of
Shenyang Military Command (Shenyang, Liaoning, China). Written
informed consent was obtained from each patient. A total of 47
primary tumor and corresponding normal tissue specimens were
obtained from the Second Military Medical University affiliated to
Changhai Hospital (Shanghai, China) between September 2007 and
September 2009. Immediately after surgical resection, the tissue
samples were stored in liquid nitrogen. Tumor tissues containing
>70% tumor cells were used as primary tumor samples, and the
corresponding adjacent normal tissues without any tumor cell
infiltration were selected as normal tissue samples. Genomic DNA
from the tissues was extracted using the phenol/chloroform method
and ethanol precipitation.
Sodium bisulfite modification
Genomic DNA (1 µg) was processed using the EZ DNA
Methylation™ kit (Zymo Research, Orange, CA, USA) according to the
manufacturer's instructions. The bisulfite-modified DNA was
subsequently suspended in deionized water (20 µl) for immediate use
or stored at −80°C.
Bisulfite-specific PCR (BSP) and DNA
sequencing
GSH2 promoter methylation detection was conducted
using custom primers designed to specifically amplify the
bisulfite-converted DNA of the GSH2 TRR (Shanghai Shenggong Biology
Engineering Technology Service, Ltd., Shanghai, China). The primers
used were as follows: Forward, 5′-GTTTAAAGGGAGGCGATTAGATAG-3′ and
reverse, 5′-TTCTCTCTCCAACTCCAAAAATTA-3′. For PCR analysis,
bisulfite modified DNA (2 µl from each sample) was combined in 25
µl of reaction mixture (Takara Biotechnology Co., Ltd., Dalian,
China) containing 1X PCR buffer, 2.0 mM MgCl2, 2.5 mM
dNTP, 1 mM primer and 800 U/l EX Taq DNA HS. The reaction mixture
was preheated at 95°C for 5 min and amplified using a touchdown PCR
program as follows on a PCR system (TProfessional Thermocycler;
Biometra GmbH, Göttingen, Germany): 9 cycles of 95°C for 30 sec,
59°C for 30 sec (next cycle touchdown 0.5°C) and 72°C for 30 sec;
42 cycles of 95°C for 30 sec, 55°C for 30 sec, and 72°C for 30 sec;
and a final extension of 4 min at 72°C. The products subsequently
underwent direct sequencing analysis, or were cloned into the
pMD-18-T vector (Takara Biotechnology Co., Ltd.) prior to
sequencing analysis; from each sample, 10–25 clones were randomly
selected for DNA sequencing.
Quantitative methylation-specific PCR
(qMSP)
The bisulfite-treated DNA was amplified using qMSP,
in a 7500 Real-Time PCR System (Applied Biosystems, Foster City,
CA, USA), using the following primers and probes: GSH2 forward,
5′-GTTTTCGATGCGTAGGATGC-3′ and reverse,
5′-ACGTTTTTAACTAAACCTACGCGC-3′; GSH2 probe,
5′-FAM-ATCCCTCTTTAATCCT-MGB-3′; β-ACTB forward,
5′-TGGTGATGGAGGAGGTTTAGTAAGT-3′ and reverse,
5′-AACCAATAAAACCTACTCCTCCCTTAA-3′; and β-ACTB probe,
5′-FAM-TTTGTTATTGTGTGTTGGGTG-MGB-3′. The PCR amplification was
conducted as follows: Initial denaturation step of 95°C for 10 min,
then 45 cycles of 95°C for 15 sec, 70°C for 15 sec and 60°C for 60
sec. The bisulfite-treated DNA that was obtained from the Panc1
cells and was fully methylated by SssI methylase was used as a
positive control. β-actin was used as an internal control to
correct for differences in quality and quantity between
samples.
Statistical analysis
The associations between GSH2 methylation and the
clinicopathological parameters were analyzed using χ2 or
Student's t-tests with SPSS software, version 13.0 (SPSS, Inc.,
Chicago, IL, USA). P<0.05 was considered to indicate a
statistically significant difference.
Results
Methylation of the GSH2 gene TRR in
pancreatic tissues and pancreatic cancer cells
According to the National Center for Biotechnology
Information genome database, the GSH2 gene TRR features a CpG
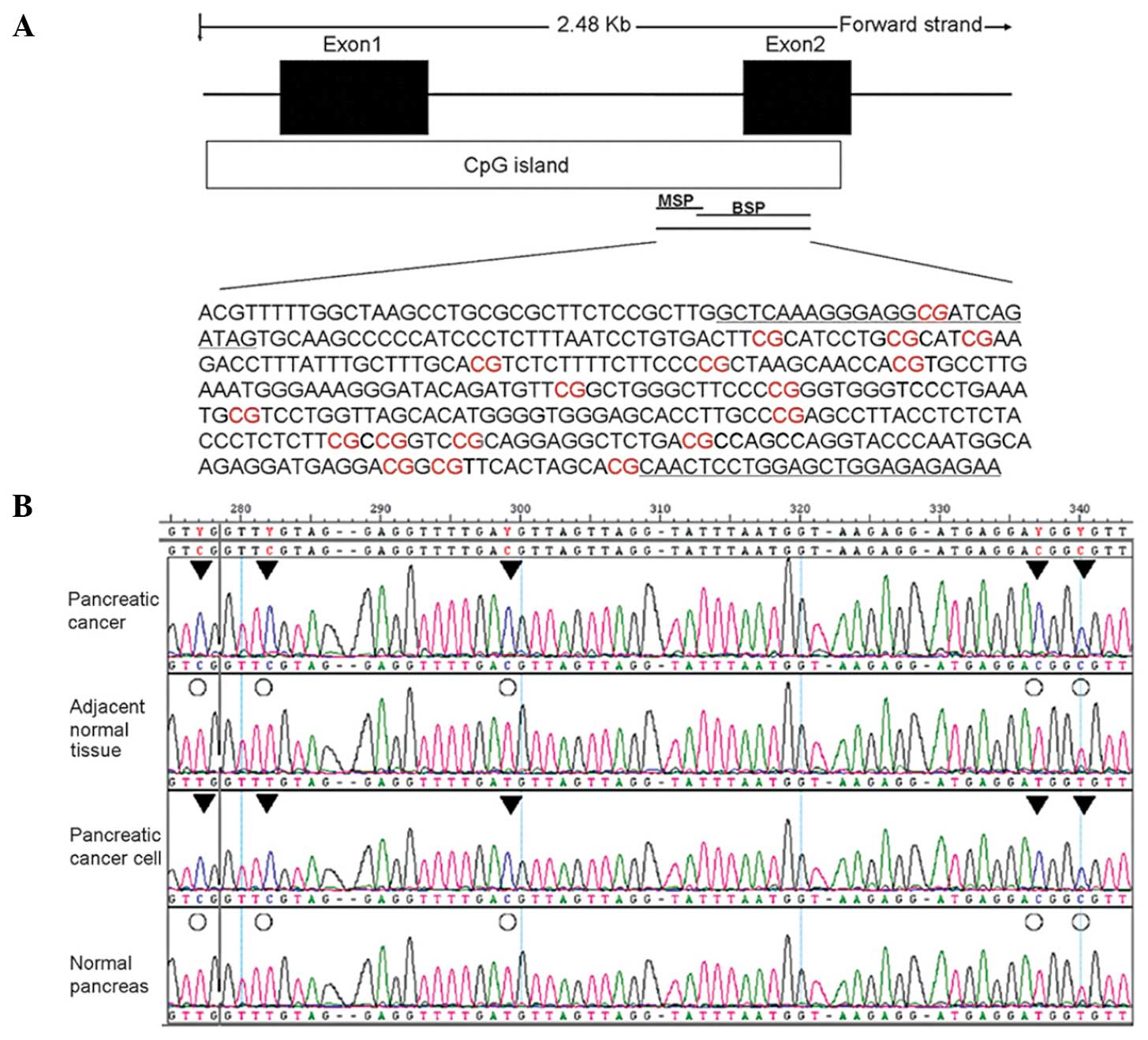
island between exons 1 and 2. BSP and MSP primers were designed to
bind the 3′ end of this region (Fig.
1A). BSP PCR-based sequencing analysis was then performed to
assess the methylation status of the GSH2 gene TRR in four tissue
groups: Two pancreatic cancer cell lines (PANC1 and PaTu8988), two
cases of pancreatic cancer, their adjacent normal tissue, and two
cases of normal pancreatic tissues (Fig.
1B).
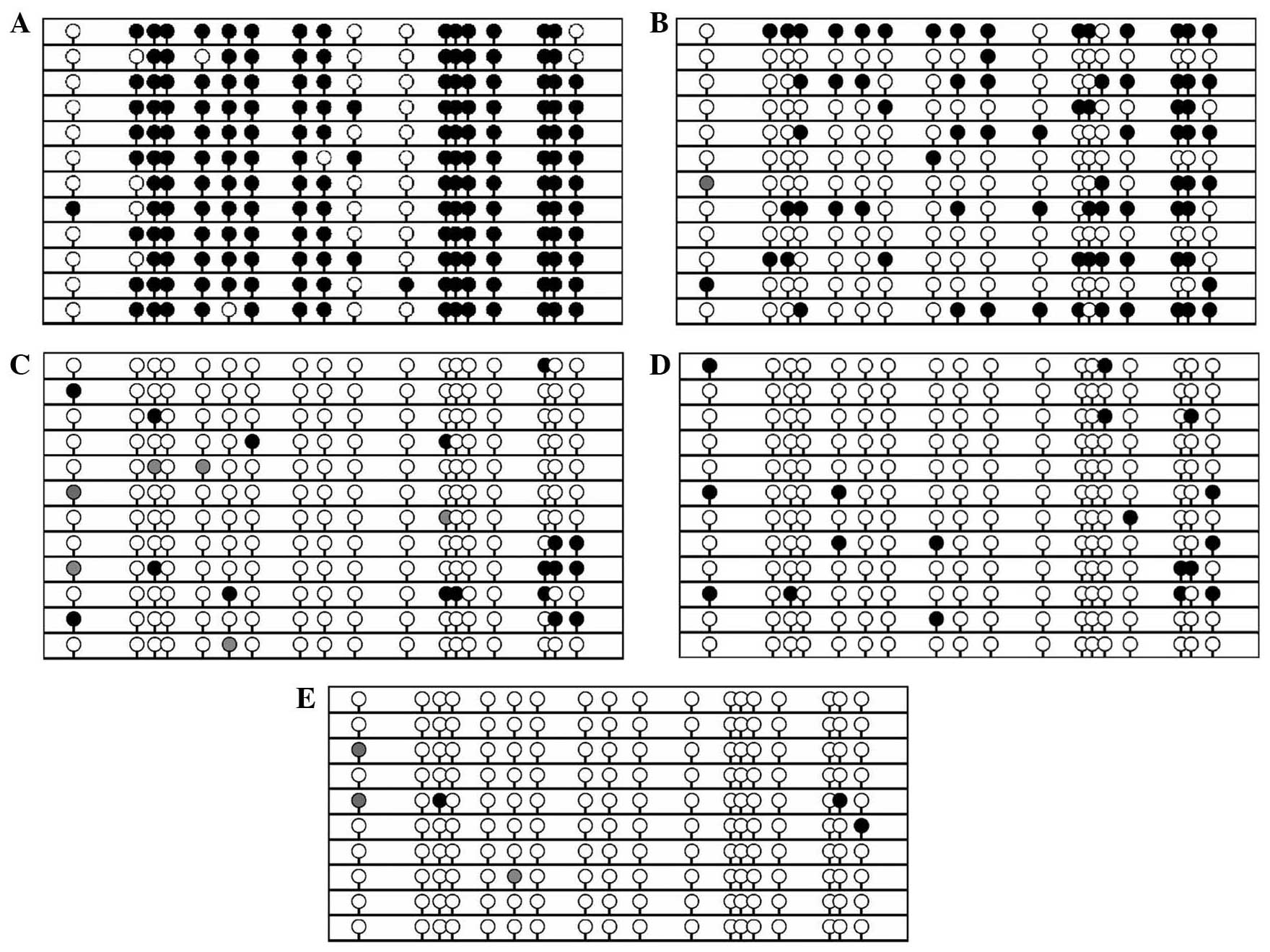
To further confirm that hypermethylation of the GSH2
gene TRR occurs in pancreatic cancer, BSP cloning-based sequencing
analysis was conducted to identify the methylation patterns in
these same samples in addition to one sample of white blood cells
from a healthy volunteer. Marked CpG methylation was identified in
the pancreatic cancer tissue and cell lines (Fig. 2A and B), but not in the normal tissue
(Fig. 2C-E). These data are
consistent with the results of the BSP PCR-based sequencing
analysis.
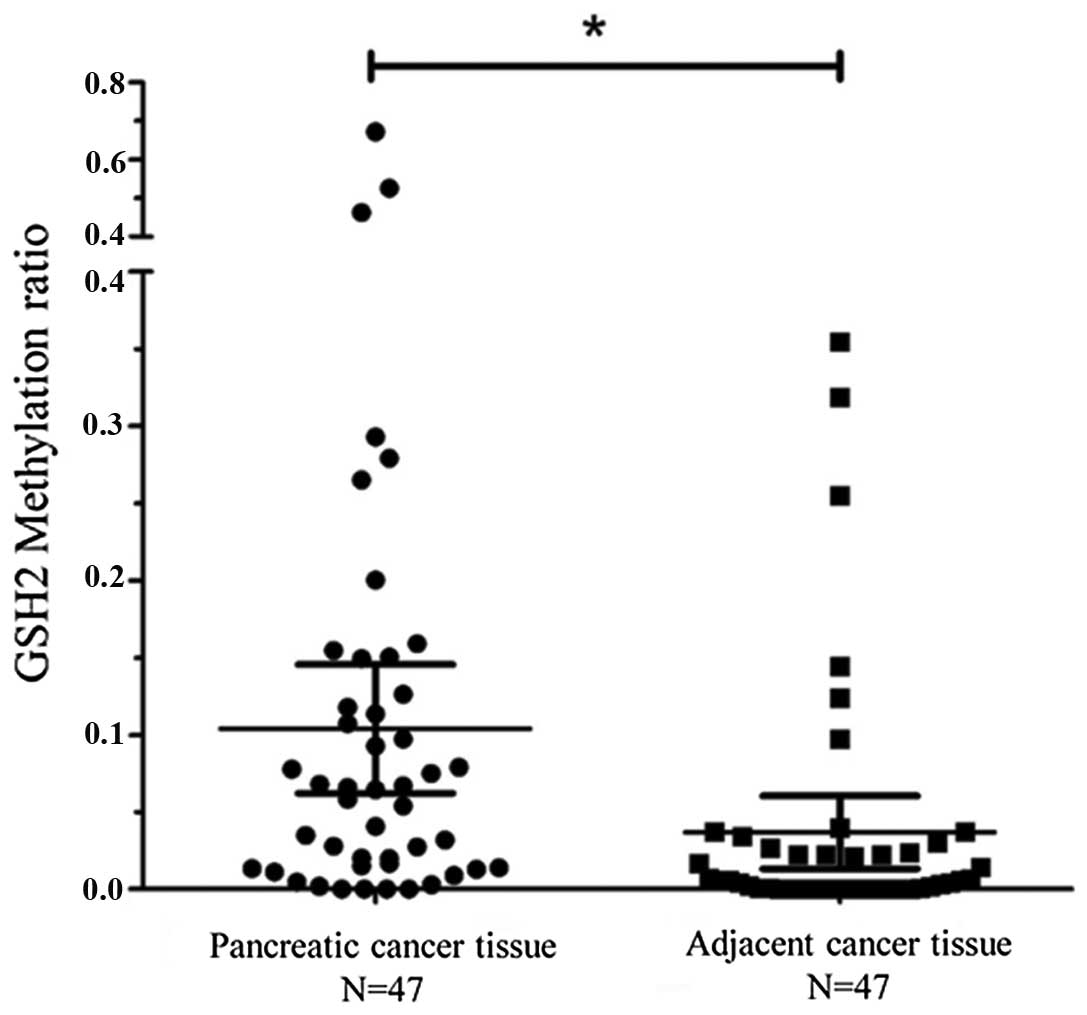
Based on these data, methylation of the GSH2
promoter was evaluated in a panel of 47 pancreatic cancer tissues
and adjacent normal tissues using qMSP (Fig. 3). The mean GSH2 expression level in
the pancreatic cancer tissues was significantly higher than that of
the adjacent normal tissues (cancer tissue: Mean, 0.10; 95%
confidence interval, 0.06–0.14; normal tissue: Mean, 0.04; 95%
confidence interval, 0.01–0.06; P=0.0089). Based on these data,
hypermethylation was defined by a value of >0.04.
Hypermethylation of the GSH2 gene was detected in 26 out of the 47
(55.3%) primary pancreatic cancer tissue samples, indicating that
GSH2 TRR hypermethylation occurs frequently in pancreatic
cancer.
Association of GSH2 gene TRR
methylation with clinicopathological parameters in patients with
pancreatic cancer
To further assess the diagnostic significance of
GSH2 methylation, the methylation results were assessed with regard
to the clinicopathological features of the patients. No significant
associations were observed between the GSH2 methylation status and
patient gender or age, tumor size, tumor location or lymph node
metastasis. However, a significant association was observed between
GSH2 methylation levels and tumor-node-metastasis stage (P=0.031;
Table I). This supports the qMSP
findings, suggesting that GSH2 methylation frequently occurs in
pancreatic cancers.
 | Table I.Clinicopathological features and GSH2
methylation in pancreatic carcinoma (n=47). |
Table I.
Clinicopathological features and GSH2
methylation in pancreatic carcinoma (n=47).
|
|
| GSH2 methylation |
|
|---|
|
|
|
|
|
|---|
| Clinicopathological
feature | Total cases | + | − | P-value |
|---|
| Patients, n | 47 | 26 | 21 |
|
| Gender, n |
|
|
| 0.805a |
| Male | 30 | 17 | 13 |
|
|
Female | 17 | 9 | 8 |
|
| Age, years | 47 |
61.31±7.47b |
58.90±7.81b | 0.288c |
| Maximal tumor size,
mm | 47 |
4.58±2.07b |
4.46±1.77b | 0.835c |
| Histology, n |
|
|
| 0.435a |
|
Well-differentiated | 34 | 20 | 14 |
|
|
Poorly-differentiated | 13 | 6 | 7 |
|
| Lymph node
metastasis, n |
|
|
| 0.160a |
|
Positive | 26 | 12 | 14 |
|
|
Negative | 21 | 14 | 7 |
|
| TNM stage, n |
|
|
| 0.031a |
| I/II | 22 | 9 | 13 |
|
|
III/IV | 25 | 18 | 7 |
|
Discussion
Pancreatic cancer is one of the most lethal human
cancers, and an effective treatment remains elusive (12). Hypermethylation of gene promoters is a
common event during carcinogenesis and tumor progression. The
present study investigated the methylation of the GSH2 promoter in
pancreatic cancer cell lines, pancreatic cancer and corresponding
adjacent normal pancreatic tissues, and normal pancreatic tissue
from disease-free subjects. The data indicated that GSH2 promoter
methylation is highly associated with pancreatic cancer. The GSH2
promoter was found to be hypermethylated in the pancreatic cancer
tissues compared with the adjacent normal tissues and normal
pancreatic tissue from healthy subjects. Furthermore, aberrant
hypermethylation of GSH2 was found to be associated with
advanced-stage disease. To the best of our knowledge, the present
study is the first to report the correlation of GSH2 promoter
hypermethylation with pancreatic cancer and tumor progression.
GSH2 is a homeobox gene that is activated as a
downstream target of the Shh pathway; this pathway is essential in
organ development and embryonic pancreatic development (7). Misregulation of Shh signaling has been
associated with a number of cancer types, including pancreatic
carcinoma (13). For example, Shh
ligand overexpression induces PanIN-1 and -2-like lesions and may
contribute to the formation of desmoplasia in pancreatic cancer
(14,15). Conversely, inhibition of Shh signaling
suppresses the self-renewal capacity of pancreatic cancer stem
cells and reverses chemotherapy resistance (16). Shh may also downregulate GSH2 in
ventral telencephalic progenitors (17), suggesting that Shh signaling is able
to suppress GSH2 under certain conditions.
The results of the current study indicated that
hypermethylation of the GSH2 promoter is a common feature of
pancreatic cancer. This may be due to the aberrant activation of
Shh signaling commonly observed in pancreatic cancers (18). Furthermore, these data indicate that
GSH2 promoter methylation may be used as a novel indicator of Shh
activation, as well as a diagnostic tool for pancreatic cancer
patients. The precise transcriptional mechanisms that control GSH2
remain unclear, however, the present findings suggest that a better
understanding of these mechanisms may inform diagnostic and
therapeutic approaches for pancreatic cancer.
In summary, the results of the current study
demonstrated that hypermethylation of the GSH2 promoter is
associated with the progression of pancreatic cancer, suggesting
that this may serve as a diagnostic and prognostic marker for
pancreatic cancer patients. Following this preliminary result,
further studies are required to investigate the molecular
mechanisms underlying the regulation of changes to GSH2
methylation, and the association between GSH2 hypermethylation and
the biological features of pancreatic cancer. In future, the
investigation of a large study population should be conducted in
order to further support the current results.
Acknowledgements
This study was supported by the China Postdoctoral
Science Foundation (grant no. 2012M521920) and a grant from the
National Natural Science Foundation of China (grant no. 81300353)
to Mr. Huang.
References
|
1
|
Landis SH, Murray T, Bolden S and Wingo
PA: Cancer statistics, 1998. CA Cancer J Clin. 48:6–29. 1998.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Li D, Xie K, Wolff R and Abbruzzese JL:
Pancreatic cancer. Lancet. 363:1049–1057. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Warshaw AL and Fernández-del Castillo C:
Pancreatic carcinoma. N Engl J Med. 326:455–465. 1992. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Chen WD, Han ZJ, Skoletsky J, et al:
Detection in fecal DNA of colon cancer-specific methylation of the
nonexpressed vimentin gene. J Natl Cancer Inst. 97:1124–1132. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Feinberg AP and Tycko B: The history of
cancer epigenetics. Nat Rev Cancer. 4:143–153. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Brune K, Hong SM, Li A, et al: Genetic and
epigenetic alterations of familial pancreatic cancers. Cancer
Epidemiol Biomarkers Prev. 17:3536–3542. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Corbin JG, Gaiano N, Machold RP, Langston
A and Fishell G: The Gsh2 homeodomain gene controls multiple
aspects of telencephalic development. Development. 127:5007–5020.
2000.PubMed/NCBI
|
|
8
|
Wu X, Rauch TA, Zhong X, et al: CpG island
hypermethylation in human astrocytomas. Cancer Res. 70:2718–2727.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Bailey JM, Mohr AM and Hollingsworth MA:
Sonic hedgehog paracrine signaling regulates metastasis and
lymphangiogenesis in pancreatic cancer. Oncogene. 28:3513–3525.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Morton JP, Mongeau ME, Klimstra DS, et al:
Sonic hedgehog acts at multiple stages during pancreatic
tumorigenesis. Proc Natl Acad Sci USA. 104:5103–5108. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Pasca di Magliano M, Sekine S, Ermilov A,
Ferris J, Dlugosz AA and Hebrok M: Hedgehog/Ras interactions
regulate early stages of pancreatic cancer. Genes Dev.
20:3161–3173. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Boj SF, Hwang CI, Baker LA, et al:
Organoid models of human and mouse ductal pancreatic cancer. Cell.
160:324–338. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Jemal A, Siegel R, Xu J and Ward E: Cancer
statistics, 2010. CA Cancer J Clin. 60:277–300. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Bailey JM, Swanson BJ, Hamada T, et al:
Sonic hedgehog promotes desmoplasia in pancreatic cancer. Clin
Cancer Res. 14:5995–6004. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Thayer SP, di Magliano MP, Heiser PW, et
al: Hedgehog is an early and late mediator of pancreatic cancer
tumorigenesis. Nature. 425:851–856. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Huang FT, Zhuan-Sun YX, Zhuang YY, et al:
Inhibition of hedgehog signaling depresses self-renewal of
pancreatic cancer stem cells and reverses chemoresistance. Int J
Oncol. 41:1707–1714. 2012.PubMed/NCBI
|
|
17
|
Xu Q, Guo L, Moore H, Waclaw RR, Campbell
K and Anderson SA: Sonic hedgehog signaling confers ventral
telencephalic progenitors with distinct cortical interneuron fates.
Neuron. 65:328–340. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Maréchal R, Bachet JB, Calomme A, et al:
Sonic hedgehog and Gli1 expression predict outcome in resected
pancreatic adenocarcinoma. Clin Cancer Res. 21:1215–1224. 2015.
View Article : Google Scholar : PubMed/NCBI
|