Introduction
Lung cancer has become one of the most serious
malignant types of tumor, and is associated with high incidence and
mortality rates worldwide. According to a report by the American
Cancer Society in 2013, the occurrence of lung cancer constitutes
14% of the total incidence of malignant tumors (1,2).
Furthermore, lung cancer is the second most common malignancy
globally after prostatic and breast cancer, and accounts for 28 and
16% of mortalities caused by malignancies in men and women,
respectively. Thus, lung cancer is the most lethal human malignancy
(3). In fact, lung cancer has even
higher incidence and mortality rates in China, with ~85% of cases
of lung cancer diagnosed as non-small cell lung cancer (NSCLC)
(2). This indicates the importance of
fundamental investigations into clinical treatment guidance for
patients with NSCLC.
Recently, significant advancements have been made in
the comprehensive treatment of lung cancer, particularly with
targeted agents. Despite this, there has been little improvement in
the 5-year survival rates of patients with lung cancer, with the
rate remaining as low as 15%. This is true even for those treated
patients with surgery, constituting ≤40% of patients (3). One of the leading factors associated
with this poor survival is the lack of effective early diagnostic
criteria. In fact, the majority of patients are already in the
aggressive stage when diagnosed with lung cancer (4). Another important factor is the lack of
efficient treatment strategies for severe cases with marked
resistance to multiple agents. On this basis, there is an urgent
requirement to identify effective diagnostic criteria for the early
diagnosis of lung cancer, as well as indexes for its prognosis,
that together may contribute to improvements in the total survival
rates.
The heterogeneous nuclear ribonucleoprotein (hnRNP)
family are important in the process of mRNA transcription, shearing
and splicing (5). The superfamily is
composed of >20 members, the majority of which bind to the
splicing sequences located within introns and exons to perform
regulatory roles (6). Among them, the
principle members are hnRNP A1, A2, A3 and B1. hnRNP A2 and B1
constitute the core, and are isomers of the same protein derived
from the hnRNP A2/B1 gene. Thus, there are similarities in the
sequence and function of these two proteins (7–9). Numerous
studies have identified abnormally high expression of hnRNP A2/B1
in a variety of tumor types (10–14).
According to previous reports, it is well-established that hnRNP
A2/B1 serves as an early diagnostic indicator of lung cancer
(15–17). However, it remains unclear whether it
is effective as a prognostic criterion.
AXL belongs to the receptor tyrosine kinase (RTK)
family of proteins (18). AXL,
together with two other RTKs (TYRO3 and MER), constitutes the TAM
tyrosine receptor sub-family. The three proteins share similar
structures and functions (19–21).
Structurally, TAM proteins are characterized by two
immunoglobulin-like extra-cellular domains and two repeating
fibronectin type III cytoplasmic kinase domains (19). They also share the same ligand, Gas6.
Previous studies indicated that the activation of TAM, as well as
the downstream signal transduction cascades, are key in various
cellular functions and behaviors, including cell survival,
proliferation, migration and adherence (18). High expression of AXL was also
identified in a variety of tumors, and AXL appears to be important
in the invasion and metastasis of lung cancer (22). More notably, He et al (23) performed RNA interference (RNAi) to
reduce the expression of hnRNP A2/B1 in Colo16 cancer cells. Using
high-throughput gene chip screening, marked differences were
observed in the expression of 123 downstream target genes,
including AXL, indicating a potential interaction between hnRNP
A2/B1 and AXL.
The present study aimed to determine the expression
of hnRNP A2/B1 and AXL in NSCLC and paracancerous lung control
samples, as well as perform a prognostic analysis of the collected
clinical data to explore any potential association. In addition,
RNAi was performed to silence the expression of hnRNP A2/B1, on
which basis molecular and cellular functions of hnRNP A2/B1 were
evaluated to determine its mechanism in NSCLC, possibly by means of
AXL mediation. Written informed consent was obtained from all
patients.
Materials and methods
Tissue microarray
The tissue microarray was purchased from the
National Biological Chip Center (Shanghai, China) and constructed
using 150 resected NSCLC and paracancerous lung tissues samples
(two sets of chips with serial sections). Matched cancerous and
paracancerous samples were collected from the First Hospital of
China Medical University (Shenyang, China) between 2004 and 2007
during surgery for pulmonary lobectomy or total pneumonectomy.
Following the exclusion of cases with incomplete data, such as
gender, age, tumor size, histological type, TNM American Joint
Committee on Cancer classification (24), degree of differentiation, lymph node
metastasis and prognostic data, 134 cases were available for
prognostic analysis. The most recent follow-up time was July 2012.
The study was approved by the ethics committee of the First
Affiliated Hospital of China Medical University.
Reagents
Mouse monoclonal anti-human hnRNP A2/B1 (cat no.
ab6102) and rabbit polyclonal anti-human AXL (cat no. ab72069)
antibodies were purchased from Abcam (Cambridge, UK), and
monoclonal mouse anti-human β-actin (cat no. SC-47778) antibodies
were purchased from Santa Cruz Biotechnology, Inc. (Dallas, TX,
USA). Additionally, the UltraSensitive™ SP IHC and MaxVision™ DAB
kits were purchased from Maxim Biotech, Inc. (Rockville, MD,
USA).
Immunohistochemical
streptavidin-peroxidase analysis
The tissue chips were dewaxed with dimethylbenzene
two times (15 min each), then washed twice with 100% ethanol (5 min
each), once with 95% ethanol (2 min each), once with 85% ethanol (2
min each), once with 75% ethanol (2 min each) and three times with
distilled water (3 min each). Endogenous peroxidase was blocked
with liquid-A of the UltraSensitive SP IHC kit for 30 min and then
washed three times with phosphate-buffered saline (PBS; 3 min
each). Antigen retrieval was performed at a high pressure using
citric acid for 3 min, cooled to room temperature and washed three
times with PBS (3 min each). The non-immune serum (liquid-B,
UltraSensitive SP IHC kit) was added prior incubating the chips at
37°C for 30 min to block non-specific antigens. Excess serum was
discarded, primary hnRNP A2/B1 (1:500 dilution) and AXL (1:100
dilution) antibodies were added, and the tissue chips were stored
at 4°C overnight. Secondary antibody (solution-C, UltraSensitive SP
IHC kit) was added and incubated at 37°C for 30 min. After washing
three times with PBS (3 min each), the coloring conditions were
observed under microscopy by adding solution-D of the
UltraSensitive SP IHC kit and the DAB reagents. The reaction was
terminated and stained with hematoxylin for 3 min, differentiated
with 1% hydrochloric acid ethanol and rinsed in water for 10 min.
Dehydration with ethanol was performed in gradients prior to
clearing in xylene and mounting with neutral gum.
Classification
The results of the immunohistochemical analysis were
classified according to the following criterion: The proportion of
positive cells (<30%, 1 point; 30–60%, 2 point; >60%, 3
points) and the color of staining (colorless or light yellow, 1
point; yellow, 2 points; brown, 3 points). The final scores were
obtained by multiplying the two integrates and were used to
determine the following classifications: 1–2, negative expression;
and 3–9, positive expression. All scoring was performed by two
independent pathologists and the mean of the scores was used as the
final result.
Statistical analysis
Comparisons were performed using t-tests, and
correlation analysis between the expression of hnRNP A2/B1 and AXL
in the lung tissue samples was conducted using the Pearson's
correlation method. The clinical data was analyzed by performing a
χ2-test or Fisher's exact probability. Survival curves
were constructed according to the Kaplan-Meier method and analyzed
using log-rank tests. Furthermore, univariate and multivariate Cox
regression analysis was used to identify independent prognostic
factors. SPSS software (version 18.0; IBM SPSS, Armonk, NY, USA)
was used to perform all statistical analyses. P<0.05 was
considered to indicate a statistically significant difference.
Results
Protein expression levels of hnRNP
A2/B1 in NSCLC and paracancerous tissue samples
In the present study, a total of 134 NSCLC and
paracancerous tissue chip samples were collected.
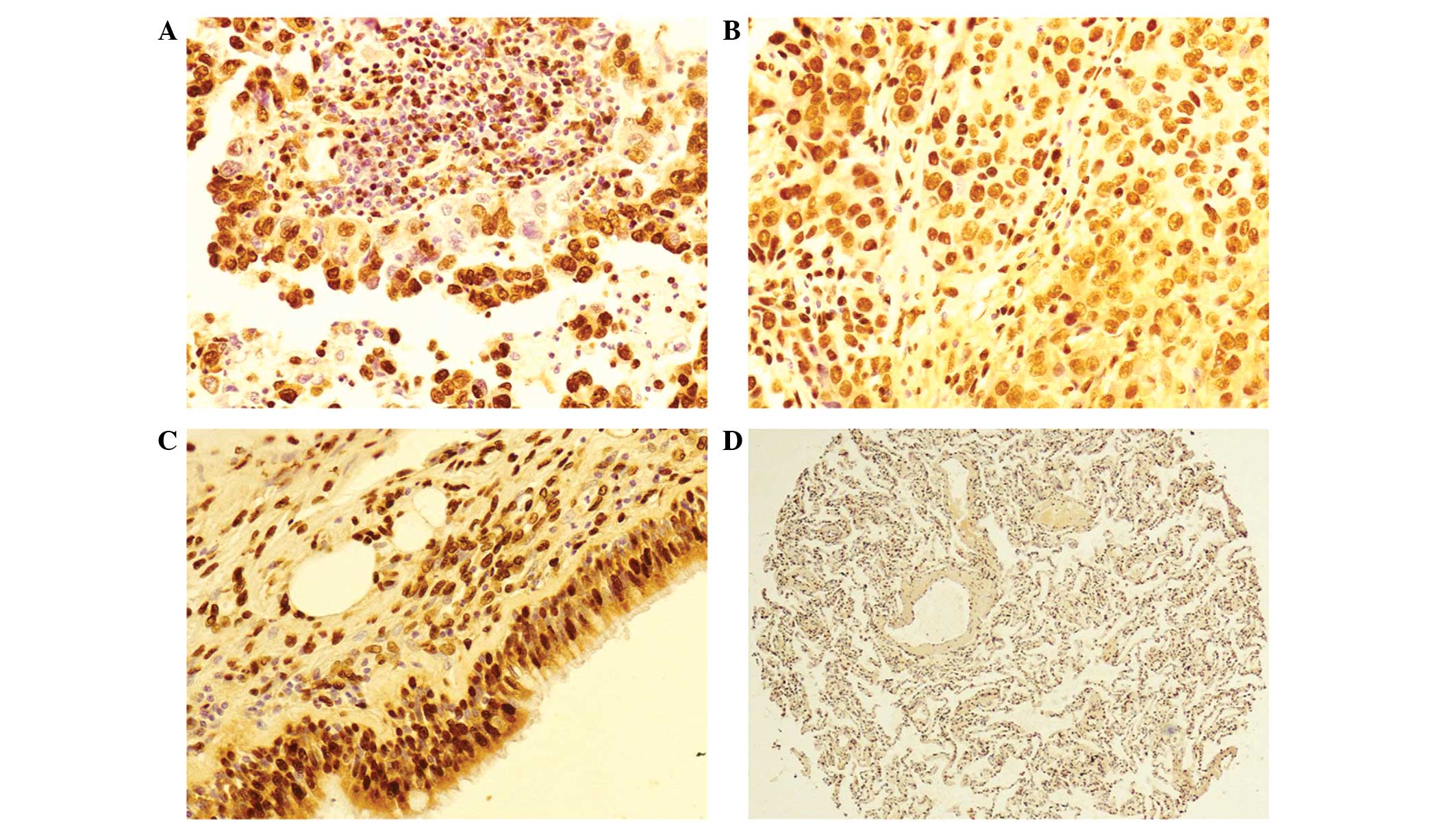
Immunohistochemical staining was used to identify that hnRNP A2/B1
protein is located in the nucleus and cytoplasm of invasive
adenocarcinoma (Fig. 1A) and highly
differentiated squamous cell carcinoma (Fig. 1B). High hnRNP A2/B1 expression was
observed in hyperplastic bronchial epithelia (Fig. 1C), however, little expression of hnRNP
A2/B1 was observed in the paracancerous lung tissues (Fig. 1D). According to statistical analysis,
there was a significantly greater rate of positive hnRNP A2/B1
expression in NSCLC compared with paracancerous lung tissues
(P<0.001; Table I).
 | Table I.Expression of hnRNP A2/B1 in NSCLC and
paracancerous lung tissues. |
Table I.
Expression of hnRNP A2/B1 in NSCLC and
paracancerous lung tissues.
| Lung tissue | Total cases, n | hnRNP A2/B1
expression, n | Positive rate, % | P-value |
|---|
|
|---|
| Negative | Positive |
|---|
| NSCLC | 134 | 47 | 87 | 67.9 | 0.000a |
| Paracancerous | 134 | 121 | 13 | 9.7 |
|
Association between hnRNP A2/B1
expression and clinicopathological factors
According to statistical analysis of the clinical
data, it was identified that the expression of hnRNP A2/B1 was
significantly correlated with the differentiation of lesions in 134
cases of NSCLC. The lower the degree of differentiation, the
greater the positive expression of hnRNP A2/B1 (P=0.001).
Furthermore, according to the TNM classification system, there was
a significant increase in positive hnRNP A2/B1 expression in phases
III/IV compared with phases I/II (P=0.007). However, hnRNP A2/B1
expression was not significantly associated with gender, age,
histological type, lymph node metastasis or tumor size (Table II).
 | Table II.Association between hnRNP A2/B1
expression and clinicopathological factors in patients with
NSCLC. |
Table II.
Association between hnRNP A2/B1
expression and clinicopathological factors in patients with
NSCLC.
| Clinicopathological
factor | Cases, n (n=134) | hnRNP A2/B1
expression | Positive rate, % | P-value |
|---|
|
|---|
| – | + |
|---|
| Gender |
|
|
|
|
|
| Male | 99 | 35 | 64 | 64.6 | 1.000 |
|
Female | 35 | 12 | 23 | 65.7 |
|
| Age, years |
|
|
|
|
|
| ≤60 | 58 | 19 | 39 | 67.2 | 0.716 |
|
>60 | 76 | 28 | 48 | 63.2 |
|
| Histological
type |
|
|
|
|
|
|
Adenocarcinoma | 67 | 22 | 45 | 67.2 | 0.718 |
| Squamous
cell carcinoma | 67 | 25 | 42 | 62.7 |
|
| Degree of
differentiation |
|
|
|
|
|
| High | 23 | 13 | 10 | 43.5 | 0.001a |
|
Middle | 73 | 29 | 44 | 60.3 |
|
| Low | 38 | 5 | 33 | 86.8 |
|
| TNM
classification |
|
|
|
|
|
| I/II | 99 | 41 | 58 | 58.6 | 0.013a |
|
III/IV | 35 | 6 | 29 | 82.9 |
|
| Lymphatic
metastasis |
|
|
|
|
|
|
Yes | 60 | 20 | 40 | 66.7 | 0.720 |
| No | 74 | 27 | 47 | 63.5 |
|
| Tumor size, cm |
|
|
|
|
|
| ≤3 | 48 | 21 | 27 | 56.3 | 0.133 |
|
>3 | 86 | 26 | 60 | 69.8 |
|
Association between hnRNP A2/B1
expression and survival
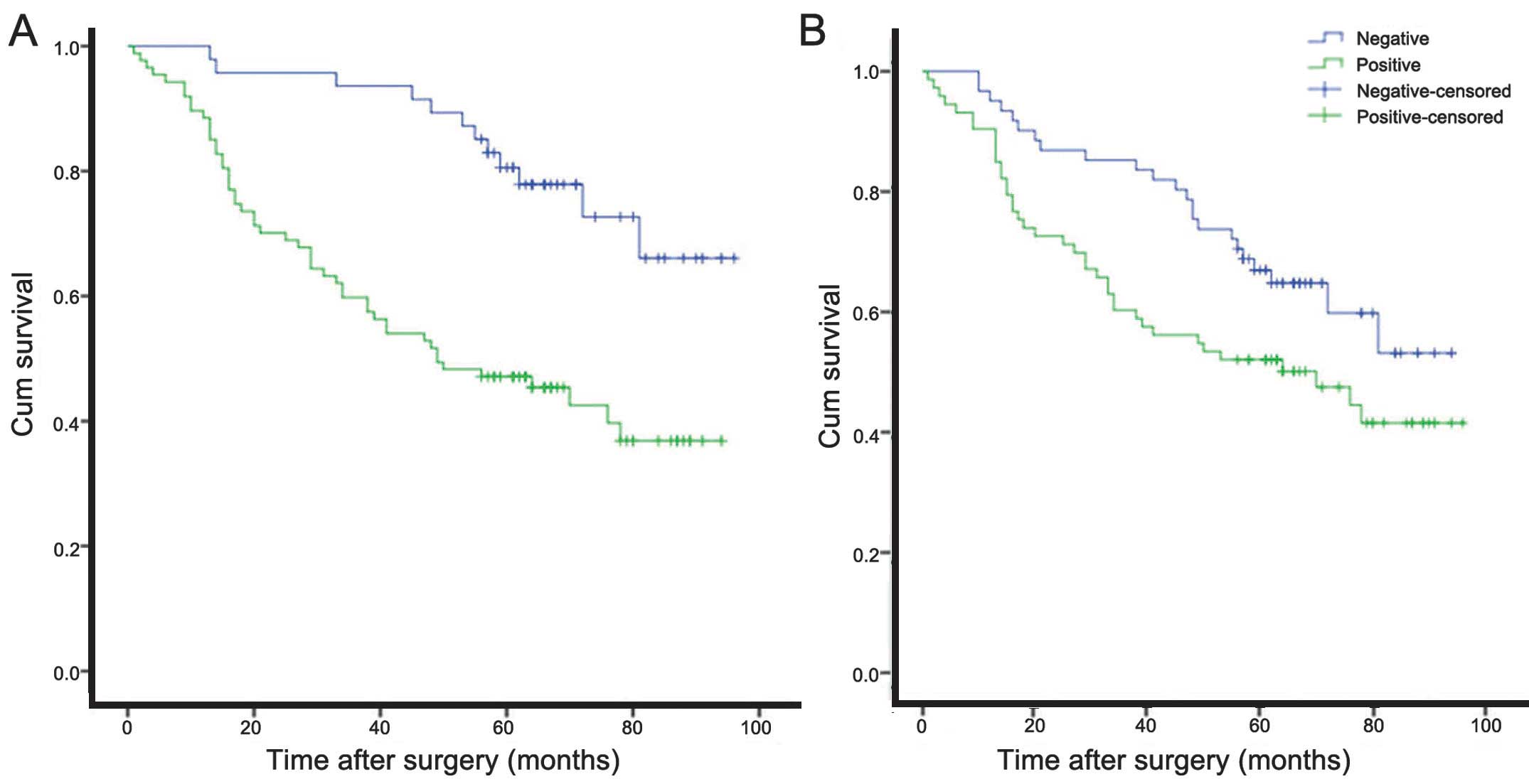
To analyze survival, Kaplan-Meier plots were
constructed for hnRNP A2/B1-positive and -negative patients
(Fig. 2A). The median survival time
was 54.7±3.8 months for hnRNP A2/B1-positive patients and 82.4±3.5
months for hnRNP A2/B1-negative patients. There was a statistically
significant difference between survival in these two groups
(log-rank test, P<0.001). Compared with other clinical factors,
such as gender, histological type and degree of differentiation,
positive expression of hnRNP A2/B1 was identified as an independent
risk factor that may influence the prognosis of patients with NSCLC
(P=0.001; Table III), according to
multivariate Cox regression analysis.
 | Table III.Univariate and multivariate
regression analysis of clinical data and prognosis with Cox
model. |
Table III.
Univariate and multivariate
regression analysis of clinical data and prognosis with Cox
model.
| Index | Univariate
regression analysis | Multivariate
regression analysis |
|---|
|
|
|---|
| 95% confidence
interval | P-value | 95% confidence
interval | P-value |
|---|
| Gender | 0.926
(0.704–1.219) | 0.584 | – | – |
| Tumor size | 0.756
(0.572–1.000) | 0.050a | – | – |
| Histological
type | 1.083
(0.844–1.390) | 0.532 | – | – |
| Degree of
differentiation | 0.997
(0.687–1.446) | 0.986 | – | – |
| TNM
classification | 1.800
(1.378–2.351) |
<0.001a | 1.558
(1.086–2.237) | 0.016a |
| Lymphatic
metastasis | 2.234
(1.396–3.867) | 0.001a | 1.490
(0.809–2.746) | 0.201 |
| Age | 1.910
(1.112–3.280) | 0.019a | 2.119
(1.228–3.655) | 0.007a |
| hnRNPA2/B
expression | 0.313
(0.166–0.588) |
<0.001a | 2.928
(1.539–5.573) | 0.001a |
Protein expression levels of AXL in
NSCLC and paracancerous tissue samples
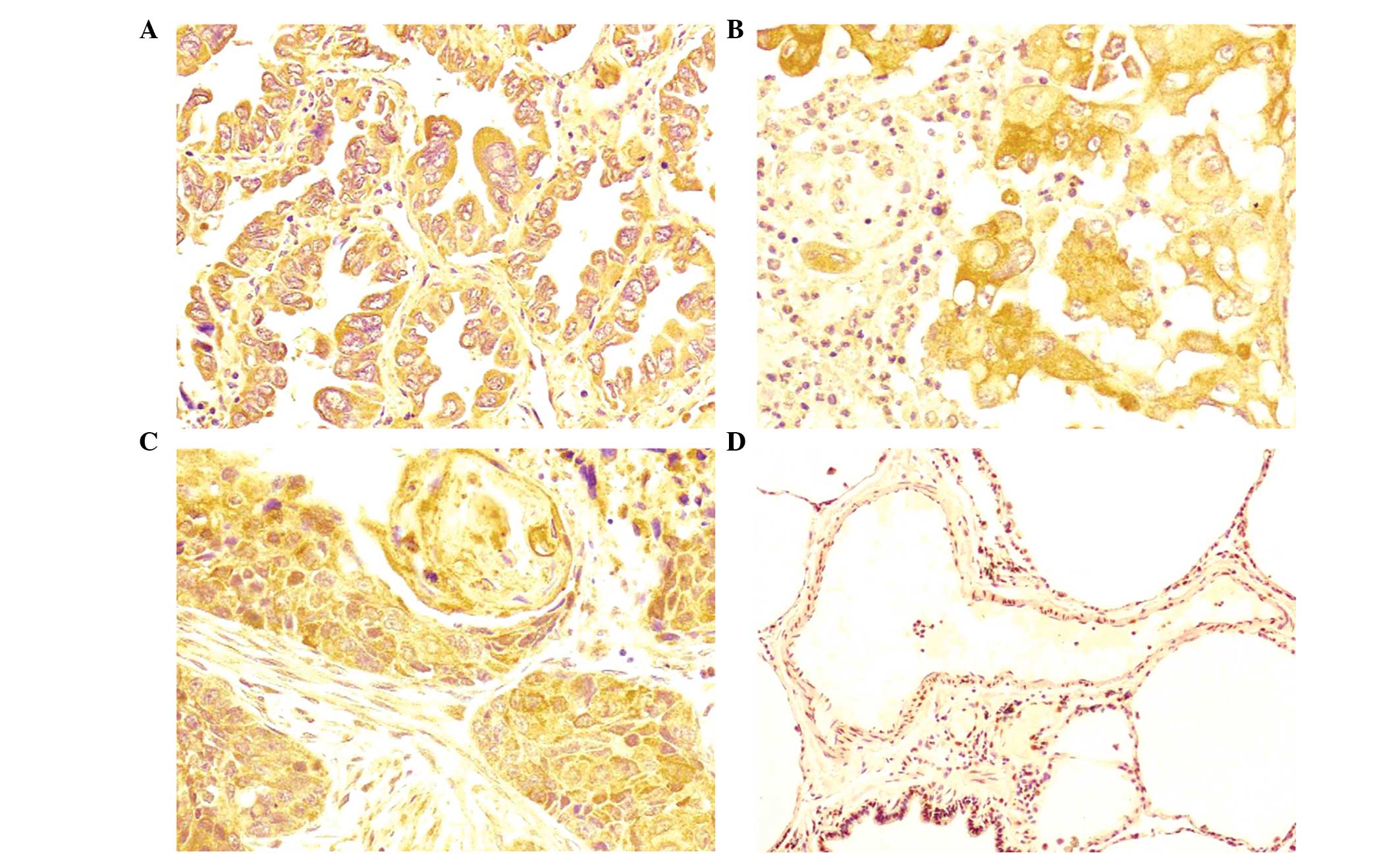
Immunohistochemical analysis of 134 NSCLC and
paracancerous tissue samples revealed that AXL protein is
predominantly located in the cytoplasm (Fig. 3), with little expression in the
nucleus. Furthermore, statistical analysis identified a
significantly greater rate of positive AXL expression in NSCLC
compared with in paracancerous lung tissues (P<0.05; Table IV).
 | Table IV.Expression of AXL in NSCLC and
paracancerous lung tissues. |
Table IV.
Expression of AXL in NSCLC and
paracancerous lung tissues.
| Lung tissue | Total cases, n | AXL expression,
n | Positive rate,
% | P-value |
|---|
|
|---|
| Negative | Positive |
|---|
| NSCLC | 134 | 61 | 73 | 54.5 | 0.001a |
| Paracancerous | 134 | 95 | 39 | 29.1 |
|
Association between AXL expression and
clinicopathological factors
Following statistical analysis of the clinical data,
it was identified that the expression of AXL is significantly
correlated with the differentiation of lesions in the 134 cases of
NSCLC investigated. The lower the degree of differentiation, the
greater the positive expression of AXL (P=0.001). According to the
TNM classification system, there was a significant increase in the
expression of AXL in TNM phases II/III/IV compared with phase I
(P=0.005). However, AXL expression was not significantly associated
with other clinical factors, such as gender, age, histological
type, lymph node metastasis and tumor size (Table V).
 | Table V.Association between AXL expression
and clinicopathological factors in patients with NSCLC. |
Table V.
Association between AXL expression
and clinicopathological factors in patients with NSCLC.
| Clinicopathological
factor | Cases, n
(n=134) | AXL expression | Positive rate,
% | P-value |
|---|
|
|---|
| – | + |
|---|
| Gender |
|
|
|
|
|
|
Male | 99 | 42 | 57 | 57.6 | 0.242 |
|
Female | 35 | 19 | 16 | 45.7 |
|
| Age, years |
|
|
|
|
|
|
≤60 | 58 | 25 | 33 | 56.9 | 0.727 |
|
>60 | 76 | 36 | 40 | 53.6 |
|
| Histological
type |
|
|
|
|
|
|
Adenocarcinoma | 67 | 31 | 36 | 53.7 | 1.000 |
|
Squamous cell carcinoma | 67 | 30 | 37 | 55.2 |
|
| Degree of
differentiation |
|
|
|
|
|
|
High | 23 | 17 | 6 | 26.1 | 0.001a |
|
Middle | 73 | 37 | 36 | 49.3 |
|
|
Low | 38 | 7 | 31 | 81.6 |
|
| TNM
classification |
|
|
|
|
|
| I | 23 | 17 | 6 | 26.1 | 0.005a |
|
II/III/IV | 111 | 44 | 67 | 60.4 |
|
| Lymphatic
metastasis |
|
|
|
|
|
|
Yes | 60 | 23 | 37 | 61.7 | 0.092 |
| No | 74 | 38 | 36 | 48.7 |
|
| Tumor size, cm |
|
|
|
|
|
| ≤3 | 48 | 26 | 22 | 45.8 | 0.167 |
|
>3 | 86 | 35 | 51 | 59.3 |
|
Association between AXL expression and
survival
To analyze survival, Kaplan-Meier plots were
constructed for AXL-positive and -negative patients (Fig. 2B). The median survival time was
58.2±4.3 months for AXL-positive patients and 71.8±3.8 months for
AXL-negative patients. There was a statistically significant
difference between survival in these two groups (log-rank test;
P=0.042). According to univariate Cox regression analysis, the
positive expression of AXL was identified as an independent risk
factor that may influence prognosis in NSCLC (P=0.045; Table VI).
 | Table VI.Univariate and multivariate Cox
regression analysis of clinical data and prognosis of patients with
NSCLC. |
Table VI.
Univariate and multivariate Cox
regression analysis of clinical data and prognosis of patients with
NSCLC.
| Index | Univariate
regression analysis | Multivariate
regression analysis |
|---|
|
|
|---|
| 95% confidence
interval | P-value | 95% confidence
interval | P-value |
|---|
| Gender | 0.926
(0.704–1.219) | 0.584 | – | – |
| Tumor size | 0.756
(0.572–1.000) | 0.050a | – | – |
| Histological
type | 1.083
(0.844–1.390) | 0.532 | – | – |
| Degree of
differentiation | 0.997
(0.687–1.446) | 0.986 | – | – |
| TNM
classification | 1.800
(1.378–2.351) |
<0.001a | 1.733
(1.219–2.464) | 0.002a |
| Lymphatic
metastasis | 2.234
(1.396–3.867) | 0.001a | 1.291
(0.707–2.357) | 0.405 |
| Age | 1.910
(1.112–3.280) | 0.019a | 2.025
(1.173–3.497) | 0.011a |
| AXL expression | 1.696
(1.012–2.841) | 0.045a | 1.406
(0.830–2.383) | 0.206 |
|
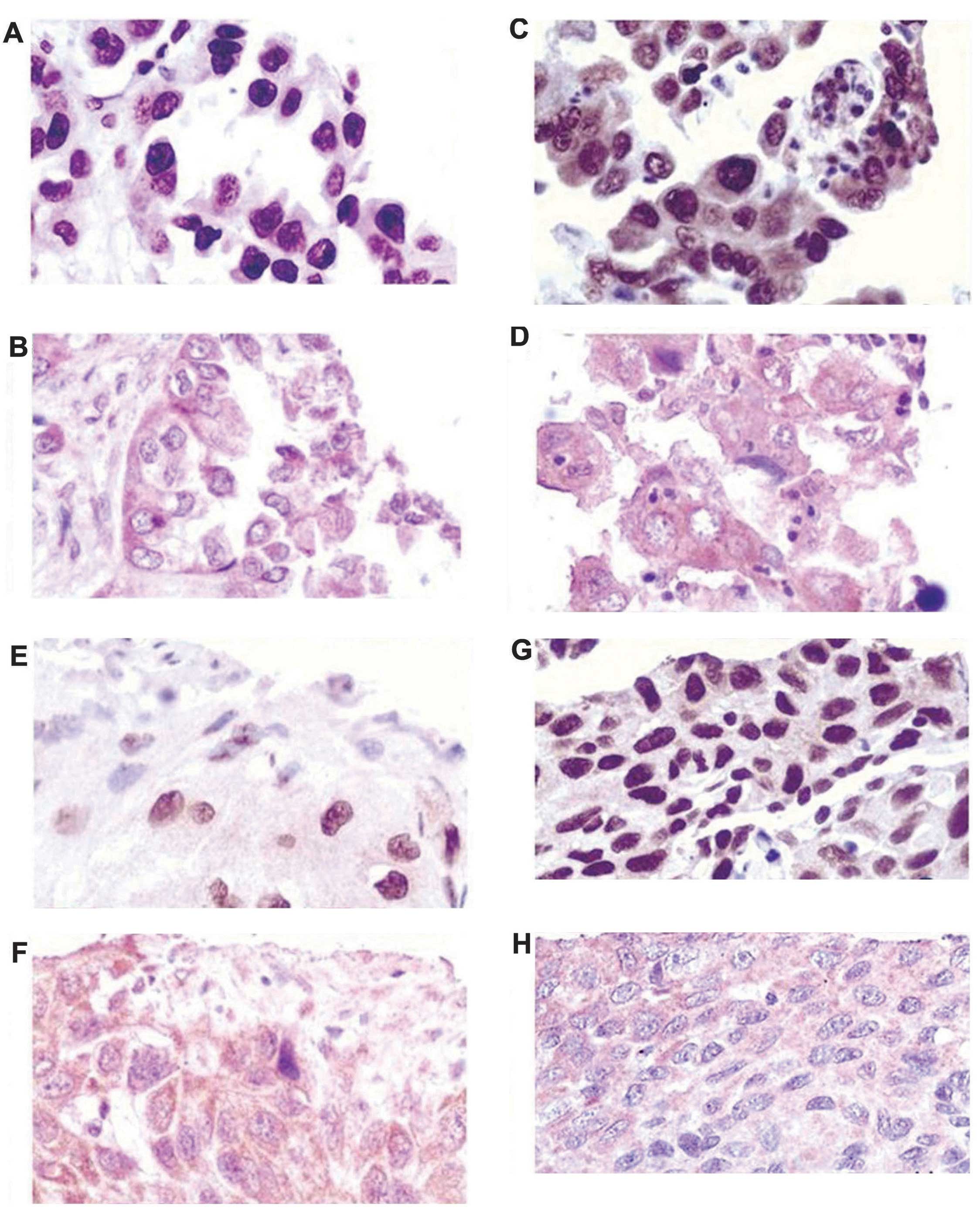
Correlation between the expression of
hnRNP A2/B1 and AXL
Pearson's correlation of the expression of hnRNP
A2/B1 and AXL identified an unexpectedly high correlation, with a
coefficient of 0.459 (P<0.001; data not shown). Thus, there was
a statistically significant correlation between the expression of
hnRNP A2/B1 and AXL in the 134 cases of NSCLC analyzed in the
current study. In addition, Fig. 4
indicates the staining patterns of hnRNP A2/B1 and AXL in different
cases of NSCLC. It was observed that the expression of hnRNP A2/B1
and AXL correlated well in the same tissue, with correlation
increasing as the degree of tissue differentiation reduced.
Discussion
In the present study, the expression of hnRNP A2/B1
was examined in NSCLC and paracancerous tissue samples. The
association between hnRNP A2/B1 expression and the prognosis of
patients with NSCLC was investigated by analyzing the statistical
correlation between hnRNP A2/B1 expression and survival, as well as
numerous clinicopathological factors.
The present study identified that the expression of
hnRNP A2/B1 was predominantly located in the nucleus of cells,
followed by the cytoplasm. This indicates the potential transfer of
hnRNP A2/B1 from the cytoplasm to the nucleus, which was previously
associated with tumor progression (25). The results of tissue microarray assays
demonstrated that the rate of positive hnRNP A2/B1 expression was
significantly higher in lung tumor than in paracancerous lung
tissues. Furthermore, analysis of clinical factors revealed that
positive expression of hnRNP A2/B1 was significantly higher in TNM
stages III–IV compared with stages I–II (P=0.013). In other words,
as NSCLC progresses, hnRNP A2/B1 expression increases. It was also
identified that the expression of hnRNP A2/B1 is highly associated
with the degree of tumor differentiation. Thus, the lower
differentiation, the higher level of positive hnRNP A2/B1
expression. This indicates that the expression of hnRNP A2/B1
increases as the NSCLC becomes more severe.
Based on the aforementioned data, it is proposed
that hnRNP A2/B1 may serve as a prognostic factor in NSCLC. This
proposal is supported by the results of the current survival
analysis. In addition to the log-rank test, Kaplan-Meier survival
curves clarified a negative correlation between positive expression
of hnRNP A2/B1 and survival rates of patients with NSCLC (P=0.000).
Thus, high positive expression of hnRNP A2/B1 indicates an
increased risk of patients with NSCLC exhibiting a poor prognosis.
By using the multivariate Cox regression model to analyze the data,
it was demonstrated that positive expression of hnRNP A2/B1 may be
an independent risk factor for the prognosis of NSCLC
(P=0.001).
He et al (23)
previously reported that differences in the expression of 123
downstream target genes in Colo16 squamous carcinoma cells
following RNAi silencing of hnRNP A2/B1 expression by means of
high-throughput gene chip screening. Among them, AXL expression was
affected, indicating a potential interaction between hnRNP A2/B1
and AXL. Similar to the results reported by Linger et al
(26), the current study revealed
that positive expression of AXL was significantly higher in lung
tumors than in paracancerous lung tissues. Additional investigation
of clinical data indicated that AXL is highly associated with the
prognosis of lung cancer, using univariate (but not multivariate)
Cox regression models. Abnormally high positive expression of AXL
may be involved in the processes of apoptosis and tumor invasion
(27), and, thus, associated with the
degree of tumor differentiation and clinical TNM stage
classification. A pair-wise t-test of protein expression
demonstrated a significant correlation between hnRNP A2/B1 and AXL
expression in the same tissue microarray (P<0.05).
In conclusion, the present study identified a
significant increase in the protein expression of hnRNP A2/B1 and
AXL in NSCLC compared with paracancerous lung tissue samples. Thus,
hnRNP A2/B1 and AXL constitute independent risk factors for the
prognosis of patients with NSCLC. In addition, the results of
immunohistochemical analyses indicated a correlation between the
expressions of hnRNP A2/B1 and AXL. Therefore, the present study
provides the basis for the potential use of hnRNP A2/B1, as well as
AXL, as clinicial prognostic criterion in NSCLC.
References
|
1
|
Siegel R, Naishadham D and Jemal A: Cancer
statistics, 2013. CA Cancer J Clin. 63:11–30. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Yang Y, Li H, Hou S, Hu B, Liu J and Wang
J: The noncoding RNA expression profile and the effect of lncRNA
AK126698 on cisplatin resistance in non-small-cell lung cancer
cell. PLoS One. 8:e653092013. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Grutters JP, Kessels AG, Pijls-Johannesma
M, De Ruysscher D, Joore MA and Lambin P: Comparison of the
effectiveness of radiotherapy with photons, protons and carbon-ions
for non-small cell lung cancer: A meta-analysis. Radiother Oncol.
95:32–40. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Vansteenkiste J, Crinò L, Dooms C,
Douillard JY, Faivre-Finn C, Lim E, Rocco G, Senan S, Van Schil P,
Veronesi G, et al: Role: Panel Members2nd ESMO Consensus Conference
on Lung Cancer: Early-stage non-small-cell lung cancer consensus on
diagnosis, treatment and follow-up. Ann Oncol. 25:1462–1474. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Han SP, Tang YH and Smith R: Functional
diversity of the hnRNPs: Past, present and perspectives. Biochem J.
430:379–392. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Majumder M, Yaman I, Gaccioli F, Zeenko
VV, Wang C, Caprara MG, Venema RC, Komar AA, Snider MD and
Hatzoglou M: The hnRNA-binding proteins hnRNP L and PTB are
required for efficient translation of the Cat-1 arginine/lysine
transporter mRNA during amino acid starvation. Mol Cell Biol.
29:2899–2912. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Hutchison S, LeBel C, Blanchette M and
Chabot B: Distinct sets of adjacent heterogeneous nuclear
ribonucleoprotein (hnRNP) A1/A2 binding sites control 5′ splice
site selection in the hnRNP A1 mRNA precursor. J Biol Chem.
277:29745–29752. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Han K, Yeo G, An P, Burge CB and Grabowski
PJ: A combinatorial code for splicing silencing: UAGG and GGGG
motifs. PLoS Biol. 3:e1582005. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Dreyfuss G, Kim VN and Kataoka N:
Messenger-RNA-binding proteins and the messages they carry. Nat Rev
Mol Cell Biol. 3:195–205. 2002. View
Article : Google Scholar : PubMed/NCBI
|
|
10
|
Chettouh H, Fartoux L, Aoudjehane L,
Wendum D, Clapéron A, Chrétien Y, Rey C, Scatton O, Soubrane O,
Conti F, et al: Mitogenic insulin receptor-A is overexpressed in
human hepatocellular carcinoma due to EGFR-mediated dysregulation
of RNA splicing factors. Cancer Res. 73:3974–3986. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Chen Z-Y, Cai L, Zhu J, Chen M, Chen J, Li
ZH, Liu XD, Wang SG, Bie P, Jiang P, et al: Fyn requires HnRNPA2B1
and Sam68 to synergistically regulate apoptosis in pancreatic
cancer. Carcinogenesis. 32:1419–1426. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Golan-Gerstl R, Cohen M, Shilo A, Suh SS,
Bakàcs A, Coppola L and Karni R: Splicing factor hnRNP A2/B1
regulates tumor suppressor gene splicing and is an oncogenic driver
in glioblastoma. Cancer Res. 71:4464–4472. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Santarosa M, Del Col L, Viel A, Bivi N,
D'Ambrosio C, Scaloni A, Tell G and Maestro R: BRCA1 modulates the
expression of hnRNPA2B1 and KHSRP. Cell Cycle. 9:4666–4673. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
He Y, Brown MA, Rothnagel JA, Saunders NA
and Smith R: Roles of heterogeneous nuclear ribonucleoproteins A
and B in cell proliferation. J Cell Sci. 118:3173–3183. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Tauler J, Zudaire E, Liu H, Shih J and
Mulshine JL: hnRNP A2/B1 modulates epithelial-mesenchymal
transition in lung cancer cell lines. Cancer Res. 70:7137–7147.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Katsimpoula S, Patrinou-Georgoula M,
Makrilia N, Dimakou K, Guialis A, Orfanidou D and Syrigos KN:
Overexpression of hnRNPA2/B1 in bronchoscopic specimens: A
potential early detection marker in lung cancer. Anticancer Res.
29:1373–1382. 2009.PubMed/NCBI
|
|
17
|
Fielding P, Turnbull L, Prime W, Walshaw M
and Field JK: Heterogeneous nuclear ribonucleoprotein A2/B1
up-regulation in bronchial lavage specimens: A clinical marker of
early lung cancer detection. Clinical Cancer Res. 5:4048–4052.
1999.
|
|
18
|
Korshunov VA: Axl-dependent signalling: A
clinical update. Clin Sci (Lond). 122:361–368. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Rothlin CV, Leighton JA and Ghosh S:
Tyro3, Axl, and Mertk receptor signaling in inflammatory bowel
disease and colitis-associated cancer. Inflamm Bowel Dis.
20:1472–1480. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Wu H, Tang H, Chen Y, Wang H and Han D:
High incidence of distal vaginal atresia in mice lacking Tyro3 RTK
subfamily. Molecular reproduction and development. 75:1775–1782.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Wang H, Chen SU, Chen Y, Wang H, Wu H,
Tang H, Xiong W, Ma J, Ge Y, Lu Q and Han D: The role of Tyro 3
subfamily receptors in the regulation of hemostasis and
megakaryocytopoiesis. Haematologica. 92:643–650. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Mudduluru G, Ceppi P, Kumarswamy R,
Scagliotti GV, Papotti M and Allgayer H: Regulation of Axl receptor
tyrosine kinase expression by miR-34a and miR-199a/b in solid
cancer. Oncogene. 30:2888–2899. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
He Y, Rothnagel JA, Epis MR, Leedman PJ
and Smith R: Downstream targets of heterogeneous nuclear
ribonucleoprotein A2 mediate cell proliferation. Mol Carcinog.
48:167–179. 2009. View
Article : Google Scholar : PubMed/NCBI
|
|
24
|
Edge SB and Compton CC: The American Joint
Committee on Cancer: The 7th edition of the AJCC cancer staging
manual and the future of TNM. Ann Surg Oncol. 17:1471–1474. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Man YG, Martinez A, Avis IM, Hong SH,
Cuttitta F, Venzon DJ and Mulshine JL: Phenotypically different
cells with heterogeneous nuclear ribonucleoprotein A2/B1
overexpression show similar genetic alterations. Am J Respir Cell
Mol Biol. 23:636–645. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Linger RM, Cohen RA, Cummings CT, Sather
S, Migdall-Wilson J, Middleton DH, Lu X, Barón AE, Franklin WA,
Merrick DT, et al: Mer or Axl receptor tyrosine kinase inhibition
promotes apoptosis, blocks growth and enhances chemosensitivity of
human non-small cell lung cancer. Oncogene. 32:3420–3431. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Zhang Z, Lee JC, Lin L, Olivas V, Au V,
LaFramboise T, Abdel-Rahman M, Wang X, Levine AD, Rho JK, et al:
Activation of the AXL kinase causes resistance to EGFR-targeted
therapy in lung cancer. Nat Genet. 44:852–860. 2012. View Article : Google Scholar : PubMed/NCBI
|