Introduction
Lung cancer is the leading cause of
cancer-associated mortalities worldwide (1). Non-small cell lung cancer (NSCLC), which
includes adenocarcinoma, squamous cell carcinoma and large cell
carcinoma, comprises 85% of lung cancer (2,3). Despite
improvements to the treatment of lung cancer, the overall 5-year
survival rate has remained poor at <15% (3,4).
Therefore, the search for effective biomarkers for early diagnosis,
prognosis and individualized medication treatment plans for NSCLC
has become a significant focus of recent research.
Sushi domain containing 2 (SUSD2) is located on
chromosome 22 and encodes an 822-amino acid type I membrane
protein, including somatomedin B, AMOP, von Willebrand factor type
D and sushi domains, which have significant roles in the mediation
of cell-cell and cell-matrix adhesion in molecules (5). Two studies by Sugahara et al
(6,7)
investigated the mouse homolog: SUSD2 (also known as mSVS-1 or
SVS-1) was found to be downregulated in activated
oncogene-v-K-ras-transformed NIH3T3 cells (Ki3T3 cells), compared
with mouse NIH3T3 cells. One of the studies (6) revealed that overexpression of SUSD2 in
HT1080 fibrosarcoma cells and HeLa cervical carcinoma cells
inhibited clonogenicity, anchorage-independent growth, migration
and invasion, via Matrigel assays. Simultaneously, Sugahara et
al (7) also suggested that SUSD2
induced apoptosis by inactivation of survival signaling component
Akt, and activation of the caspase cascade (mitochondrial pathway)
in HeLa cells. The results indicated a potential tumor suppressive
function of SUSD2. Conversely, Watson et al (5) reported that SUSD2 was expressed at high
levels in human breast cancer. The study suggested that SUSD2 may
enhance the invasive ability of breast cancer cells and potentially
contribute to a immune evasion mechanism by inducing apoptosis of
Jurkat T cells. However, to the best of our knowledge, SUSD2
expression status and its correlation with the clinicopathological
features of NSCLC have not previously been investigated.
In the present study, SUSD2 messenger RNA (mRNA) and
protein expression were evaluated in NSCLC and normal lung tissues
by SYBR Green quantitative polymerase chain reaction (qPCR) and
western blot analysis of samples obtained from 24 patients with
NSCLC. Immunohistochemical (IHC) analysis for SUSD2 was performed
on an NSCLC tissue microarray (TMA) obtained from 192 patients with
NSCLC. In addition, the potential associations between SUSD2
protein expression and the clinicopathological characteristics of
NSCLC were analyzed.
Materials and methods
Patients and tissue samples
In the present study, 24 paired NSCLC and adjacent
normal lung tissue samples were collected from patients between
June 2012 and May 2014 for reverse transcription-qPCR (RT-qPCR) and
western blot analysis. All samples were collected by resection from
patients with lung cancer at the Department of Thoracic Surgery,
Nanfang Hospital of Southern Medical University (Guangzhou, China).
The samples selected were from patients with pathologically
diagnosed cases of NSCLC, having received no chemotherapy or
radiotherapy prior to surgery. NSCLC tissue microarrays, including
160 formalin-fixed, paraffin-embedded NSCLC tissues and 32 normal
lung tissues, were purchased from Alena Biotech (Xi'an, China). The
ages of the 192 patients with NSCLC ranged from 14–81 years.
Clinicopathological features of the patients, including gender, age
at diagnosis, histological grade, clinical stage, pathology type
and pTNM status, are listed in Table
I. All cases were histologically classified as NSCLC and the
cancer stages were classified according to the American Joint
Committee on Cancer criteria (8). The
present study was approved by the Ethics Committee of the Nanfang
Hospital and conducted with written informed consent from all
patients included.
 | Table I.AUC for each clinicopathological
feature. |
Table I.
AUC for each clinicopathological
feature.
| Characteristic | AUC (95% CI) | P-value |
|---|
| Gender | 0.540
(0.438–0.643) | 0.441 |
| Age | 0.539
(0.444–0.633) | 0.405 |
| Histological
grade | 0.799
(0.715–0.883) | 0.000 |
| Clinical stage | 0.548
(0.435–0.661) | 0.427 |
| Pathology type | 0.593
(0.505–0.681) | 0.043 |
| pT status | 0.533
(0.421–0.645) | 0.581 |
| pN status | 0.601
(0.499–0.703) | 0.042 |
| pM status | 0.740
(0.572–0.908) | 0.245 |
RNA isolation and RT-qPCR
analysis
Total RNAs were isolated from 24 pairs of fresh
NSCLC and adjacent normal lung tissues using RNAiso Plus reagent
(Takara Biotechnology, Co., Ltd, Dalian, China). RNA concentration
and optical density 260/280 were assessed by using a BioPhotometer
Plus spectrophotometer (Eppendorf, Hamburg, Germany). First-strand
complementary DNA (cDNA) was prepared from total RNA using the
PrimeScript® RT reagent kit (Takara Biotechnology, Co., Ltd,
Dalian, China). The RT reaction was performed under the following
conditions: 37°C for 15 min and 85°C for 5 sec, followed by 42°C
for 2 min. The 20 µl PCR reaction mixture included 50 ng cDNA
template, 0.4 µM each of the forward and reverse primers, 10 µl 2X
SYBR Premix Ex Taq™ II, 0.4 µl ROX Reference Dye II (Takara
Biotechnology, Co., Ltd) and double distilled H2O. The
primers for the SUSD2 gene were: Forward,
5′-CTCCAATGACTGCCGCAACTA-3 and reverse,
5′-GAACATTCCTTTCAGGTCCATCC-3′. The primers for the β-actin gene
were: Forward, 5′-AGCCTCGCCTTTGCCGA-3, and reverse,
5′-CTGGTGCCTGGGGCG-3′ (9). The qPCR
reaction was performed in an ABI 7500 real-time PCR amplifier
(Applied Biosystems Life Technologies, Foster City, CA, USA) with
an initial denaturing temperature of 95°C for 30 sec, followed by
40 cycles of 95°C for 5 sec and 60°C for 30 sec. The Ct values were
acquired using the 7500 system SDS software, version 2.0.1 (Applied
Biosystems Life Technologies) with manual thresholds. β-actin was
used as an internal control gene to normalize PCR to the quantity
of RNA added to the RT reactions (10). Fold-changes between NSCLC and normal
tissue pairs were analyzed by calculating the 2−ΔΔCt
values (11). Each PCR reaction was
repeated in triplicate for stable results.
Western blot analysis
Total proteins from 24 pairs of fresh NSCLC and
adjacent normal tissues were extracted using
radioimmunoprecipitation assay buffer containing 1 mM
phenylmethylsulfonyl fluoride (Beyotime Institute of Biotechnology,
Haimen, China). Western blotting was performed according to the
method described by Lee et al (11). Proteins were separated on NuPAGE 4–12%
bis-Tris polyacrylamide gels (Invitrogen Life Technologies,
Carlsbad, CA, USA) and then electrophoretically transferred to
polyvinylidene fluoride membranes (EMD Millipore, Billerica, MA,
USA) by a Transblot SD Cell semi-dry transfer machine (Bio-Rad
Laboratories, Inc., Hercules, CA, USA). The membranes were then
incubated for 1 h at room temperature, with a 1:1,000 dilution of
rabbit polyclonal anti-SUSD2 antibody (Sigma-Aldrich, St. Louis,
MO, USA). Subsequently, horseradish peroxidase (HRP)-conjugated
goat anti-rabbit immunoglobulin G (Multisciences, Hangzhou, China)
was applied at a dilution of 1:2,000 for 2 h at room temperature.
Anti-β-actin antibody was used as a loading control and the signals
were visualized using an enhanced chemiluminescence detection kit
(GE Healthcare Life Sciences, Little Chalfont, UK).
IHC
NSCLC TMAs (LC1921; Alenabio, Xi'an, China) were
constructed with 160 formalin-fixed, paraffin-embedded NSCLC
tissues and 32 normal lung tissues. The TMA slides were
deparaffinized in xylene and rehydrated in graded ethanol. The
sections were subsequently immersed in 3% hydrogen peroxide
solution for 10 min to block endogenous peroxidase activity.
Antigen retrieval was performed by microwave heating with sodium
citrate buffer (pH 6.0) at 100°C for 20 min. The TMA slides were
blocked with 5% normal goat serum at room temperature for 1 h,
followed by incubation with polyclonal rabbit anti-SUSD2 antibody
(1:200; Sigma-Aldrich) at 4°C overnight, and polymer
peroxidase-labeled anti-rabbit secondary antibody (ZSGB-Bio,
Beijing, China) at room temperature for 45 min (12). The TMA slides were stained using the
diaminobenzidene HRP Color Development kit (Beyotime Institute of
Biotechnology), and then counterstained with hematoxylin (Chemical
reagent factory, Guangzhou, China). A negative control was
performed without the primary antibodies, and a known IHC-positive
slide (Alenabio) was used as a positive control (13,14).
IHC evaluation
The intensity of SUSD2 protein staining of the 192
lung samples was scored using a semiquantitative scale as
previously described (15). Briefly,
with regard to distribution of SUSD2 protein, the scores were
expressed as a percentage of positive tumor cells over the total
tumor cells, in 5% increments (0–100%). Two investigators graded
the immunohistochemical expression of SUSD2 and the scores were
accepted if the scores determined by the two investigators agreed.
Otherwise, values were re-estimated until a consensus was
reached.
Selection of cut-off score and
statistical analysis
Receiver operating characteristic (ROC) curve
analysis was used to determine an optimal cut-off score for the
positive expression of SUSD2 using the 0,1-criterion (15). The ROC curves were generated by
plotting the sensitivity and specificity for each outcome at
various SUSD2 scores. The corresponding area under the ROC curve
(AUC) and P-value were analyzed using SPSS 18.0 software (SPSS,
Inc., Chicago, IL, USA). The score closest to the point (0.0, 1.0)
on the curve, with maximum sensitivity and specificity, was
selected as the cut-off value (15).
Dichotomization of the clinicopathological features for ROC
analysis were as follow: Gender (male vs. female), age (≤55 vs.
>55 years), histology grade (I–II vs. III), clinical stage (I–II
vs. III–IV), pathology type (adenocarcinoma vs. squamous cell
carcinoma), pT stage (T1-T2 vs. T3-T4), pN stage (N0 vs. N1-N2) and
pM stage (M0 vs. M1). Statistical analyses were performed using
SPSS 18.0 software. The association of SUSD2 expression with the
clinicopathological features of NSCLC patients was analyzed by the
χ2 test. ∆Ct and grey value of lung cancer tissue
samples and nomal tissues was tested by paired-sample Student's
t-test. P<0.05 was considered to indicate a statistically
significant difference (two-tailed test) (16).
Results
SUSD2 mRNA and protein expression
levels are downregulated in NSCLC tissues
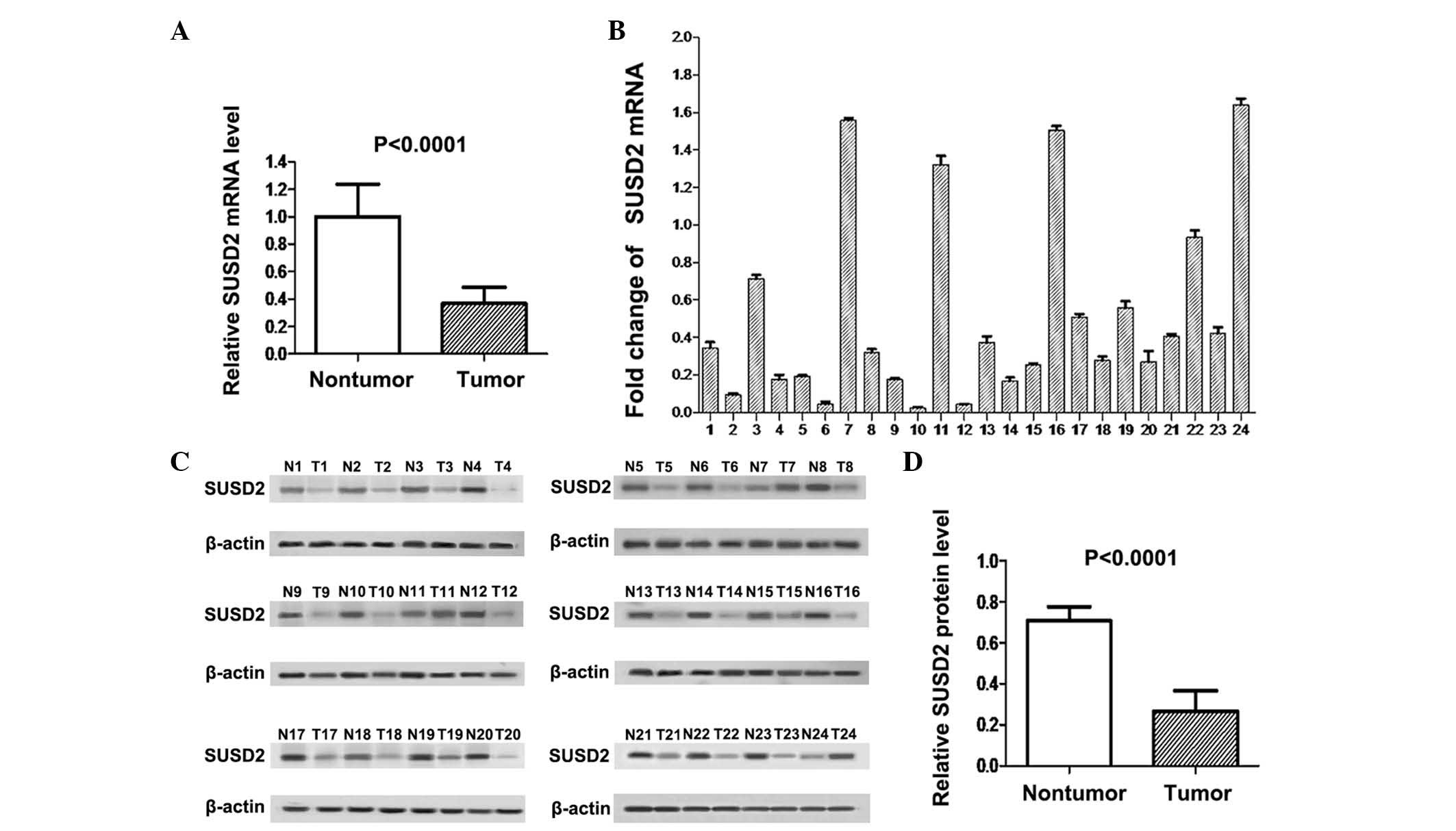
As shown in Fig. 1A,
SUSD2 mRNA expression levels in cancer tissues were significantly
lower than those in adjacent normal tissues (P<0.0001). RT-qPCR
analysis indicated that SUSD2 mRNA expression was downregulated in
NSCLC tissues compared with adjacent normal lung tissues in 20/24
sample pairs (Fig. 1B). Western blot
analysis demonstrated that expression of the SUSD2 protein was also
downregulated in 21/24 NSCLC tissues compared with their adjacent
normal counterparts (Fig. 1C). The
difference in SUSD2 protein expression between the two groups was
statistically significant, as indicated by paired t-test
(P<0.0001; Fig. 1D).
Positive rate of SUSD2 expression is
downregulated in NSCLC tissues
The subcellular localization and expression of SUSD2
protein were observed and scored by IHC analysis of the TMA, which
included 160 NSCLC and 32 normal lung tissues. The variation in
SUSD2 expression levels observed in the NSCLC samples varied
between 0 and 100%. As indicated in Fig.
2, positive SUSD2 immunohistochemical staining was
predominantly observed on the cell membrane, while lower levels of
staining were also found in the cytoplasm.
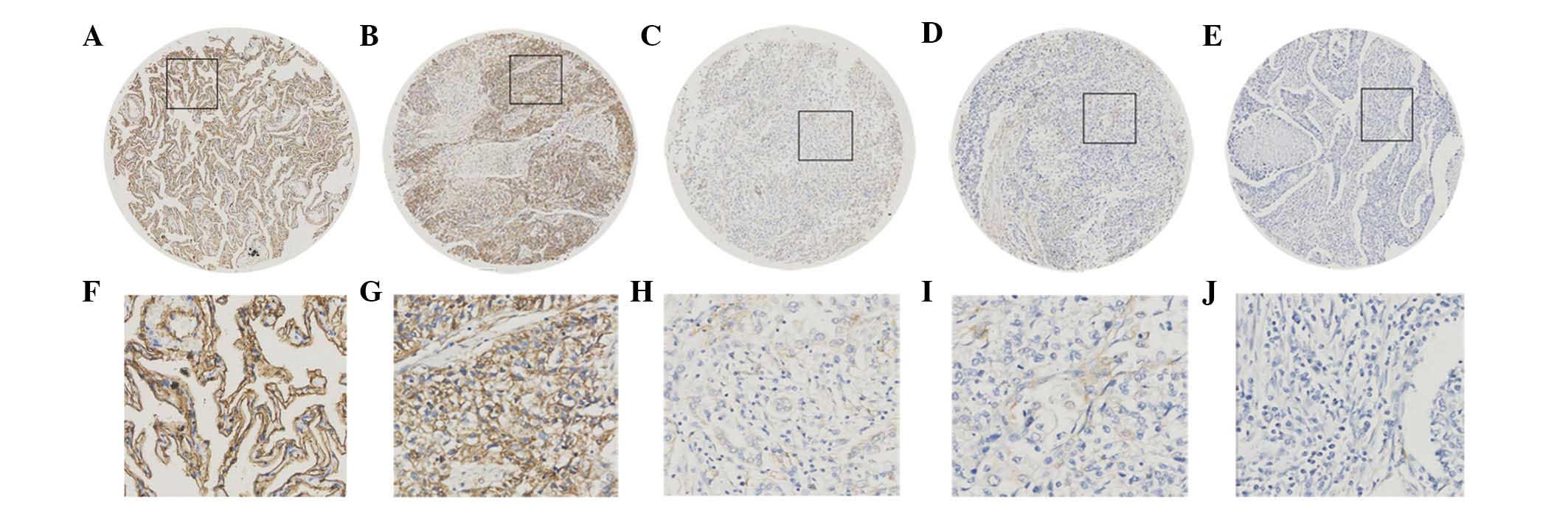 | Figure 2.Expression of SUSD2 protein in normal
lung and NSCLC tissues by immunohistochemical analysis of the
tissue microarray. (A) Positive expression of SUSD2 in normal lung
tissue, in which ~90% of the lung cells demonstrated marked
membrane staining of SUSD2, while lesser staining was also observed
in the cytoplasm (magnification, ×100). (B) Positive expression of
SUSD2 in NSCLC (case 13), in which >90% of tumor cells exhibited
positive staining (magnification, ×100). (C) Negative expression of
SUSD2 in NSCLC (case 168), with <35% positive tumor cells
(magnification, ×100). (D) Negative expression of SUSD2 in NSCLC
(case 74), with <20% positive tumor cells (magnification, ×100).
(E) Negative expression of SUSD2 in NSCLC (case 161), with <5%
positive tumor cells (magnification, ×100). (F-J) Higher
magnification (×400) of the black squares outlined in A-E,
respectively. SUSD2, sushi domain containing 2; NSCLC, non-small
cell lung cancer. |
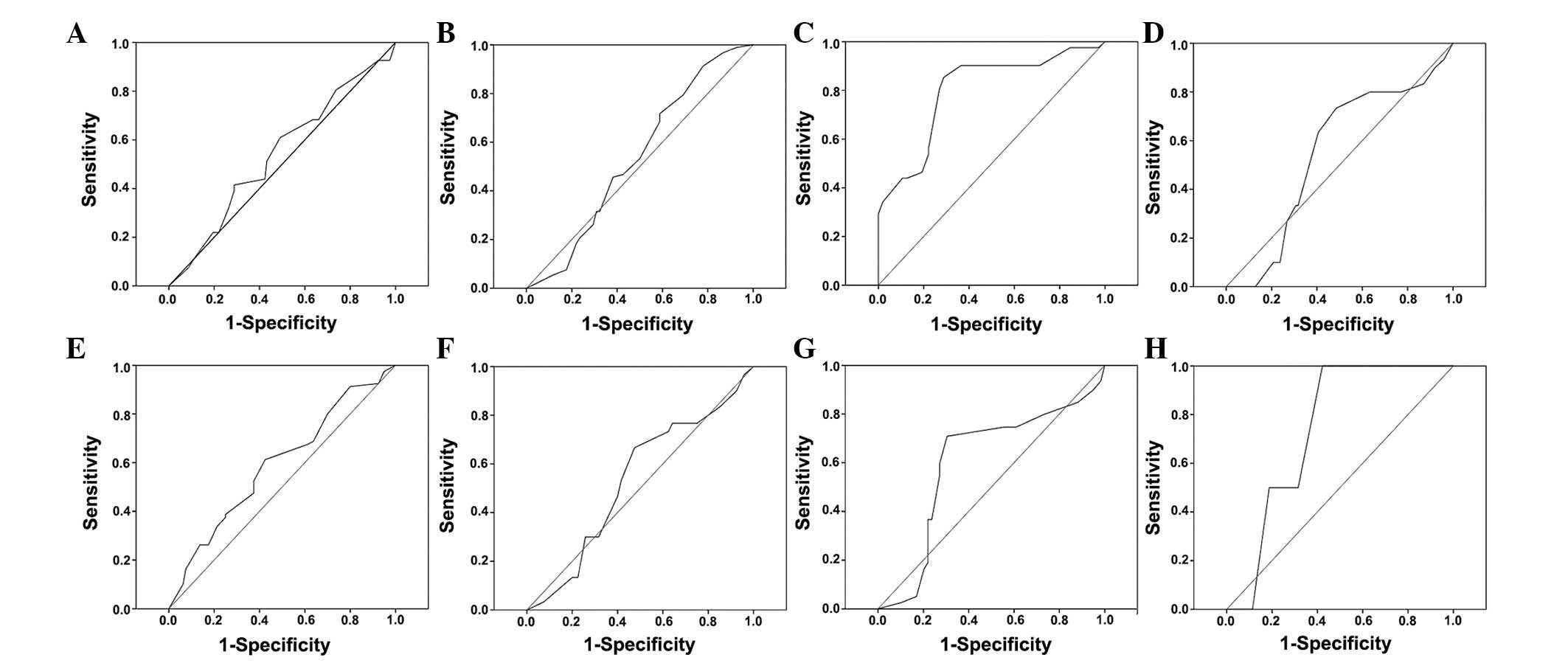
ROC curves for the clinicopathological
characteristics at various SUSD2 scores were plotted (Fig. 3). The optimal cut-off score for SUSD2
was determined by the ROC curve for histological grade, which was
closest to the point (0.0, 1.0). According to the cut-off score,
tissues were defined as SUSD2 positive when the SUSD2 expression
percentage was >47.5%. Based on the SUSD2 scores for each sample
and the cut-off score determined by ROC curve analysis, the
positive rate of SUSD2 in cancer and normal tissues were 55%
(88/160) and 100% (32/32), respectively, which indicated a
statistically significant difference (χ2=23.040;
P<0.000).
Correlation between SUSD2 expression
and clinicopathological characteristics of patients with NSCLC
In order to reveal the clinical significance of
SUSD2 protein expression levels in the NSCLC tissues, the
correlation between SUSD2 protein expression in the cancer tissues
and various clinicopathological parameters was analyzed (Table II). The result indicated that reduced
expression of SUSD2 in NSCLC patients was correlated with poor
histological grade (χ2=41.764; P<0.000), advanced
clinical stage (χ2=10.790; P=0.013), higher pT
(χ2=9.070; P=0.028) and positive regional lymph node
metastasis (χ2=15.399; P=0.002). No correlation was
observed between SUSD2 expression and other features analyzed,
including gender, age, pathology type or pM status (P>0.05).
 | Table II.Association between SUSD2 expression
and clinicopathological features of patients with non-small cell
lung cancer. |
Table II.
Association between SUSD2 expression
and clinicopathological features of patients with non-small cell
lung cancer.
|
| SUSD2 staining |
|
|
|---|
|
|
|
|
|
|---|
| Clinicopathological
feature | Negative, n (%) | Positive, n (%) | Total, n | P-valuea |
|---|
| Gender |
|
|
| 0.375 |
| Male | 51 (43.2) | 67 (56.8) | 118 |
|
|
Female | 21 (51.2) | 20 (48.8) | 41 |
|
| Age at surgery,
years |
|
|
| 0.607 |
| ≤55 | 29 (42.6) | 39 (57.4) | 68 |
|
|
>55 | 43 (46.7) | 49 (53.3) | 92 |
|
| Histological
grade |
|
|
| 0.000 |
| 1 | 1 (6.7) | 14 (93.3) | 15 |
|
| 2 | 26 (31.7) | 56 (68.3) | 82 |
|
| 3 | 35 (85.4) | 6
(14.6) | 41 |
|
| Clinical stage |
|
|
| 0.013 |
| I | 14 (29.2) | 34 (70.8) | 48 |
|
| II | 27 (50.9) | 26 (49.1) | 53 |
|
| III | 17 (60.7) | 11 (39.3) | 28 |
|
| IV | 2 (100) | 0 (0.0) |
2 |
|
| Pathology |
|
|
| 0.057 |
|
Adenocarcinoma | 30 (37.5) | 50 (62.5) | 80 |
|
| Squamous
cell carcinoma | 42 (52.5) | 38 (47.5) | 80 |
|
| pT status |
|
|
| 0.028 |
| T1 | 1 (7.7) | 12 (92.3) | 13 |
|
| T2 | 49 (45.8) | 58 (54.2) | 107 |
|
| T3 | 15 (51.7) | 14 (48.3) | 29 |
|
| T4 | 1 (100) | 0 (0.0) |
1 |
|
| Lymph node
metastasis |
|
|
| 0.002 |
| N0 | 16 (27.1) | 43 (72.9) | 59 |
|
| N1 | 40 (60.6) | 26 (39.4) | 66 |
|
| N2 | 6 (50) | 6 (50) | 12 |
|
| N3 | 1 (100) | 0 (0.0) |
1 |
|
| Distant
metastasis |
|
|
| 0.121 |
| No | 67 (45) | 82 (55) | 149 |
|
|
Yes | 2
(100) | 0
(0.0) |
2 |
|
Discussion
Worldwide, lung cancer has demonstrated a
significant rising trend in terms of morbidity and mortality in
recent years, and has become a significant hazard to human health
and life (15). Due to the
difficulties associated with the early detection of lung cancer,
the majority of patients with NSCLC present with advanced stage
cancer at diagnosis (17).
Conventional platinum-based chemotherapy and radiation have low
efficacy against this malignancy due to its frequent recurrence and
distant metastasis, and the overall 5-year survival rate of
patients with NSCLC remains at <15% (18,19).
Therefore, considerable efforts are required to explore novel
biomarkers and improve the early diagnosis and targeted therapeutic
interventions of NSCLC.
SUSD2 is a transmembrane protein that is widely
expressed in human tissues. Two previous studies (6,7) have been
published describing the mouse homolog SUSD2 as a potential tumor
suppressor. By contrast, Watson et al (5) reported that SUSD2 was overexpressed in
breast cancer, and indicated that SUSD2 enhanced the invasion of
breast cancer cells. Accelerated tumor formation and decreased
survival rates in mice with tumors expressing SUSD2 were observed
in a syngeneic mouse model (5).
However, the underlying physiopathological role of SUSD2 in human
NSCLC has remained to be elucidated. In the present study, the
expression levels of SUSD2 mRNA and protein were analyzed by
RT-qPCR and western blot analysis in paired NSCLC and adjacent
normal lung tissue samples. The results revealed that the
expression of SUSD2 mRNA and protein was decreased in the majority
of cancer tissues, when compared with their paired adjacent normal
lung tissues (P<0.001). Furthermore, IHC analysis of SUSD2 on a
TMA, including 160 NSCLC and 32 normal lung tissues, was also
conducted on various pathological types, histological grades and
clinical stages of NSCLC. In the present study, reduced expression
of SUSD2 was frequently observed in NSCLC tissues, while all normal
lung tissues exhibited marked staining of SUSD2. This was in
accordance with the results of the RT-qPCR and western blot
analyses.
In order to reveal the prognostic value of SUSD2
expression in NSCLC tissues, the correlation between SUSD2 protein
levels and clinicopathological characteristics was analyzed. The
results indicated that reduced expression of SUSD2 was correlated
with poor histological grade, advanced clinical stage, higher T
status and higher N status. These results indicated that low
expression levels of SUSD2 were correlated with the aggressive
behavior of NSCLC, which suggested the potential value of SUSD2
protein expression level evaluation in the determination of NSCLC
progression. However, to fully elucidate the specific function of
SUSD2 in NSCLC, further investigations with larger cohorts are
required.
In conclusion, the current study demonstrated that
SUSD2 was downregulated in human NSCLC tissues and was closely
correlated with poor differentiation and aggressive behavior of
NSCLC, indicating a potential role of the SUSD2 protein in the
pathogenesis of NSCLC.
Acknowledgements
The present study was supported by grants from the
Guangdong Supporting Grant for Outstanding Talent (no. C1030925)
and the Natural Science Foundation of Guangdong Province (no.
S2013010016631).
References
|
1
|
Brody H: Lung cancer. Nature. 513:S12014.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Salgia R: Prognostic significance of
angiogenesis and angiogenic growth factors in nonsmall cell lung
cancer. Cancer. 117:3889–3899. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Siegel R, Naishadham D and Jemal A: Cancer
statistics for Hispanics/Latinos, 2012. CA Cancer J Clin.
62:283–298. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Govindan R, Page N, Morgensztern D, Read
W, Tierney R, Vlahiotis A, Spitznagel EL and Piccirillo J: Changing
epidemiology of small-cell lung cancer in the United States over
the last 30 years: Analysis of the surveillance, epidemiologic and
end results database. J Clin Oncol. 24:4539–4544. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Watson AP, Evans RL and Egland KA:
Multiple functions of sushi domain containing 2 (SUSD2) in breast
tumorigenesis. Mol Cancer Res. 11:74–85. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Sugahara T, Yamashita Y, Shinomi M,
Yamanoha B, Iseki H, Takeda A, Okazaki Y, Hayashizaki Y, Kawai K,
Suemizu H and Andoh T: Isolation of a novel mouse gene,
mSVS-1/SUSD2, reversing tumorigenic phenotypes of cancer cells in
vitro. Cancer Sci. 98:900–908. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Sugahara T, Yamashita Y, Shinomi M, Isobe
Y, Yamanoha B, Iseki H, Takeda A, Okazaki Y, Kawai K, Suemizu H and
Andoh T: von Willebrand factor type D domain mutant of SVS-1/SUSD2,
vWd(m), induces apoptosis in HeLa cells. Cancer Sci. 98:909–915.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Chheang S and Brown K: Lung Cancer
staging: Clinical and radiologic perspectives. Semin Intervent
Radiol. 30:99–113. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Kreuzer KA, Lass U, Landt O, Nitsche A,
Laser J, Ellerbrok H, Pauli G, Huhn D and Schmidt CA: Highly
sensitive and specific fluorescence reverse transcription-PCR assay
for the pseudogene-free detection of beta-actin transcripts as
quantitative reference. Clin Chem. 45:297–300. 1999.PubMed/NCBI
|
|
10
|
Baine MJ, Mallya K and Batra SK:
Quantitative real-time PCR expression analysis of peripheral blood
mononuclear cells in pancreatic cancer patients. Methods Mol Biol.
980:157–173. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2 (-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Lee YJ, Kim DH, Lee SH, Kim DW, Nam HS and
Cho MK: Expression of the c-Met proteins in malignant skin cancers.
Ann Dermatol. 23:33–38. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Shi M, Yu DH, Chen Y, Zhao CY, Zhang J,
Liu QH, Ni CR and Zhu MH: Expression of fibroblast activation
protein in human pancreatic adenocarcinoma and its
clinicopathological significance. World J Gastroenterol.
18:840–846. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Shan LH, Sun WG, Han W, Qi L, Yang C, Chai
CC, Yao K, Zhou QF, Wu HM, Wang LF and Liu JR: Roles of fibroblasts
from the interface zone in invasion, migration, proliferation and
apoptosis of gastric adenocarcinoma. J Clin Pathol. 65:888–895.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Shi R, Zhao Z, Zhou H, Wei M, Ma WL, Zhou
JY and Tan WL: Reduced expression of PinX1 correlates to
progressive features in patients with prostate cancer. Cancer Cell
Int. 14:462014. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Wang G, Wu J and Song H: LRIG2 expression
and prognosis in non-small cell lung cancer. Oncol Lett. 8:667–672.
2014.PubMed/NCBI
|
|
17
|
Saintigny P and Burger JA: Recent advances
in non-small cell lung cancer biology and clinical management.
Discov Med. 13:287–297. 2012.PubMed/NCBI
|
|
18
|
Konishi J, Yamazaki K, Azuma M, Kinoshita
I, Dosaka-Akita H and Nishimura M: B7-H1 expression on non-small
cell lung cancer cells and its relationship with tumor-infiltrating
lymphocytes and their PD-1 expression. Clin Clin Cancer Res.
10:5094–5100. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Molina JR, Yang P, Cassivi SD, Schild SE
and Adjei AA: Non-small cell lung cancer: Epidemiology, risk
factors, treatment and survivorship. Mayo Clin Proc. 83:584–594.
2008. View Article : Google Scholar : PubMed/NCBI
|