Introduction
Renal cell carcinoma (RCC) is among the most common
types of cancer and accounts for ~2–3% of all malignancies
(1). The incidence of RCC
continuously increases each year, with an estimated 61,560 new
cases of RCC and 14,080 RCC-associated mortalities predicted to
occur in the United States in 2015 (2). Despite advances in diagnostic
techniques, 25–30% of patients present with metastatic disease
(3). The prognosis is poor for such
patients, with subsequent chemotherapy and radiotherapy treatment
regimes yielding ineffective results (4). Tumourigenesis and progression are
multistep processes that are affected by changes in gene
expression. Therefore, understanding the gene expression changes
that occur in RCC may improve the diagnosis, treatment and
prevention of RCC.
B-cell translocation gene 1 (BTG1) is a member of
the BTG/transducer of Erb (TOB) family. This family comprises six
members; BTG1, BTG2/TIS21/PC3, BTG3, BTG4/PC3B, TOB1 and TOB2. The
BTG/TOB family proteins are composed of two highly conserved and
characteristic domains, Box A and Box B, in the N-terminal region.
In addition, these proteins are involved in regulating cell cycle
progression, inhibiting proliferation, promoting apoptosis and
stimulating cellular differentiation in multiple cell types
(5). As BTG1 exhibits these
characteristics, it is considered to be a tumour suppressor gene
(6). Previous studies have identified
that BTG1 enhances the antiproliferative function of homeobox
B9-mediated transcription (7), while
overexpression of BTG1 induces increased apoptosis in NIH 3T3 cells
(8). However, the functions of BTG1
and its precise molecular mechanisms in RCC remain unclear. It has
been shown that BTG1 interacts with protein arginine
N-methyltransferase 1 (PRMT1) in vitro (9). PRMT1 then catalyses the formation of
ω-monomethylarginine and asymmetric dimethylarginine. This arginine
methylation regulates transcription or affects cytokine signalling
pathways (10). However, whether BTG1
functions in RCC via its effect on PRMT1 remains unknown.
Therefore, the present study examined BTG1
expression in RCC tissues and cells, and investigated the function
of BTG1 in cell proliferation, cell cycle distribution and
apoptosis in vitro. In addition, it was investigated whether
the functions of BTG1 are attributable to interactions with
PRMT1.
Materials and methods
Tissues
RCC and corresponding para-carcinoma tissue samples
were obtained from 20 patients, who underwent nephrectomy at the
Affiliated Zhongda Hospital of Southeast University (Nanjing,
China) between June 2007 and June 2010. Histological diagnoses were
established following analysis of standard hematoxylin and
eosin-stained sections by two senior pathologists experienced in
RCC diagnosis. All patients were diagnosed with RCC. Approval was
obtained from the ethics committee of the Affiliated Zhongda
Hospital of Southeast University and samples were collected
following receipt of written informed consent from all
patients.
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR)
Total RNA was extracted from the specimens using
TRIzol® reagent (Invitrogen Life Technologies, Carlsbad, CA, USA).
Complementary (c)DNA synthesis was performed using Avian
Myeloblastosis Virus Reverse Transcriptase and random primers
(Takara Biotechnology Co., Ltd., Dalian, China), and the RT-qPCR
reactions were performed using a 7300 Real-Time RT-PCR system
(Applied Biosystems Life Technologies, Foster City, CA, USA). The
amplification steps consisted of 95°C for 1 min, 40 cycles at 95°C
for 15 s and 60°C for 30 s, followed by 72°C for 10 min. The primer
sequences for BTG1 were as follows: F 5′-ATCTCCAAGTTTCTCCGCACC-3′
and R 5′-CAACGGTAACCCGATCCCTT-3′. Subsequently, the expression BTG1
mRNA was calculated relative to GAPDH mRNA expression levels using
the 2−ΔΔCt method.
Immunohistochemistry
All surgical samples were fixed in 10% buffered
formaldehyde solution and embedded in paraffin. Paraffin sections
(4-µm thick) were then reacted with monoclonal antibodies against
BTG1 (mouse anti-human; 1:75 dilution; cat. no. ab50991; Abcam,
Cambridge, UK). The antibody was replaced by phosphate-buffered
saline (PBS) as a negative control.
Immunohistochemical evaluation
The BTG1 immunohistochemistry results were scored
using semi-quantitative immunoreactivity scores (IRSs) for all
sections. The intensity of staining and percentage of
positively-stained cells were noted. The intensity of staining was
scored as follows: No staining, 0; mild staining, 1; moderate
staining, 2; and strong staining, 3. The percentage of positive
cells was scored as follows: No positive cells, 0; <5% positive
cells 1; 5–25% positive cells, 2; 26–50% positive cells, 3; and
>50% positive cells, 4. The overall IRSs were calculated as
follows: IRS = percentage of positive cells × intensity of
staining. Expression was then classified as negative (–; IRS, 0),
weak (positive, +; IRS, 1–2), moderate (double positive, ++; IRS,
3–4) or strong (triple positive, +++; IRS, 4–12).
Plasmid construction
cDNA encoding human BTG1 was generated by RT-PCR and
subcloned into the EcoRI and MluI restriction sites of the pCI-neo
expression vector (Promega Corporation, Madison, Wisconsin, USA).
The plasmid sequence was confirmed using sequencing.
Cell culture and transfection
The 786-O and HK-2 cells were obtained from the Cell
Resource Centre, Institute of Basic Medical Sciences, Chinese
Academy of Medical Sciences (Shanghai, China). The cells were
cultured in RPMI 1640 (GE Healthcare Life Sciences, Beijing, China)
supplemented with 50 U/ml penicillin, 50 mg/ml streptomycin
(Generay Biotech Ltd., Shanghai, China) and 10% fetal bovine serum
(GE Healthcare Life Sciences, Carlsbad, USA) in an atmosphere of 5%
CO2 at 37°C. Cells were transfected with the
pCI-neo/BTG1 vector using Lipofectamine 2000 (Invitrogen Life
Technologies), according to the manufacturer's instructions, and
eosin Y disodium trihydrate (Santa Cruz Biotechnology, Inc.,
Dallas, TX, USA) was added to inhibit PRMT1 in a proportion of the
BTG1-overexpressed 786-O cells. The 786-O cells were transfected
with blank pCI-neo vector as the control.
Western blot
Whole cell extracts (786-O RCC and HK-2 control
cells) were separated by 10% SDS-PAGE and transferred onto
polyvinylidene fluoride membranes. Antigen retrieval was performed
in heated citrate buffer (PH 6.0). Blots were blocked with 5%
non-fat milk at room temperature for 1 h and incubated with the
monoclonal mouse anti-human BTG1 (1:500 dilution; cat. no. ab50991;
Abcam, Cambridge, UK) antibody in blocking buffer overnight at a
temperature of 4°C. The blots were then washed and incubated with
horseradish peroxidase-labelled goat anti-mouse IgG secondary
antibody (1:3,000 dilution; cat. no. ZB-2301; Zhongshan
Goldenbridge Biotechnology Co., Ltd., Beijing, China) and
visualised using enhanced chemiluminescence (Beyotime Institute of
Biotechnology, Haimen, China).
Cell viability and clonability
assays
Cell viability was determined at 24, 48 and 72 h by
performing an MTT assay. In brief, 786-O cells transfected for
>48 h were seeded into 96-well plates (3,000 cells/plate), and
cultured for 24, 48 and 72 h. Each well was supplemented with 20 µl
MTT (5 mg/ml) and incubated at 37°C for 4 h. The supernatant was
removed and the purple precipitates of formazan were dissolved in
200 µl DMSO (Sigma-Aldrich, St. Louis, MO, USA). Absorbance was
then read at a wavelength of 570 nm using an automatic multi-well
MK3 spectrophotometer (Thermo Fisher Scientific, Inc., Waltham, MA,
USA). For the clonability assay, cells were counted >48 h after
transfection, seeded into 6-well plates at a low density (3,000
cells/plate) and cultured for 9–14 days until visible colonies
appeared. Cells were subsequently stained with methyl violet and
the number of colonies was counted under observation of a Olympus
CKX41 light microscope (Olympus Corporation, Tokyo, Japan).
Flow cytometry
Fluorescence-activated cell-sorting analysis was
performed 72 h after transfection. The cells were harvested, washed
with cold PBS and resuspended. Cells were then stained with
propidium iodide (PI) for cell cycle analysis, and PI plus
Annexin-V-fluorescein isothiocyanate (FITC) for apoptosis analysis
using an Annexin V-FITC/PI Apoptosis kit (UBio Biological
Technology Co., Ltd, Ji'nan, China), according to the
manufacturer's instructions.
Co-immunoprecipitation
Total proteins were extracted from 786-O cells using
cell lysis buffer for western blotting and immunoprecipitation
(Beyotime Institute of Biotechnology) was performed using protease
inhibitors (Amresco LLC, Solon, OH, USA). The primary antibody used
was rabbit anti-human PRMT1 polyclonal antibody (1:100 dilution,
cat. no. sc-130851; Santa Cruz Biotechnology, Inc.) and the
secondary antibody used was mouse anti-BTG1 monoclonal antibody
(Abcam). For co-immunoprecipitation, total cellular protein was
incubated with 10 µl anti-PRMT1 antibody for 1 h at 4°C.
Subsequently, 20 µl Protein A/G PLUS-Agarose was added and the
capped tubes were rocked overnight at 4°C. Beads were then pelleted
by centrifugation at ~600 × g. The supernatant was removed, and the
beads were washed, boiled in 1X SDS sample buffer and subjected to
SDS-PAGE gel electrophoresis and western blotting with the
anti-BTG1 antibody.
Immunostaining and confocal laser
microscopic analysis
Cells were grown in culture dishes with a bottom
well. Monolayers were washed with PBS, fixed with immunostaining
fix solution (Beyotime Institute of Biotechnology) for 1 h at
routine temperature, washed with Tris-buffered saline and Triton
X-100 (2.42 g Tris, 8 g NaCl, 0.1% Triton X-100) three times, and
blocked with immunostaining blocking buffer (Beyotime Institute of
Biotechnology) for 1 h. The monolayers were incubated overnight
with the BTG1 and PRMT1 antibodies at a temperature of 4°C. After
washing, cells were incubated with cyanine 3-conjugated anti-rabbit
antibody and FITC-conjugated anti-mouse antibody (Beyotime
Institute of Biotechnology) for 1 h at a routine temperature.
Finally, specimens were examined under a confocal laser scanning
microscope (LSM 710; Zeiss, Oberkochen, Germany).
Statistical analysis
All quantitative data represent a mean value of at
least triplicate samples and all statistical analyses were
performed using Statistical Package of the Social Sciences software
(version 16.0; SPSS, Inc., Chicago, IL, USA). Data are presented as
the mean ± standard error of the mean. All calculations were
performed using GraphPad Prism 5 software (GraphPad Software, Inc.,
La Jolla, CA, USA) and scanned images of western blots were
quantified using ImageJ 2X software (National Institutes of Health,
Bethesda, MD, USA). Group means were compared by performing
Student's t-test. P<0.05 was considered to indicate a
statistically significant difference.
Results
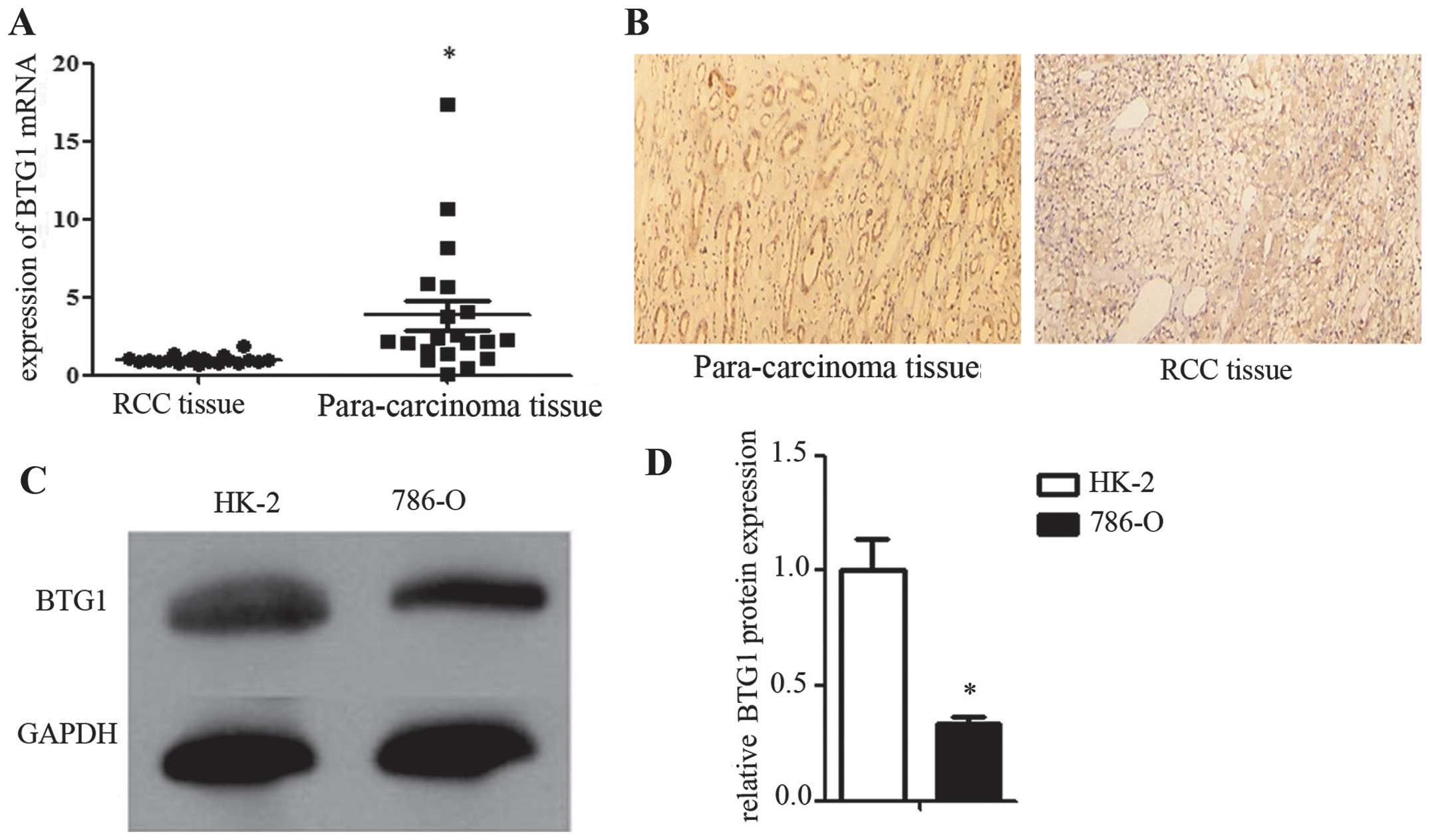
Low BTG1 expression in RCC tissues and
cells
In order to investigate BTG1 expression in RCC, BTG1
mRNA expression was measured using RT-qPCR, and BTG1 protein
expression was determined using immunohistochemistry, in 20 RCC and
20 corresponding para-carcinoma tissue samples. BTG1 expression was
significantly lower in the RCC tissues compared with that in the
corresponding para-carcinoma tissues (P<0.05; Fig. 1A and 1B). In order to construct a
reliable in vitro model for investigating the mechanism of
action of BTG1 in RCC, BTG1 expression was examined by western blot
analysis in HK-2 (control) and 786-O (RCC) cells. BTG1 expression
was significantly lower in the 786-O cells compared with that in
HK-2 cells (P<0.05; Fig. 1C and
1D).
In vitro effects of BTG1 in RCC
In order to investigate the function of BTG1 in RCC,
a BTG1 overexpression plasmid was transfected into 786-O cells.
Western blot analysis identified a significant increase in BTG1
protein expression in the 786-O cells following transfection
(P<0.05; Fig. 2A). Additionally,
BTG1 overexpression significantly inhibited 786-O cell
proliferation, as shown by MTT and colony formation assays
(P<0.05; Fig. 2B and 2C).
Furthermore, flow cytometry assays revealed a significant increase
in cell apoptosis and G0/G1 arrest in 786-O cells, following
transfection (P<0.05; Fig. 2D and
2E).
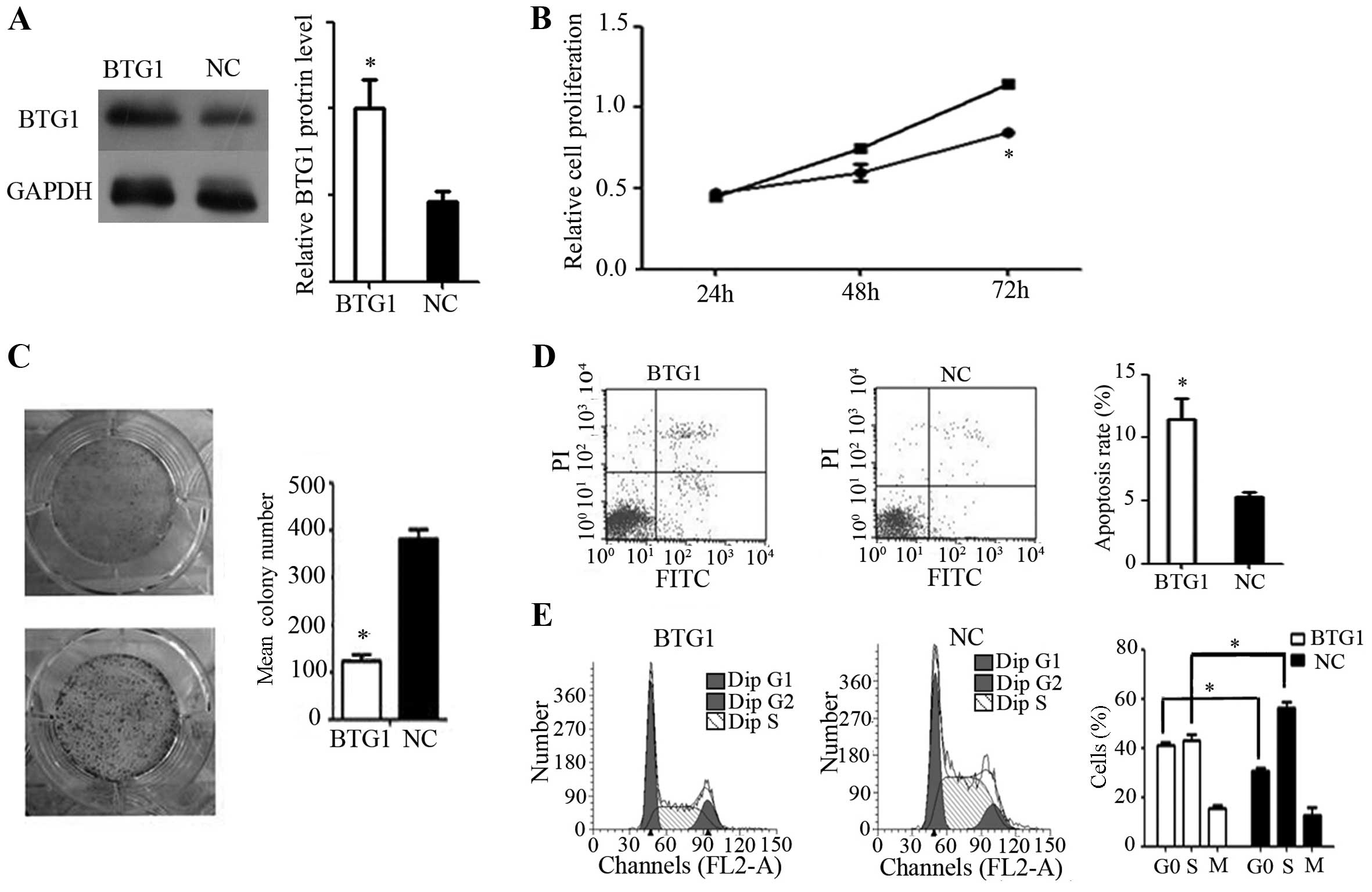 | Figure 2.Effect of BTG1 on 786-O cells in
vitro. (A) Western blotting demonstrated increased BTG1
expression following transfection. *P<0.05, vs. NC. (B) Cell
proliferation rate, showing that BTG1 overexpression significantly
inhibited 786-O proliferation, as determined by an MTT assay.
*P<0.05, vs. cells not overexpressing BTG1. (C) Colony forming
assay, demonstrating reduced colony-forming efficiency in 786-O
cells with forced BTG1 expression. *P<0.05, vs. NC. (D) Flow
cytometry, demonstrating a higher rate of apoptosis in 786-O cells
with forced BTG1 expression, as evaluated by flow cytometry.
*P<0.05, vs. NC. (E) Cell cycle analysis of BTG1-overexpressing
786-O cells, showing an increase in the proportion of cells in the
G0/G1 phase. *P<0.05. BTG1, B-cell translocation gene 1; NC,
negative control; PI, propidium iodide. |
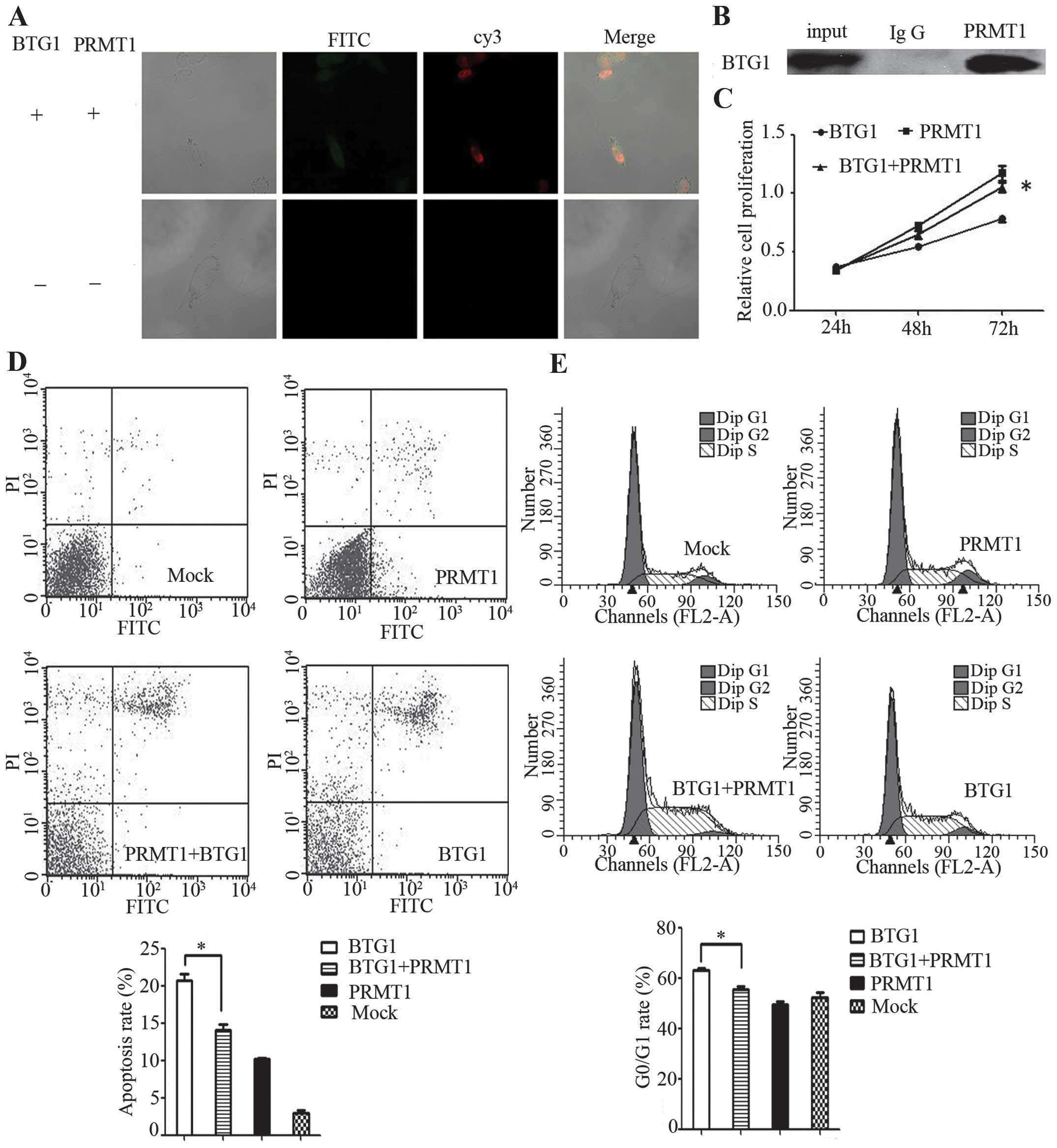
BTG1 protein functions via PRMT1
In order to clarify whether BTG1 functions via
PRMT1, the interactions between BTG1 and PRMT1 were demonstrated by
immunofluorescence and co-immunoprecipitation experiments in 786-O
cells. BTG1 interacted with PRMT1 in 786-O cells (Fig. 3A and 3B). Eosin Y disodium trihydrate
was added to inhibit PRMT1 in BTG1-overexpressing 786-O cells. This
inhibition resulted in significantly reduced proliferation, G0/G1
phase arrest and apoptosis in the BTG1-overexpressing 786-O cells
(P<0.05; Fig. 3C–E), indicating
that BTG1 may function by interacting with PRMT1.
Discussion
BTG1 was initially identified in B lymphoblastic
leukaemia and its expression appears to be highest in the G0/G1
phases of the cell cycle (11). Weak
BTG1 expression has recently been reported in various other types
of cancer, including thyroid, lung and breast (12–15). In
the present study, weak BTG1 expression was also demonstrated in
RCC tissue samples by performing qRT-PCR, western blotting and
immunohistochemistry.
Tumour development and progression are associated
with uncontrolled proliferation and apoptosis. Zhu et al
(16) and Sun et al (17) identified that BTG1 induces G0/G1 cell
cycle arrest, promotes apoptosis and inhibits cell proliferation in
breast and non-small cell lung cancer (NSCLC), respectively.
Furthermore, Sun et al (18)
determined that BTG1 expression was significantly correlated with
lymph node metastasis, clinical stage, histological grade and
survival in NSCLC cells. The present study explored the functions
of BTG1 in RCC, and obtained similar results, with respect to the
effect of BTG1 expression on cell cycle arrest, apoptosis promotion
and growth inhibition. These data indicate that BTG1 may have a
common antitumour function in various types of cancer.
The antiproliferative activity of BTG1 is controlled
by the conserved Box A domain (11).
Doidge et al (18)
demonstrated that the antiproliferative activity of BTG1 is
mediated through its interactions with the Caf1a and Caf1b
deadenylase enzymes, and the role of BTG1 in the regulation of mRNA
abundance and translation is dependent on Caf1a/Caf1b. However, in
the present study, BTG1 appeared to function by interacting with
PRMT1. In mammals, PRMT1 is the primary arginine asymmetric
dimethylation enzyme, accounting for >90% of asymmetric
dimethylation enzymes (19). Arginine
methylation is a common post-translational modification that alters
the stability of chromatin and affects the binding of
transcriptional factors, thereby regulating gene expression without
changing the original nucleotide sequence. Arginine methylation has
critical functions in gene transcription, mRNA splicing, DNA
repair, protein cellular localisation and signalling (20), and PRMT1 specifically exhibits key
functions in breast cancer cell apoptosis and osteosarcoma cell
proliferation (21–23). The present study investigated the
interaction between BTG1 and PRMT1 in RCC, and it was shown that
certain functions of BTG1 are suppressed by the inhibition of
PRMT1. Thus, it is hypothesized that BTG1 functions by interacting
with PRMT1 in RCC, as well as through other signalling
pathways.
In conclusion, the present study demonstrated weak
expression of BTG1 in RCC, and showed that BTG1 may inhibit cell
growth and promote cell apoptosis by interacting with PRMT1.
However, the underlying mechanism of action of PRMT1 in RCC
requires further investigation.
Acknowledgements
The present study was supported by the National
Natural Science Foundation of China (grant nos. 81370849, 81300472
and 81202034), the Natural Science Foundation of Jiangsu Province
(grant nos. BL2013032 and BK2012336), Nanjing City (grant no.
201201053) and Southeast University (grant no. 3290002402) and the
Science Foundation of Ministry of Education of China (grant no.
20120092120071).
References
|
1
|
Jemal A, Bray F, Center MM, Ferlay J, Ward
E and Forman D: Global cancer statistics. CA Cancer J Clin.
61:69–90. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Siegel R, Miller K and Jemal A: Cancer
statistics, 2015. CA Cancer J Clin. 65:5–29. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Gupta K, Miller JD, Li JZ, Russell MW and
Charbonneau C: Epidemiologic and socioeconomic burden of metastatic
renal cell carcinoma (mRCC): A literature review. Cancer Treat Rev.
34:193–205. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Ruys AT, Tanis PJ, Naftegaal ID, van
Duijvendijk P, Verhoef C, Porte RJ and van Gulik TM: Surgical
treatment of renal cell cancer liver metastases: A population-based
study. Ann Surg Oncol. 18:1932–1938. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Guéhenneux F, Duret L, Callanan MB, et al:
Cloning of the mouse BTG3 gene and definition of a new gene family
(the BTG family) involved in the negative control of the cell
cycle. Leukemia. 11:370–375. 1997. View Article : Google Scholar
|
|
6
|
Rouault JP, Prévôt D, Berthet C, Birot AM,
Billaud M, Magaud JP and Corbo L: Interaction of BTG1 and
p53-regulated BTG2 gene products with mCaf1, the murine homolog of
a component of the yeast CCR4 transcriptional regulatory complex. J
Biol Chem. 273:22563–22569. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Prévôt D, Voeltzel T, Birot AM, Morel AP,
Rostan MC, Magaud JP and Corbo L: The leukemia-associated protein
Btg1 and the p53-regulated protein Btg2 interact with the
homeoprotein Hoxb9 and enhance its transcriptional activation. J
Biol Chem. 275:147–153. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Corjay MH, Kearney MA, Munzer DA, Diamond
SM and Stoltenborg JK: Antiproliferative gene BTG1 is highly
expressed in apoptotic cells in macrophage-rich areas of advanced
lesions in Watanabe heritable hyperlipidemic rabbit and human. Lab
Invest. 78:847–858. 1998.PubMed/NCBI
|
|
9
|
Berthet C, Guéhenneux F, Revol V, et al:
Interaction of PRMT1 with BTG/TOB proteins in cell signalling:
UMolecular analysis and functional aspects. Genes Cells. 7:29–39.
2002. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Bedford MT and Richard S: Arginine
methylation an emerging regulator of protein function. Mol Cell.
18:263–272. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Rouault JP, Rimokh R, Tessa C, et al:
BTG1, a member of a new family of antiproliferative genes. EMBO J.
11:1663–1670. 1992.PubMed/NCBI
|
|
12
|
Ito Y, Suzuki T, Yoshida H, et al:
Phosphorylation and inactivation of Tob contributes to the
progression of papillary carcinoma of the thyroid. Cancer Lett.
220:237–242. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Iwanaga K, Sueoka N, Sato A, et al:
Alteration of expression or phosphorylation status of tob, a novel
tumor suppressor gene product, is an early event in lung cancer.
Cancer Lett. 202:71–79. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Yoneda M, Suzuki T, Nakamura T, et al:
Deficiency of antiproliferative family protein Ana correlates with
development of lung adenocarcinoma. Cancer Sci. 100:225–232. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Kawakubo H, Brachtel E, Hayashida T, et
al: Loss of B-cell translocation gene-2 in estrogen
receptor-positive breast carcinoma is associated with tumor grade
and overexpression of cyclin d1 protein. Cancer Res. 66:7075–7082.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Zhu R, Zou ST, Wan JM, Li W, Li XL and Zhu
W: BTG1 inhibits breast cancer cell growth through induction of
cell cycle arrest and apoptosis. Oncol Rep. 30:2137–2144.
2013.PubMed/NCBI
|
|
17
|
Sun GG, Lu YF, Cheng YJ and Hu WN: The
expression of BTG1 is downregulated in NSCLC and possibly
associated with tumor metastasis. Tumour Biol. 35:2949–2957. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Doidge R, Mittal S, Aslam A and Winkler
GS: The anti-proliferative activity of BTG/TOB proteins is mediated
via the Caf1a (CNOT7) and Caf1b (CNOT8) deadenylase subunits of the
Ccr4-not complex. PLoS One. 7:e513312012. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Dhar S, Vemulapalli V, Patananan AN, et
al: Loss of the major Type I arginine methyltransferase PRMT1
causes substrate scavenging by other PRMTs. Sci Rep. 3:13112013.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Zhang R, Li X, Liang Z, et al: Theoretical
insights into catalytic mechanism of protein arginine
methyltransferase 1. PLoS One. 8:e724242013. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Yu Z, Chen T, Hébert J, Li E and Richard
S: A mouse PRMT1 null allele defines an essential role for arginine
methylation in genome maintenance and cell proliferation. Mol Cell
Biol. 29:2982–2996. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Yoshimatsu M, Toyokawa G, Hayami S, et al:
Dysregulation of PRMT1 and PRMT6, Type I arginine
methyltransferases, is involved in various types of human cancers.
Int J Cancer. 128:562–573. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Cho JH, Lee MK, Yoon KW, Lee J, Cho SG and
Choi EJ: Arginine methylation-dependent regulation of ASK1
signaling by PRMT1. Cell Death Differ. 19:859–870. 2012. View Article : Google Scholar : PubMed/NCBI
|