Introduction
Esophageal cancer is one of the most common
malignant tumors in China. Each year there are ~288,000 new cases,
leading to 208,000 fatalities; squamous cell carcinoma (SCC)
accounts for 90% of these cases (1).
Esophageal cancer is also characterized by an insidious onset. The
majority of patients are already at an advanced stage when they are
first diagnosed, and the 5-year survival rate following surgery is
only 25%. However, at an early disease stage, the 5-year survival
rate following surgery is as high as 80–90% (2). Therefore, early diagnosis and treatment
are crucial. Existing examination methods for diagnosing esophageal
SCC (ESCC), including esophageal cytology, X-ray, computed
tomography examination and endoscopic ultrasound, cannot achieve an
early diagnosis (3). It is abundantly
clear that finding the ideal ESCC marker with a high specificity
and sensitivity is a pressing issue.
microRNAs (miRNAs/miRs) are small non-coding RNAs of
19–24 nucleotides in length that are involved in cancer development
(4). Recent studies have shown that
miRNA expression is stable enough to be detectable in the blood and
to serve as a useful cancer biomarker. For example, it has been
shown that it is possible to improve the potential early diagnosis
and treatment of ovarian cancer lacking clinical signs. It was
demonstrated that the expression levels of miR-21, miR-92, miR-93,
miR-126 and miR-29a in the serum of ovarian cancer patients were
significantly upregulated, whereas those of miR-155, miR-127 and
miR-99b were significantly reduced (5,6). In
patients with diffuse large B-cell lymphoma, the serum miR-21
expression level was significantly increased, and the upregulation
of miR-21 expression was also correlated with a higher
recurrence-free survival rate (7,8). These
results suggest that the detection of the specific expression of
miRNAs not only assists in the early diagnosis of cancer, but also
in the determination of the patient's prognosis. These features
point to the superior application of miRNAs as biological
markers.
Therefore, in the present study, the Agilent
MicroRNA Microarray Profiling System and bioinformatics methods
were used for the analysis and identification of specific miRNAs in
the serum of ESCC patients. Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR) was applied to verify the
microarray results. The goal of the study was to investigate the
feasibility and value of serum microRNAs as biological markers for
the prediction of the biological behavior and prognosis of
ESCC.
Patients and methods
Participants
Peripheral blood samples were collected from 78
esophageal cancer patients at the First Affiliated Hospital of
Xi'an Jiaotong University School of Medicine (Xi'an, Shaanxi,
China) between June 2012 and December 2013. None of the patients
had received any treatments prior to sample collection, including
surgery, radiotherapy or chemotherapy. All cases were
pathologically confirmed as ESCC, and none had a history of any
other type of cancer. The cohort consisted of 57 males and 21
females, with ages ranging from 53–79 years and a mean age of 58.55
years. Blood samples were also collected from 23 age and gender
matched healthy volunteers. All samples were collected after
obtaining informed consent from the participant. Written informed
consent was obtained from all patients prior to the study, and the
study was approved by the Ethics Committee of the First Affiliated
Hospital of Xi'an Jiaotong University.
Collection of serum samples
In each case, 2 ml of collected peripheral blood was
quickly transferred to an ordinary EDTA-free tube, and allowed to
settle at room temperature (22–25°C) for 15–30 min, until
completely coagulated. Next, 0.6–1 ml of supernatant was aspirated
and transferred to a clean 1.5-ml centrifuge tube, and the
following steps were completed within 1 h (at room temperature) or
2 h (at 4°C): Centrifugation at 820 × g for 10 min at 4°C;
collection of supernatant and transferal to a clean 1.5-ml
centrifuge tube; centrifugation at 16,000 × g for 10 min at 4°C;
aspiration of the supernatant and transferal to another clean
centrifuge tube; and storage at −80°C for future use.
Extraction of total RNA from
serum
The serum samples prepared as aforementioned were
collected (400 µl each) and total RNA was extracted from the serum
using the mirVana™ PARIS™ kit (Applied Biosystems Life
Technologies, Foster City, CA, USA) according to the manufacturer's
instructions. RNA (1 µl) was used for quantitative analysis using a
NanoDrop ND-2000 (Thermo Fisher Scientific, Inc., Waltham, MA,
USA), and an Agilent Bioanalyzer 2100 (Agilent Technologies Inc.,
Santa Clara, CA, USA) was used to examine RNA integrity.
Detection of serum miRNA expression
using the Agilent MicroRNA Microarray Profiling System
Total RNA (100 ng) was extracted from the sera of 9
ESCC patients and 9 healthy volunteers, and used for
dephosphorylation, Cy3 labeling and hybridization with a miRNA
microarray using the miRNA Complete Labeling and Hyb kit (Agilent
Technologies Inc.), according to the manufacturer's instructions.
Following hybridization, the microarray was washed with the washing
buffer in the Gene Expression Wash Buffer kit (Agilent Technologies
Inc.. Finally, an Agilent G2505C Scanner (Agilent Technologies
Inc.) was used to scan the microarray, and GeneSpring GX11.0
software (Agilent Technologies Inc.) was used for signal
processing. The Quantile method was applied for data
normalization.
RT-qPCR for validation of microarray
results
Total serum RNA (60 ng) from the remaining 69 ESCC
patients and 14 healthy volunteers was subjected to RT reactions
using the TaqMan miRNA Reverse Transcription kit (Applied
Biosystems Life Technologies). The reaction conditions were as
follows: 30 min at 16°C, 30 min at 42°C and 5 min at 85°C; the
reaction preparation was then maintained at 4°C. Specific
fluorescent TaqMan Probe was used for the RT-qPCR assay to detect
the plasma miRNA content. In accordance with previous studies,
miR-1228 was used as the internal control (9,10). The PCR
conditions were as follows: 96°C for 5 min, followed by 50 cycles
at 95°C for 15 sec, and then 60°C for 1 min. The relative
expression level of each miRNA was expressed using the
2−ΔΔCt method, where ΔCt = CtmiRNA -
Ctmi1228. Each experiment was repeated three times.
Statistical methods
GeneSpring GX 11.0 software was used to analyze the
microarray data. Fold-change analysis and t-tests were
performed for differential gene expression analysis (screening
criteria, fold-change >2; P<0.01). MedCalc software was used
for the Mann-Whitney U and Kruskal-Wallis tests of the quantitative
fluorescence, qPCR results. The receiver operator characteristic
curves (ROC) were used to evaluate the sensitivity and specificity
of the expression levels of different miRNAs in the diagnosis of
ESCC. Multivariate logistic regression analysis was performed to
find the optimal combination of miRNAs. P<0.05 was considered to
indicate a statistically significant difference.
Results
Serum miRNA expression, as detected by
miRNA microarray
A total of 42 ESCC-specific serum miRNAs were
obtained from the miRNA microarray results using the screening
criteria of a fold-change of >2 and P<0.05 for the
t-test. To improve the likelihood of differential gene
expression, the screening criteria were increased to a fold-change
of >8 and P<0.01 for the t-test; a total of 10
ESCC-specific serum miRNAs were then obtained (Table I).
 | Table I.Differentially-expressed miRNAs
(P<0.01) in esophageal squamous cell carcinoma patients with
>8-fold change selected from microarray data analysis. |
Table I.
Differentially-expressed miRNAs
(P<0.01) in esophageal squamous cell carcinoma patients with
>8-fold change selected from microarray data analysis.
| Name | P-value | Fold-change | Regulation |
|---|
| hsa-miR-365 | 0.001805756 |
8.365508 | Up |
| hsa-miR-92b | 0.002613297 | 15.273841 | Up |
| hsa-miR-602 | 0.005954986 | 13.051751 | Up |
| hsa-miR-518b | 0.004742492 |
9.271833 | Down |
| hsa-miR-767-3p | 0.007218791 | 10.837473 | Up |
| hsa-miR-451 | 0.001277682 | 15.483809 | Up |
| hsa-miR-563 | 0.001073018 | 12.695153 | Up |
| hsa-miR-129 | 0.001711119 |
9.499698 | Up |
| hsa-miR-18b* | 0.003863498 |
8.473071 | Up |
| hsa-miR-548 | 0.002055900 |
9.823999 | Up |
Validation of microarray results using
RT-qPCR
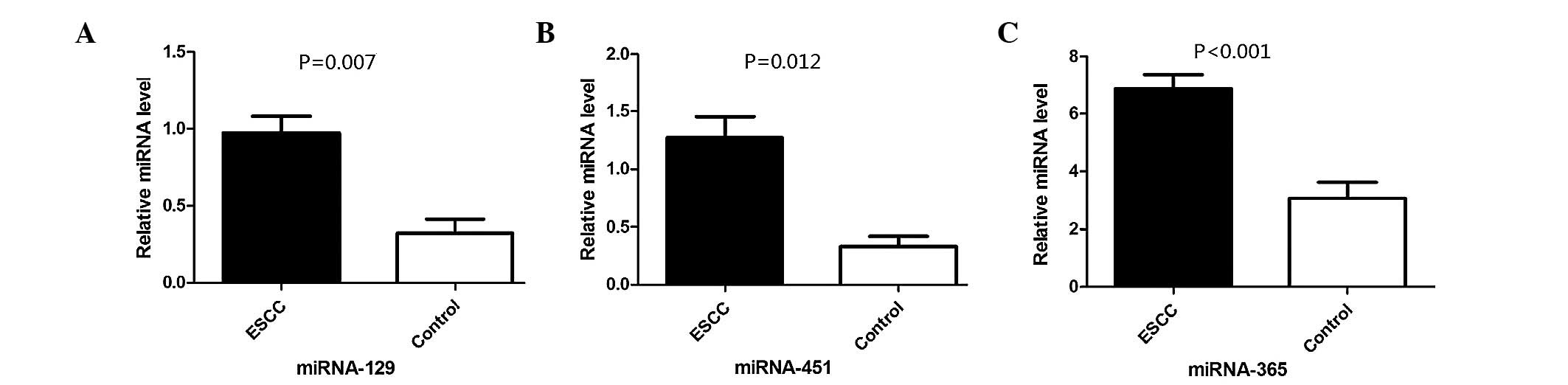
RT-qPCR showed consistent results with regard to the
expression levels of miR-129, miR-451 and miR-365, and the
microarray results; in addition, compared with the healthy
controls, the expression levels of these miRNAs were significantly
different (P=0.007, P=0.012 and P<0.001, respectively) (Fig. 1). For miR-92b, miR-563 and miR-767-3p,
although the directionality of the changes in the expression levels
was consistent with the microarray results, the expression levels
of these miRNAs were not significantly different from that of the
healthy volunteers (P>0.05). The miR-518b and miR-548 expression
levels, as detected with quantitative fluorescence qPCR, were
inconsistent with those obtained from the microarray analysis. Due
to poor specificity of the melting curve, data on the expression
levels of miR-602 and miR-18b* was not obtained.
Value of miR-129, miR-451 and miR-365
in ESCC diagnosis
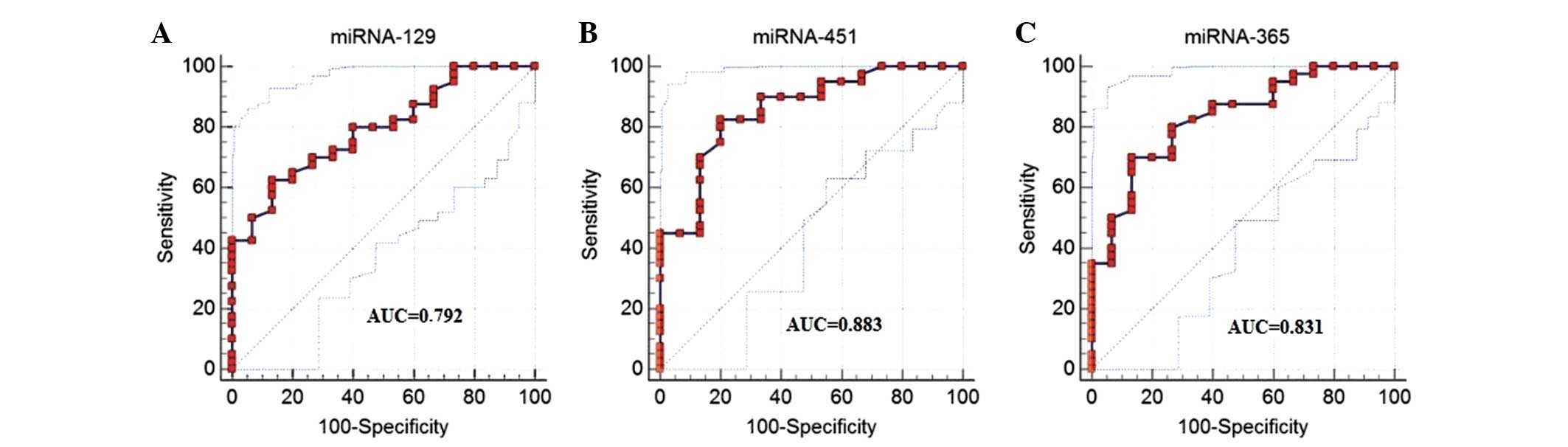
The ROC analysis results showed that by using the
relative expression of miR-129 alone (cutoff, 0.386), the
sensitivity was 78.8%, the specificity was 73.3% and the area under
the curve (AUC) was 0.792. Using the relative expression of miR-451
alone (cutoff, 0.568), the sensitivity was 82.5%, the specificity
was 79.0% and the AUC was 0.882. Using the relative expression of
miR-365 alone (cutoff value, 5.06), the sensitivity for ESCC
diagnosis was 80.56% and the specificity was 86.7%, with an AUC of
0.831 (Fig. 2). Multivariate logistic
regression analysis showed that miR-365 could serve as a potential
diagnostic marker for ESCC (P=0.002), but that adding miR-451 or
miR-129 could not further improve the diagnostic efficiency.
Correlation between serum miRNA
expression levels and TNM stage
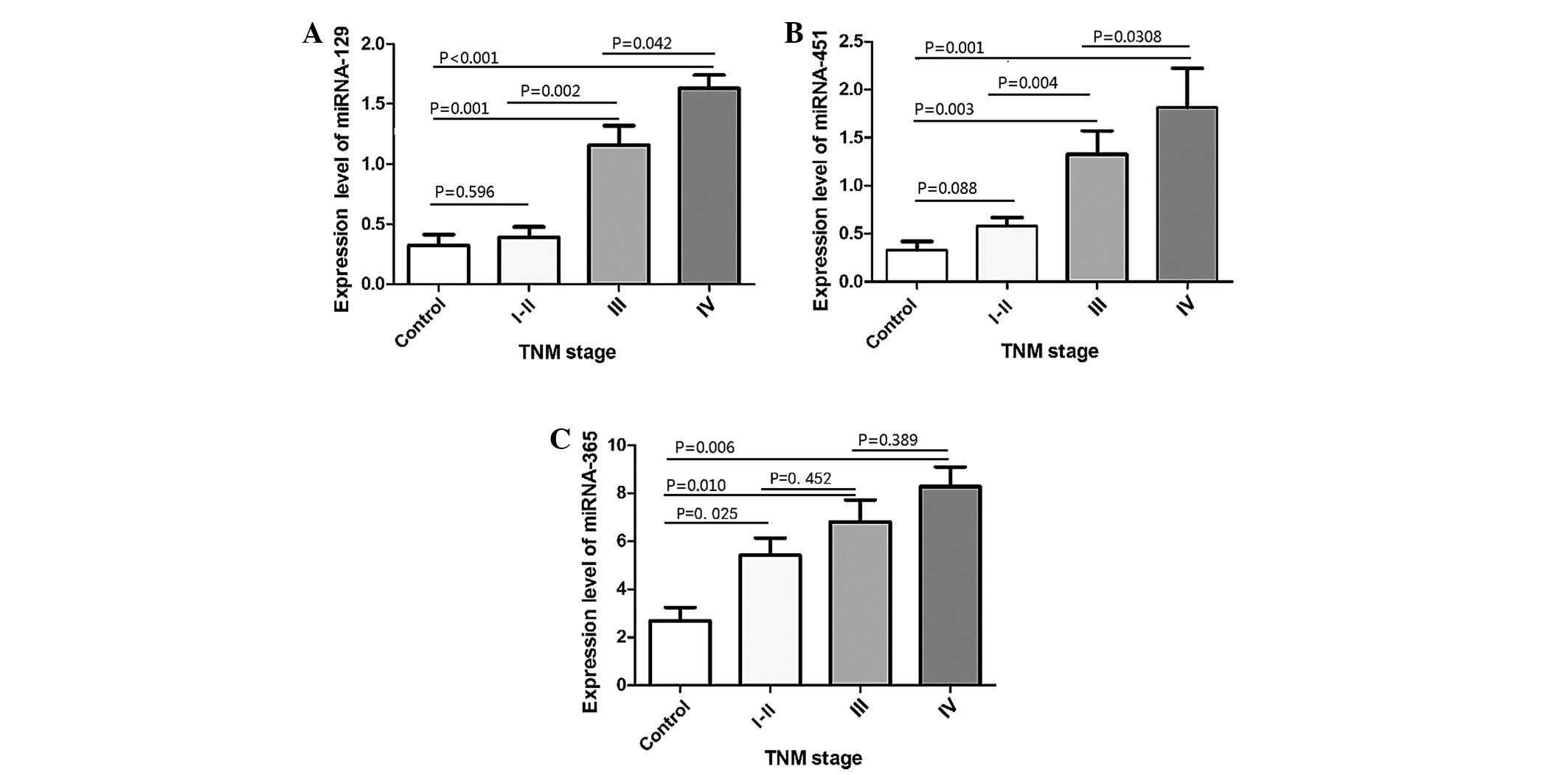
It was found that even in the early stages of ESCC,
miR-365 expression was already significantly elevated compared with
the healthy controls (stage I–II, P=0.025; stage III, P=0.001;
stage IV, P=0.001; Mann-Whitney U test); however, its expression
did not differ significantly among different TNM stages (stage II
vs. III, P=0.452; stage III vs. IV, P=0.389). The miR-451 and
miR-129 expression levels did not increase significantly compared
with the normal controls in early-stage ESCC, but did increase
significantly at stages III and IV. In particular, the expression
level of miRNA-129 differed significantly among different stages
(stage II vs. III, P=0.002; stage III vs. IV, P=0.042). However,
the expression level of miR-451 did not differ significantly
between stages III and IV (stage II vs. III, P=0.004; stage III vs.
IV, P=0.308) (Fig. 3).
Discussion
The present study examined and analyzed the serum
miRNA expression profile of ESCC patients using miRNA microarray
technology and subsequent validation with quantitative
fluorescence-based, qPCR. It was found that the expression of 3
miRNAs, miR-129, miR-451 and miR-365, was significantly elevated
compared with the healthy controls. Further multivariate regression
analysis revealed that miR-365 can serve as a biomarker for the
early diagnosis of ESCC, with a sensitivity of 80.56%, a
specificity of 86.7% and an AUC of 0.831. The miR-365 expression
level was significantly increased in the serum of TNM stage I and
II ESCC patients, but such specific upregulation was not correlated
with the TNM stage; this suggested that miR-365 may not be a
suitable indicator of ESCC progression. The upregulation of miR-451
and miR-129 in the early stages of ESCC was not significant, but
became statistically significant at stages III and IV. In
particular, the expression level of miRNA-129 differed
significantly between the different stages, suggesting that it may
serve as an independent monitoring indicator for the prognosis of
ESCC.
To date, there have no studies on the expression of
the 3 miRNAs, miR-129, miR-451 and miR-365, in the serum of cancer
patients. A recent study showed that circulating miRNAs were
selectively secreted via a cellular selection mechanism and that
this selection mechanism was associated with the degree of
malignancy (11). It was also
demonstrated that living cells secreted miRNAs capable of
inhibiting each other (12). With
regard to miR-365, overexpression was detected in these cells and
the clinical specimens of cutaneous squamous cell carcinoma. In
vivo experiments have shown that the HaCaTpre-miR-365-2 cell
line, which overexpresses miR-365, can induce the formation of
subcutaneous tumors, and that the use of the anti-miR-365
oligonucleotide Antagomir-365 can inhibit the formation of skin
tumors in vivo, inducing tumor cell cycle arrest at the G
phase and apoptosis (13). These
results suggest that miR-365 acts as an oncomiR in cutaneous
squamous cell carcinoma in vitro and in vivo. In the
present study, an elevation in the expression of miR-365 in the
sera of stage I and II ESCC patients was found, which was possibly
due to the fact that miR-365 is associated with the early formation
of esophageal cancer. Ogawa et al (14) utilized RT-PCR to identify 7 miRNAs in
ESCC cells, and found miRNA-129 overexpression in these cells.
Analysis of the post-operative survival data subsequently showed
that high levels of miRNA-129 were negatively correlated with the
post-operative survival time of the patients, and multi-factor
regression analysis also suggested that miRNA-129 was an
independent prognostic factor. In the present study, a significant
elevation in the expression of miRNA-129 in the serum from stage
III and IV ESCC patients was observed, and it was deduced that a
high expression level of miRNA-129 may be associated with the tumor
burden, and that miRNA-129 could potentially serve as an
independent factor for the prognostic monitoring of esophageal
cancer. Recent studies have found that miR-451 is not only
expressed abnormally in tumor tissues, but that it is also involved
in the biological activity of certain tumors and the genetic
regulation of their drug resistance (15–18). The
current literature illustrates the downregulation of miR-451 in
gastric, colon, esophageal and non-small-cell lung cancer, and its
upregulation in head and neck squamous cell carcinoma and
multidrug-resistant ovarian cancer (19–21). Wang
et al found that the upregulation of miR-451 expression in
EC9706 esophageal cancer cells effectively inhibited proliferation
and invasiveness, and promoted apoptosis, possibly through the
phosphoinositide 3-kinase/Akt signaling pathway (22). Therefore, we hypothesize that the body
may produce miR-451 to inhibit the proliferation of advanced
esophageal cancer cells, leading to an increase in miR-451 levels
in the circulating blood.
It has been previously reported that miRNA-21 can
serve as a marker for the diagnostic and prognostic monitoring of
esophageal cancer (23–25). However, miRNA-21 expression was also
shown to be elevated in other types of tumors and diseases, such as
colorectal, lung and gastric cancer (26–28), and
thus cannot in fact serve as a specific marker for esophageal
cancer. In addition, there have been several studies on the
esophageal cancer diagnosis using a combination of multiple miRNAs.
For example, Zhang et al (29)
applied Solexa sequencing to compare the serum miRNA expression
profile of ESCC patients against that of healthy controls, and
found that 25 miRNAs exhibited significantly elevated expression.
Furthermore, RT-qPCR revealed significant increases in 7 miRNAs
(miR-l0a, 22, 100, 148b, 223, 133a and 127–3p), in the serum of the
ESCC patients. Cluster analysis illustrated that the combination of
these 7 miRNAs could distinguish 89.6% of early ESCC patients from
normal controls, showing miRNAs to be of relatively significant
value in the early diagnosis of ESCC (29). The present study failed to replicate
the results of previous studies; that this may be due to
differences in experimental design, material selection and
detection techniques between the studies. In the current study, the
relevant miRNAs were screened and identified from an miRNA
microarray containing 2,026 miRNAs, and these were then validated
using RT-qPCR. Hence, the present results should be quite reliable.
Thus, these miRNAs exhibit a relatively high importance in terms of
their research value.
Certain limitations must be considered when
interpreting the results of the present study. First, the sample
size was small. Second, the abundance of the miRNAs in the serum
was too low to be accurately quantified by RT-qPCR, therefore,
certain potential relevant markers could not be assessed. Third,
there have been a few studies on the expression of miR-365, miR-451
and miR-129 in the serum or on the mechanisms subserving their
functions, and this remains unclear. Therefore, in the next stage
of our studies, the sample size will be expanded and complete
follow-up examinations of the patients will be performed so as to
understand the association between miRNA expression and patient
survival. In addition, future studies will characterize the
expression of these miRNAs in ESCC tissues and cells, and
investigate the mechanisms underlying their functions in ESCC. Such
studies should aid in the determination of the reason for their
elevation in the serum.
In conclusion, the present study found that the
expression levels of miR-129, miR-451 and miR-365 were
significantly increased in the serum of ESCC patients compared with
healthy subjects. Specifically, we propose that miR-365 be used as
a potential marker for early ESCC diagnosis, while miRNA-129, which
showed expression levels associated with tumor stage, may serve as
an ESCC prognostic indicator. For the purposes of the early
detection and diagnosis of ESCC, and to improve patient survival,
larger studies must be performed to screen and verify combinations
of ESCC-specific serum biomarkers; these markers could then be used
as a sensitive, specific, non-invasive and simple method for tumor
detection in the early diagnosis and prognostic monitoring of
ESCC.
Acknowledgements
This study was financially supported by the National
Natural Science Foundation of China (grant no. 30972962).
References
|
1
|
Chen W, Zheng R, Zhang S, Zhao P, Zeng H,
Zou X and He J: Annual report on status of cancer in China, 2010.
Chin J Cancer Res. 26:48–58. 2014.PubMed/NCBI
|
|
2
|
Wang GQ, Jiao GG, Chang FB, et al:
Long-term results of operation for 420 patients with early squamous
cell esophageal carcinoma discovered by screening. Ann Throac surg.
77:1740–1744. 2004. View Article : Google Scholar
|
|
3
|
Campbell F, Lauwers GY and Williams GT:
Tumors of the esophagus and stomach. Diagnostic Histopathology of
Tumors. Fletcher CDM: (3rd). Churchill Livingstone Elsevier.
(London). 328–329. 2007.
|
|
4
|
Bartel DP: MicroRNAs: Genomics,
biogenesis, mechanism and function. Cell. 116:281–297. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Resinick KE, Alder H, Hagan JP, et al: The
detection of differentially expressed microRNA from the serum of
ovarian cancer patients using a novel real-time PCR platform.
Gynecol Oncol. 112:55–59. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Taylor DD and Gercel-Taylor C: MicroRNA
signatures of tumor-derived exosomes as diagnostic biomarkers of
ovarian cancer. Gynecol Oncol. 110:13–21. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Lawrie CH, Gal S, Dunlop HM, et al:
Detection of elevated levels of tumor-associated microRNAs in serum
of patients with diffuse large B-cell lymphoma. Br J Haematol.
141:672–675. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Chin LJ and Slack FJ: A truth serum for
cancer-microRNAs have major potential as cancer biomarkers. Cell
Res. 18:983–984. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Zhou J, Yu L, Gao X, et al: A Plasma
MicroRNA panel to diagnose hepatitis B virus related hepatocellular
carcinoma. J Clin Oncol. 29:4781–4788. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Hu J, Wang Z, Liao BY, et al: Human
miR-1228 as a stable endogenous control for the quantification of
circulating microRNAs in cancer patients. Int J Cancer.
135:1187–1194. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Pigati L, Yaddanapudi SC, Iyengar R, Kim
DJ, Hearn SA, Danforth D, Hastings ML and Duelli DM: Selective
release of microRNA species from normal and malignant mammary
epithelial cells. PLoS One. 5:e135152010. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Iguchi H, Kosaka N and Ochiya T: Secretory
microRNAs as a versatile communication tool. Commun Integr Biol.
3:478–481. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Zhou M, Liu W, Ma S, et al: A novel
onco-miR-365 induces cutaneous squamous cell carcinoma.
Carcinogenesis. 34:1653–1659. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Ogawa R, Ishiguro H, Kuwabara Y, et al:
Expression profilinf of micro-RNAs in human esophageal squamous
cell carcinoma using RT-PCR. Med Mol Morphol. 42:102–109. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Pan X, Wang R and Wang ZX: The potential
role of miR-451 in cancer diagnosis, prognosis, and therapy. Mol
Cancer Ther. 12:1153–1162. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Kovalchuk O, Filkowski J, Meservy J, et
al: Involvement of microRNA-451 in resistance of the MCF-7 breast
cancer cells to chemotherapeutic drug doxorubicin. Mol Cancer Ther.
7:2152–2159. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Zhu H, Wu H, Liu X, et al: Role of
MicroRNA miR-27a and miR-451 in the regulation of
MDR1/P-glycoprotein expression in human cancer cells. Biochem
Pharmacol. 76:582–588. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Wang XC, Tian LL, Jiang XY, et al: The
expression and function of miRNA-451 in non-small cell lung cancer.
Cancer Lett. 311:203–209. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Banders E, Bitarte N, Arias F, et al:
MicroRNA-451 regulates macrophage migration inhibitory factor
production and proliferation of gastrointestinal cancer cells. Clin
Cancer Res. 15:2281–2290. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Wang R, Wang ZX, Yang JS, Pan X, De W and
Chen LB: MicroRNA-451 functions as a tumor suppressor in human
non-small cell lung cancer by targeting ras-related protein 14
(RAB14). Oncogene. 30:2644–2658. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Hui AB, Lenarduzzi M, Krushel T, et al:
Comprehensive MicroRNA profiling for head and neck squamous cell
carcinomas. Clin Cancer Res. 16:1129–1139. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Wang T, Zang WQ, Li M, Wang N, Zheng YL
and Zhao GQ: Effect of miR-451 on the biological behavior of
esophageal carcinoma cell line EC9706. Dig Dis Sci. 58:706–714.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Tanaka Y, Kamohara H, Kinoshita K, et al:
Clinical impact of serum exosomal microRNA-21 as a clinical
biomarker in human esophageal squamous cell carcinoma. Cancer.
119:1159–1167. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Cai EH, Gao YX, Wei ZZ, Chen WY, Yu P and
Li K: Serum miR-21 expression in human esophageal squamous cell
carcinomas. Asian Pac J Cancer Prev. 13:1563–1567. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Kurashige J, Kamohara H, Watanabe M, et
al: Serum microRNA-21 is a novel biomarker in patients with
esophageal squamous cell carcinoma. J Surg Oncol. 106:188–192.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Basati G, Razavi A Emami, Abdi S and
Mirzaei A: Elevated level of microRNA-21 in the serum of patients
with colorectal cancer. Med Oncol. 31:2052014. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Yang X, Guo Y, Du Y, et al: Serum
microRNA-21 as a diagnostic marker for lung carcinoma: A systematic
review and meta-analysis. PLoS One. 9:e974602014. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Song J, Bai Z, Zhang J, et al: Serum
microRNA-21 levels are related to tumor size in gastric cancer
patients but cannot predict prognosis. Oncol Lett. 6:1733–1737.
2013.PubMed/NCBI
|
|
29
|
Zhang C, Wang C, Chen X, et al: Expression
profile of microRNAs in serum: A fingerprint for esophageal
squamous cell carcinoma. Clin Chem. 56:1871–1879. 2010. View Article : Google Scholar : PubMed/NCBI
|