Introduction
Colorectal cancer (CRC) is the third and second most
common type of cancer in males and females, respectively (1). In 2008, >1.2 million cases of CRC
were diagnosed, and 608,700 associated mortalities were recorded
(1). Patients with CRC have different
prognoses depending on the tumor stage, which is commonly
classified by the American Joint Committee on Cancer (AJCC)
tumor-node-metastasis (TNM) staging system (2). For example, patients with distant
metastasis have a poor 5-year survival rate (12%), while patients
with localized disease have good prognoses (>90%) (3). However, the AJCC staging system is
mainly concerned with the depth of cancer invasion, the involvement
of the lymph nodes and the status of the metastasis, but not with
the specific biological properties of CRC (4). Thus far, few biological markers have
been validated as diagnostic criteria. However, certain molecules
involved in the pathogenesis of CRC may lead to more accurate
diagnoses and improved efficacy of comprehensive therapies
(5).
Autophagy, also known as cellular digestion, is a
highly conserved cellular catabolic pathway involved in the
degradation and recycling of superfluous or damaged proteins and
organelles via double-membrane vesicles termed autophagosomes. This
process enables cells, organs and entire organisms to endure
various stress conditions, including limited availability of
nutrients, reduced energy supply or low levels of oxygen (6,7). Numerous
autophagy-associated genes (ATG) have been identified in yeast.
Unc-51-like kinase 1 (ULK1) is a core mammalian homologue of a
yeast ATG. In mammalian cells, ULK1 forms a stable complex to sense
nutrient signals for autophagy activation (8). Under conditions of glucose starvation,
the activated AMP-activated protein kinase (AMPK) regulates the
ULK1 complex by direct interaction as follows: AMPK phosphorylates
and activates ULK1, which leads to autophagy induction (9,10). When
glucose levels are sufficient, high levels of mammalian target of
rapamycin complex 1 prevent ULK1 activation (11,12). The
role of autophagy in cancer is complex, and varies depending on the
type of tumor. For example, high expression levels of ULK1 have
been demonstrated to be a biomarker of poor prognosis in patients
with esophageal squamous cell carcinoma (13); and in operable breast cancer, low
expression levels of ULK1 are associated with tumor progression and
an adverse prognosis (14). Thus far,
the association between the expression of ULK1 and the pathogenesis
of CRC remains unclear.
To the best of our knowledge, the characterization
of the expression of ULK1 in human CRC tissues, and its association
with the clinicopathological variables of CRC are reported for the
first time in the present study.
Materials and methods
Patients and follow-up
The current study was approved by the Institute
Research Medical Ethics Committee of Sun Yat-sen University
(Guangzhou, China). A total of 339 patients with CRC, who underwent
initial surgical resection in the Department of Gastrointestinal
Surgery of the First Affiliated Hospital of Sun Yat-sen University
between January 2003 and December 2005, were enrolled in the
present study. Written informed consent was obtained from each
patient. Patients that had received preoperative chemo- or
radiotherapy were excluded. The cancer tissues were surgically
resected and histopathologically confirmed by hematoxylin and eosin
(H&E; Beyotime Institute of Biotechnology, Haimen, China)
staining.
During the follow-up, the patients were evaluated at
the hospital or contacted by telephone or letter every 3 months in
the first year, every 6 months in the second year and annually
thereafter. The following data were collected from the patients for
further investigation: General information, preoperative
information, details of the surgery, pathology reports, TNM stage,
and results of the follow-ups. The tumor stage was defined
according to the AJCC TNM stage system of CRC (7th
edition) (2). For follow-up purposes,
the primary end point was the overall survival (OS), defined as the
time from surgery to mortality due to any cause; and the secondary
end point was the disease-free survival (DFS), defined as the time
from surgery to the first event of recurrence, metastasis or
mortality.
Tissue microarray (TMA) construction
and immunohistochemical (IHC) staining
TMA was constructed using paraffin-embedded tissue
blocks of cancer tissues following screening of the H&E-stained
slides for optimal position: For each specimen, 2 cores of 1-mm
diameter were excised from the representative areas and deposited
on a recipient paraffin block using a Minicore tissue array
(ALPHELYS, Plaisir, France). The blocks were subsequently cut into
5-µm sections on silanized glass slides (Beyotime Institute of
Biotechnology)for IHC staining.
IHC staining was performed with an Envision system,
according to the manufacturer's instructions (Dako, Glostrup,
Denmark): The slides were deparaffinized in dimethylbenzene and
rehydrated with graded alcohol (100%, 95%, 75%) prior to antigen
retrieval with sodium citrate (Sangon Biotech Co., Ltd., Shanghai,
China) and blocking of the endogenous peroxidase activity with 0.3%
hydrogen peroxide (Sangon Biotech Co., Ltd.). The slides were then
incubated overnight at 4°C with a polyclonal rabbit ULK1 antibody
(1:2,000; #ab65056; Abcam, Cambridge, UK), followed by a 30-min
incubation period at room temperature with a polyclonal goat
anti-rabbit secondary antibody (#A0208; Beyotime Institute of
Biotechnology). Next, the slides were rinsed with
phosphate-buffered saline, incubated with 3,3′-diaminobenzidine for
1 min, counterstained with hematoxylin, dehydrated using graded
alcohol (90%, 95%, 100%) and mounted. A negative control was
obtained by replacing the primary antibody with a normal murine IgG
(Beyotime Institute of Biotechnology), whereas positively staining
slides were used as positive controls.
Evaluation of IHC analysis and
selection of cutpoint value
Following IHC staining, digital images of each spot
in the slides were captured at a magnification of x200 using a
DMI4000B inverted research microscope (Leica Microsystems GmbH,
Wetzlar, Germany). In order to measure the expression levels of
ULK1, the tumor area was selected and then analyzed with the TMAJ
software (TMA Core Facility, Johns Hopkins University, Baltimore,
MD, USA), which discriminated the immunostained area by hue,
saturation and brightness color range. As a result, a numerical
value known as the ULK1 expression index was obtained, which
corresponds to the density of the expression of ULK1 multiplied by
the intensity of the staining. The mean ULK1 expression index for
each of the duplicate cores was used for further statistical
analysis.
In order to optimize the cutpoint of the ULK1
expression index according to the clinicopathological data of the
patients, an open source software termed X-tile program, version
3.6.1 (School of Medicine, Yale University, New Haven, CT, USA) was
used (15,16). The X-tile program divided the cohorts
randomly into a matched training and validation set. Statistical
significance was assessed by the log-rank analysis method, using
the cutpoint derived from a training set to parse a separate
validation set. The X-tile plots determined an optimal cutpoint
value, and the correction for the minimum P statistics was
calculated using the Miller-Siegmund P-value correction (17).
Statistical analysis
All the statistical analyses were performed with
SPSS software, version 16 (SPSS, Inc., Chicago, IL, USA). The
correlation between the expression levels of ULK1 and the
clinicopathological features of CRC was analyzed using the
χ2 test. The Kaplan-Meier method was used to estimate
the OS and DFS. Furthermore, a multivariate Cox proportional hazard
regression model was constructed using the enter method with an
entry criterion of P<0.05 and a removal criterion of P>0.10.
P<0.05 (2-sided) was considered to indicate a statistically
significant difference.
Results
Characteristics of the eligible
patients
As presented in Table
I, a total of 339 patients with CRC (184 males and 155 females;
mean age, 59.1 years) were enrolled for IHC analysis in the present
study. Based on the AJCC TNM staging system, there were 51, 142,
118 and 28 patients with stages I, II, III and IV CRC,
respectively. The locations of the tumors were the colon and rectum
in 167 and 172 patients, respectively. Following a median follow-up
period of 60 months, 90 patients deceased. Table I indicates the characteristics of the
eligible patients.
 | Table I.Characteristics of 339 patients with
colorectal cancer (mean age, 59.10±0.74 years). |
Table I.
Characteristics of 339 patients with
colorectal cancer (mean age, 59.10±0.74 years).
| Variables | Frequency, n (%) |
|---|
| Gender |
|
| Male | 184 (54.3%) |
|
Female | 155 (45.7%) |
| TNM stage |
|
| I | 51 (15.0%) |
| II | 142 (41.9%) |
| III | 118 (34.8%) |
| IV | 28 (8.3%) |
| Tumor location |
|
|
Colon | 167 (49.3%) |
|
Rectum | 172 (50.7%) |
| ULK1
expression levels |
|
| Low | 68 (20.1%) |
| High | 271 (79.9%) |
Correlation between ULK1 and
clinicopathological variables
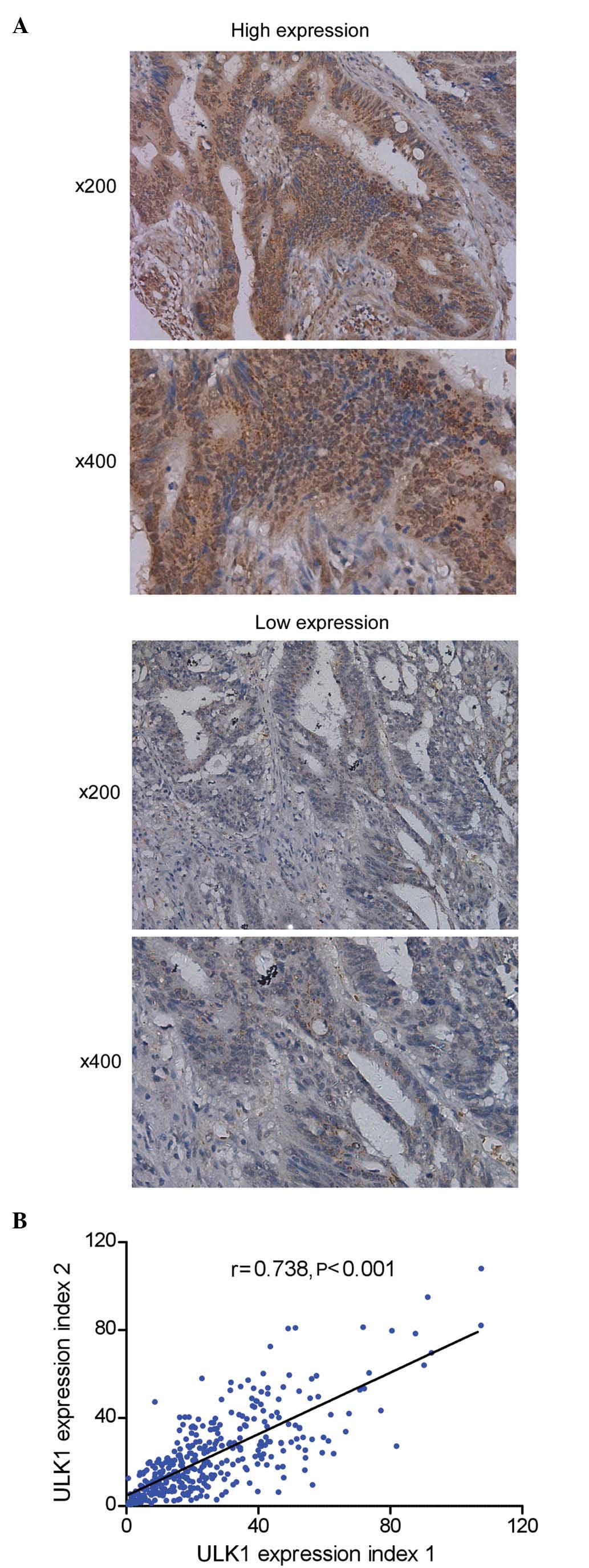
Representative images of tissues expressing ULK1 are
presented in Fig. 1A. ULK1 was
detected in the cytoplasm of cancer cells. To assess the
homogeneity and reliability of the IHC results, the data derived
from the TMAJ software were further analyzed by the Pearson's
product-moment correlation coefficient, which demonstrated a
significant correlation (r=0.738; P<0.001) between the
expression levels of ULK1 and the clinicopathological variables of
the patients with CRC (Fig. 1B).
According to the analysis performed by the X-tile program, high and
low expression levels of ULK1 were observed in 271 and 68 patients,
respectively.
High expression levels of ULK1 were significantly
correlated with gender (P=0.031), poor tumor differentiation
(P=0.029), high expression levels of preoperative carcinoembryonic
antigen (CEA) (P=0.067) and advanced TNM stage (P=0.085). Table II indicates the correlation between
the expression levels of ULK1 and the clinicopathological
characteristics of patients with CRC.
 | Table II.Correlation between the expression
levels of ULK1 and the clinicopathological characteristics of
patients with colorectal cancer. |
Table II.
Correlation between the expression
levels of ULK1 and the clinicopathological characteristics of
patients with colorectal cancer.
|
|
| ULK1 expression
levels |
|
|---|
|
|
|
|
|
|---|
| Variable | Cases | Low | High | P-value |
|---|
| Gender |
|
|
|
|
|
Female/male | 155/184 | 39/29 | 116/155 | 0.031 |
| Age, years |
|
|
|
|
|
<60/≥60 | 164/175 | 30/38 | 134/137 | 0.432 |
| CEA, ng/ml |
|
|
|
|
|
<5/≥5 | 233/106 | 53/15 | 180/91 | 0.067 |
| CA199, U/ml |
|
|
|
|
|
<37.5/≥37.5 | 265/74 | 57/11 | 208/63 | 0.207 |
| pT |
|
|
|
|
|
T1+T2/T3+T4 | 62/277 | 17/51 | 45/226 | 0.109 |
| pN |
|
|
|
|
|
N0/N1+N2 | 206/133 | 47/21 | 159/112 | 0.115 |
| pM |
|
|
|
|
|
M0/M1 | 311/28 | 66/2 | 245/26 | 0.125 |
| TNM stage |
|
|
|
|
|
I+II/III+IV | 193/146 | 45/23 | 148/123 | 0.085 |
| Tumor
differentiation |
|
|
|
|
|
Well+moderately/poorly | 301/38 | 66/2 | 235/36 | 0.029 |
Association between ULK1 and survival
status
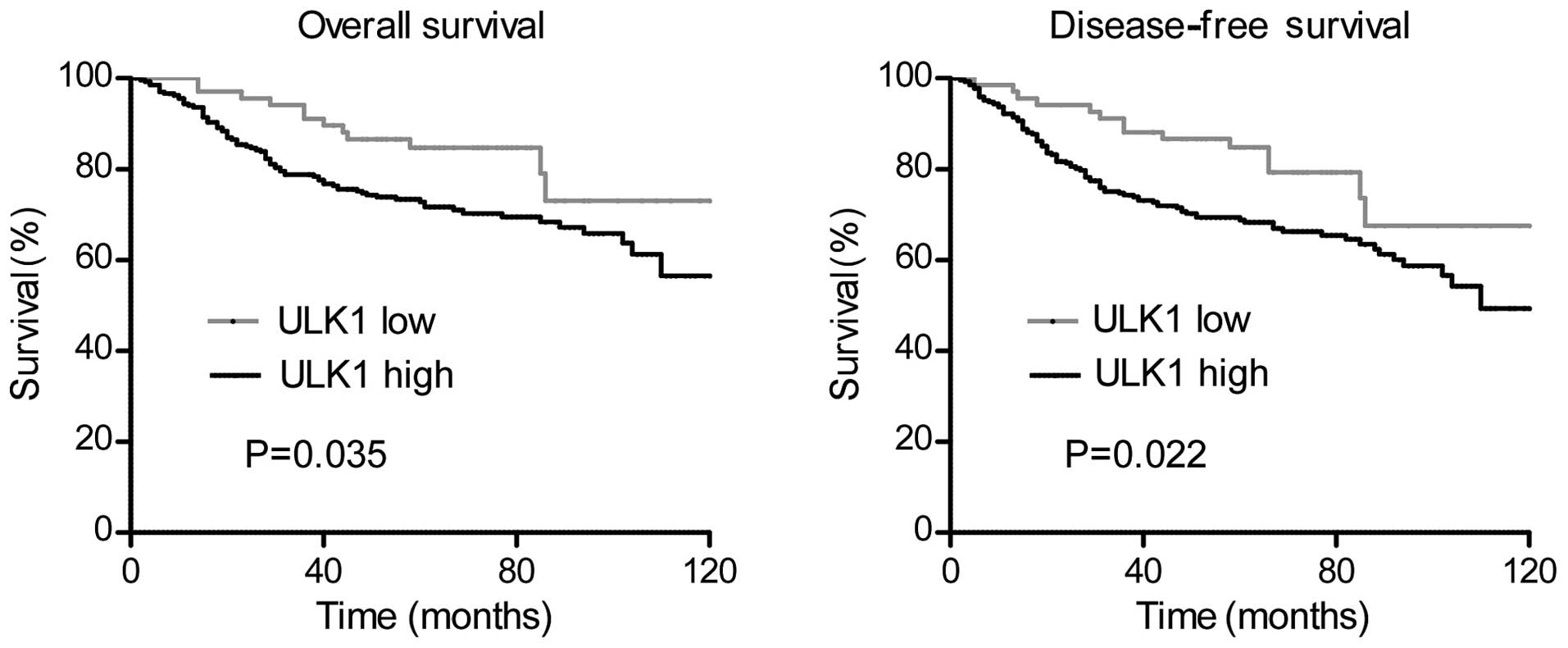
The Kaplan-Meier analysis and the univariate Cox
proportional hazard regression model were used to investigate the
impact of the expression levels of ULK1 on the OS (Table III) and DFS (Table IV) of patients with CRC. The results
demonstrated that high expression levels of ULK1 were significantly
correlated with adverse OS (log-rank test, P=0.035) and DFS
(log-rank test, P=0.022). Other clinicopathological variables,
including preoperative levels of CEA and carbohydrate antigen 19-9
(CA199) (P<0.001 and P=0.004, respectively), TNM stage
(P<0.001), differentiation (P=0.001) and postoperative
recurrence or metastasis (P<0.001) were also observed to be
significant indicators of the OS of patients with CRC, whereas
preoperative levels of CEA and CA199 (P<0.001 and P=0.002,
respectively), TNM stage (P<0.001) and differentiation
(P<0.001) were observed to significantly affect the DFS of
patients with CRC.
 | Table III.Univariate and multivariate analyses
of variables related to overall survival in patients with
colorectal cancer. |
Table III.
Univariate and multivariate analyses
of variables related to overall survival in patients with
colorectal cancer.
|
| Univariate
analysis | Multivariate
analysis |
|---|
|
|
|
|
|---|
| Variable | HR | 95% CI | P-value | HR | 95% CI | P-value |
|---|
| Gender |
|
|
|
|
|
|
|
Male/female | 1.083 | 0.719–1.631 | 0.704 |
|
|
|
| Age, years |
|
|
|
|
|
|
|
≥60/<60 | 1.443 | 0.953–2.186 | 0.084 |
|
|
|
| CEA, ng/ml |
|
|
|
|
|
|
|
≥5/<5 | 2.206 | 1.452–3.351 | <0.001 | 1.769 | 1.126–2.777 | 0.013 |
| CA199, U/ml |
|
|
|
|
|
|
|
≥37.5/<37.5 | 1.932 | 1.236–3.020 | 0.004 | 1.274 | 0.787–2.063 | 0.324 |
| TNM stage |
|
|
|
|
|
|
|
III+IV/I+II | 2.980 | 1.953–4.546 | <0.001 | 2.294 | 1.465–3.590 | <0.001 |
|
Differentiation |
|
|
|
|
|
|
|
Poorly/well+moderately | 2.476 | 1.478–4.151 | 0.001 | 1.597 | 0. 929–2.743 | 0.090 |
| Postoperative
recurrence or metastasis |
|
|
|
|
|
|
Present/absent | 3.483 | 2.209–5.492 | <0.001 | 2.274 | 1.407–3.676 | 0.001 |
| ULK1 expression
levels |
|
|
|
|
|
|
|
High/low | 1.923 | 1.048–3.526 | 0.035 | 1.535 | 0.828–2.846 | 0.173 |
 | Table IV.Univariate and multivariate analyses
of variables related to disease-free survival in patients with
colorectal cancer. |
Table IV.
Univariate and multivariate analyses
of variables related to disease-free survival in patients with
colorectal cancer.
|
| Univariate
analysis | Multivariate
analysis |
|---|
|
|
|
|
|---|
| Variable | HR | 95% CI | P-value | HR | 95% CI | P-value |
|---|
| Gender |
|
|
|
|
|
|
|
Female/male | 1.084 | 0.743–1.583 | 0.675 |
|
|
|
| Age, years |
|
|
|
|
|
|
|
<60/≥60 | 1.248 | 0.854–1.824 | 0.251 |
|
|
|
| CEA, ng/ml |
|
|
|
|
|
|
|
<5/≥5 | 2.126 | 1.440–3.137 | <0.001 | 1.836 | 1.202–2.805 | 0.005 |
| CA199, U/ml |
|
|
|
|
|
|
|
|
<37.5/≥37.5 | 1.939 | 1.278–2.943 | 0.002 | 1.345 | 0.856–2.115 | 0.198 |
| TNM stage |
|
|
|
|
|
|
|
I+II/III+IV | 2.923 | 1.982–4.310 | <0.001 | 2.495 | 1.677–3.714 | <0.001 |
|
Differentiation |
|
|
|
|
|
|
|
|
Well+moderately/poorly | 2.649 | 1.657–4.236 | <0.001 | 1.914 | 1.173–3.125 | 0.009 |
| ULK1 expression
levels |
|
|
|
|
|
|
|
Low/high | 1.930 | 1.101–3.384 | 0.022 | 1.576 | 0.891–2.786 | 0.118 |
Additionally, multivariate analysis was performed
using the Cox proportional hazards model for all of the variables
observed to be significant in the univariate analysis. The results
indicated that the preoperative levels of CEA and the TNM stage
remained significant independent adverse prognostic factors for OS
(P=0.013 and <0.001, respectively) and DFS (P=0.005 and
<0.001, respectively), and that the postoperative recurrence or
metastasis was an independent prognostic factor for OS (P=0.001).
However, the expression levels of ULK1 were not observed to be an
independent prognostic factor for OS or DFS (P=0.173 and 0.118,
respectively). The association between the variables and the
survival status is presented in Tables
III and IV. The survival curves
obtained according to the expression levels of ULK1 in patients
with CRC are represented in Fig.
2.
Discussion
CRC is a common malignancy worldwide (1). Despite the improvements in diagnostic
techniques and therapeutic methods, CRC remains a serious challenge
(18). Currently, the treatment and
prognosis of patients with CRC depends on the AJCC TNM stage
classification system, according to the results of pathological
analysis (2). However, this staging
system is not sufficiently reliable. Different studies have
demonstrated that the OS of patients with CRC of stage IIb was
poorer than those of stage IIIa (19–21). As an
alternative to the AJCC TNM staging system, an increasing number of
molecular biomarkers have been defined as useful prognostic and
predictive factors in CRC (22),
including KARS and BRAF, which are commonly used in clinical
practice (23,24).
Autophagy is a cellular dynamic process involved in
the regulation of carcinogenesis and the response to anticancer
therapy (25,26). ULK1, as a core ATG, is involved in the
initiation of autophagy (27). The
role of ULK1 in different tumors varies with the type of cancer
(13,14). To the best of our knowledge, the
present study reports for the first time that the expression levels
of ULK1 increase with the magnitude of cancer progression in tissue
specimens derived from patients with CRC, according to the results
of IHC staining.
In the current study, high expression levels of ULK1
were observed to be significantly associated with female gender and
poor tumor differentiation, and were slightly associated with high
expression levels of preoperative CEA and advanced TNM stage. In
the survival analysis, high expression levels of ULK1 were
significantly correlated with adverse OS and DFS in the univariate
analysis, but not in the multivariate analysis. These results
suggest that the expression levels of ULK1 are of clinical value to
assess the prognosis and survival of patients with CRC.
The majority of cancer cells grow preferentially in
the presence of abundant nutrients and oxygen, which facilitate a
high proliferation rate (13).
However, CRC is an example of a common solid tumor with a limited
blood supply, and the resulting metabolic stress becomes more
severe as the cancer develops (28).
Since the autophagy process maintains cellular homeostasis and
degrades toxic cytoplasmic constituents, it also enables malignant
cells to endure various stress conditions, including limited
availability of nutrients, reduced energy supply or low levels of
oxygen (6,7). Consistently, the cancer cells may
express high levels of ULK1 in order to activate the initiation of
autophagy and survive. This may explain why high expression levels
of ULK1 in patients with CRC indicate a poor prognosis. However,
further studies are required to clarify the exact mechanism of
action.
In conclusion, the current study demonstrates that
high expression levels of ULK1 may be a useful independent
biomarker for predicting the poor prognosis of patients with
CRC.
Acknowledgements
The current work was supported by the National
Natural Science Foundation of China (No. 81300367) and the Natural
Science Foundation of Guangdong Province (No. S2013010014186).
References
|
1
|
Jemal A, Bray F, Center MM, Ferlay J, Ward
E and Forman D: Global cancer statistics. CA Cancer J Clin.
61:69–90. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Edge SB and Compton CC: The American Joint
Committee on Cancer: The 7th edition of the AJCC cancer staging
manual and the future of TNM. Ann Surg Oncol. 17:1471–1474. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Siegel R, Ward E, Brawley O and Jemal A:
Cancer statistics, 2011: The impact of eliminating socioeconomic
and racial disparities on premature cancer deaths. CA Cancer J
Clin. 61:212–236. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Nagtegaal ID and Quirke P: Revised
staging: Is it really better, or do we not know? J Clin Oncol.
28:e397–e400. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Graziano F and Cascinu S: Prognostic
molecular markers for planning adjuvant chemotherapy trials in
Dukes' B colorectal cancer patients: How much evidence is enough?
Ann Oncol. 14:1026–1038. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Zhao M and Klionsky DJ: AMPK-dependent
phosphorylation of ULK1 induces autophagy. Cell Metab. 13:119–120.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Mizushima N, Levine B, Cuervo AM and
Klionsky DJ: Autophagy fights disease through cellular
self-digestion. Nature. 451:1069–1075. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Wong PM, Puente C, Ganley IG and Jiang X:
The ULK1 complex: Sensing nutrient signals for autophagy
activation. Autophagy. 9:124–137. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Kim J, Kundu M, Viollet B and Guan KL:
AMPK and mTOR regulate autophagy through direct phosphorylation of
Ulk1. Nat Cell Biol. 13:132–141. 2011. View
Article : Google Scholar : PubMed/NCBI
|
|
10
|
Lee JW, Park S, Takahashi Y and Wang HG:
The association of AMPK with ULK1 regulates autophagy. PLoS One.
5:e153942010. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Hosokawa N, Hara T, Kaizuka T, Kishi C,
Takamura A, Miura Y, Iemura S, Natsume T, Takehana K, Yamada N, et
al: Nutrient-dependent mTORC1 association with the
ULK1-Atg13-FIP200 complex required for autophagy. Mol Biol Cell.
20:1981–1991. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Jung CH, Jun CB, Ro SH, Kim YM, Otto NM,
Cao J, Kundu M and Kim DH: ULK-Atg13-FIP200 complexes mediate mTOR
signaling to the autophagy machinery. Mol Biol Cell. 20:1992–2003.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Jiang S, Li Y, Zhu YH, Wu XQ, Tang J, Li
Z, Feng GK, Deng R, Li DD, Luo RZ, et al: Intensive expression of
UNC-51-like kinase 1 is a novel biomarker of poor prognosis in
patients with esophageal squamous cell carcinoma. Cancer Sci.
102:1568–1575. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Tang J, Deng R, Luo RZ, Shen GP, Cai MY,
Du ZM, Jiang S, Yang MT, Fu JH and Zhu XF: Low expression of ULK1
is associated with operable breast cancer progression and is an
adverse prognostic marker of survival for patients. Breast Cancer
Res Treat. 134:549–560. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Cai MY, Hou JH, Rao HL, Luo RZ, Li M, Pei
XQ, Lin MC, Guan XY, Kung HF, Zeng YX and Xie D: High expression of
H3K27me3 in human hepatocellular carcinomas correlates closely with
vascular invasion and predicts worse prognosis in patients. Mol
Med. 17:12–20. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Camp RL, Dolled-Filhart M and Rimm DL:
X-tile: A new bio-informatics tool for biomarker assessment and
outcome-based cut-point optimization. Clin Cancer Res.
10:7252–7259. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Raeside DE: Monte Carlo principles and
applications. Phys Med Biol. 21:181–197. 1976. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Brezden-Masley C and Polenz C: Current
practices and challenges of adjuvant chemotherapy in patients with
colorectal cancer. Surg Oncol Clin N Am. 23:49–58. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
O'Connell JB, Maggard MA and Ko CY: Colon
cancer survival rates with the new American Joint Committee on
Cancer sixth edition staging. J Natl Cancer Inst. 96:1420–1425.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Oh HS, Chung HJ, Kim HK and Choi JS:
Differences in overall survival when colorectal cancer patients are
stratified into new TNM staging strategy. Cancer Res Treat.
39:61–64. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Kim KH, Yang SS, Yoon YS, Lim SB, Yu CS
and Kim JC: Validation of the seventh edition of the American Joint
Committee on Cancer tumor-node-metastasis (AJCC TNM) staging in
patients with stage II and stage III colorectal carcinoma: Analysis
of 2511 cases from a medical centre in Korea. Colorectal Dis.
13:e220–e226. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Ross JS: Biomarker-based selection of
therapy for colorectal cancer. Biomarkers Med. 5:319–332. 2011.
View Article : Google Scholar
|
|
23
|
Rizzo S, Bronte G, Fanale D, Corsini L,
Silvestris N, Santini D, Gulotta G, Bazan V, Gebbia N, Fulfaro F
and Russo A: Prognostic vs predictive molecular biomarkers in
colorectal cancer: Is KRAS and BRAF wild type status required for
anti-EGFR therapy? Cancer Treat Rev. 36:(Sul 3). S56–S61. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Yokota T: Are KRAS/BRAF mutations potent
prognostic and/or predictive biomarkers in colorectal cancers?
Anticancer Agents Med Chem. 12:163–171. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Hönscheid P, Datta K and Muders MH:
Autophagy: Detection, regulation and its role in cancer and therapy
response. Int J Radiat Biol. 90:628–635. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Schmukler E, Kloog Y and Pinkas-Kramarski
R: Ras and autophagy in cancer development and therapy. Oncotarget.
5:577–586. 2014.PubMed/NCBI
|
|
27
|
Mizushima N: The role of the Atg1/ULK1
complex in autophagy regulation. Curr Opin Cell Biol. 22:132–139.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Mathonnet M, Perraud A, Christou N, Akil
H, Melin C, Battu S, Jauberteau MO and Denizot Y: Hallmarks in
colorectal cancer: Angiogenesis and cancer stem-like cells. World J
Gastroenterol. 20:4189–4196. 2014. View Article : Google Scholar : PubMed/NCBI
|