Introduction
Lung cancer is the most common cancer worldwide, in
terms of incidence and mortality, as it comprises 17% of total
novel cancer cases and 23% of the total global cancer mortalities
(1). In order to improve the survival
rate, it is important to diagnose and surgically excise lung cancer
at an early stage of disease (2).
Therefore, it is necessary to identify biomarkers with the
potential to facilitate tumor diagnosis, particularly in the early
stages of the disease. The cancer stem cell (CSC) theory proposes
that tumors contain a small subpopulation of CSCs, which are
responsible for tumor growth, invasion and metastasis (2). CSCs and normal tissue stem cells share
important characteristics, including self-renewal, multipotency and
unlimited proliferation, and potentially possess overlapping
molecular mechanisms (3,4). Currently, a considerable number of stem
cell-associated markers have been identified (2,3). Numerous
studies have revealed that Sal-like protein 4 (SALL4) and
leucine-rich repeat-containing G-protein coupled receptor 5 (LGR5)
are also involved in the tumorigenesis, development and metastasis
of various tumors (5–7), and these proteins are expected to become
potential diagnostic markers and therapeutic targets in cancer.
SALL4, a homolog of the Drosophila homeotic
gene spalt, is a zinc-finger transcription factor that is required
for the proliferation and maintenance of pluripotency through
interaction with OCT3/4, sex determining region Y-box 2 and NANOG
(8,9).
SALL4 is also highly expressed in embryonic stem cells (8–12).
Notably, SALL4 is also overexpressed in various types of human
hematopoietic malignancies, including acute myelocytic and
lymphocytic leukemia (5,13). In addition, SALL4 upregulates the
expression of the oncogene Bmi-1 in human hematopoietic stem cells
and leukemia cells (14). Numerous
other studies have also revealed the oncogenic potential of SALL4
(5,6,15),
indicating that SALL4 may be used as a diagnostic marker in human
malignancies. However, there have been no studies investigating the
sensitivity and specificity of SALL4 expression in the diagnosis of
non-germ cell tumors (GCTs). The present study analyzed the
significance of SALL4 expression in lung cancer cells.
LGR5, also termed G-protein coupled receptor 49 or
G-protein coupled receptor 67, is a protein that is encoded by the
LGR5 gene in humans (16,17). LGR5 is a member of the G-protein
coupled receptor class A orphan receptor proteins. LGR5 is
expressed across a diverse range of tissues, including the muscle,
placenta, spinal cord and brain, and particularly acts as a
biomarker of adult stem cells in certain tissues, such as adipose
tissue and skeletal muscles (18).
LGR5 was first identified by Hsu et al in Drosophila
(18) and acts as a CSC marker for
colorectal carcinoma (CRC). LGR5 is not only used as a biomarker,
but is also likely to play an important role in maintaining the
undifferentiated state of CSCs. Numerous studies have revealed that
LGR5 acts as a CSC marker (19–21).
In the present study, the expression of SALL4 and
LGR5 was examined in lung cancer and non-cancerous lung tissues
using immunohistochemistry and the clinical significance and
diagnostic value of these two tumor markers in lung cancer was
evaluated.
Materials and methods
Lung cancer specimens
Tissue chips containing 135 specimens of lung cancer
tissue, 5 specimens of normal lung tissue and 5 specimens of
pneumonia-infected tissues were purchased from Guilin Fanpu Biotech
(Guilin, Guangxi, China).
The age range of the patients that the tissues were
obtained from was 31–78 years. In total, 102 patients were male and
33 patients were female. In addition, 91 patients were diagnosed
with lymph node metastasis. The types of lung cancer that the
present patients were diagnosed with are reported in Table I. Written informed consent was
obtained from the patients, and the study was approved by the
ethics committee of Guilin Medical University (Guilin, Guangxi,
China).
 | Table I.Pathological types of lung cancer
investigated in the present study. |
Table I.
Pathological types of lung cancer
investigated in the present study.
| Types of lung
cancer | Cases, n |
|---|
| Squamous cell
carcinoma | 64 |
| Adenocarcinoma | 35 |
| Papillary
adenocarcinoma | 9 |
| Adenosquamous
carcinoma | 9 |
| Small cell
carcinoma | 7 |
| Pulmonary arterial
carcinoma | 7 |
| Metastatic
carcinoma | 2 |
| Carcinoid tumor | 1 |
| Undifferential
tumor | 1 |
Immunohistochemical staining
The paraffin-embedded specimens were sliced into 4
µm sections. The slides were dried at 37°C overnight and the tissue
sections were baked at 60°C for 2 h. The slides were then
deparaffinized with xylene and rehydrated using ethanol and
immersed in 3% hydrogen peroxide for 10 min to block endogenous
peroxidase activity. An antigen retrieval process was accomplished
by pressure cooking the slides at 120°C and pressure of 103 kPa (15
psi) for 3 min in Tris/EDTA (pH 8.0). The slides were then
incubated with primary rabbit anti-human polyclonal antibodies
against SALL4 (cat no. GTX109983; GeneTex, Inc., Irvine, CA, USA)
and LGR5 (cat no. 21833-1-AP; ProteinTech, Chicago, IL, USA) for 1
h at room temperature in a moist chamber with saturated humidity.
The dilutions used for the SALL4 and LGR5 antibodies were 1:100 and
1:600, respectively. The specimens were stained with
3,3′-diaminobenzidine subsequent to incubation with the secondary
antibody (goat anti-rabbit IgG; 1:400 dilution; cat no. sc-2040;
Santa Cruz Biotechnology, Inc., Dallas, TX, USA) for 30 min.
Finally, the sections were counterstained with hematoxylin,
dehydrated and mounted. Confirmed biopsy specimens were used as a
positive control. The slides treated with non-specific serum were
used as a negative control.
Criteria for evaluation
The SALL4 protein was located in the cytoplasm and
cell membrane. Positive cytoplasmic staining for SALL4 was
indicated by yellow staining with cell shading. The LGR5 protein
was located in the cytoplasm and nucleus. LGR5 expression was
indicated by brown staining. In total, 5 microscopic fields in each
slide were randomly selected. Positive cells were counted under
high magnification (magnification, ×400) and the results were
expressed according to the following criteria: 1, Strong positive,
>75% tumor cells stained brown; 2, mild positive, 25–75% of
tumor cells stained medium brown; 3, weak positive, <25% of
tumor cells stained pale yellow; and 4, absent, no expression.
All slides were independently reviewed by an
experienced pathologist and researcher, without knowledge of the
clinical data. Inconsistent cases were assessed again together.
Statistical analysis
Statistical analysis of results was performed using
the SPSS 17.0 statistical package (SPSS, Inc., Chicago, IL, USA).
The expression rate of stem cell-associated markers in lung cancer
and the difference between the expression rates in the benign lung
tissue specimens were analyzed using Fisher's exact test. The
association between the expression of stem cell-associated markers
in lung cancer and clinicopathological factors was analyzed using
the χ2 test or Fisher's exact test, and the Mann-Whitney
U test or Kruskal-Wallis H test. The correlation between two stem
cell-associated markers was analyzed using Spearman's rank
correlation coefficient. P<0.05 was considered to indicate a
statistically significant difference.
Results
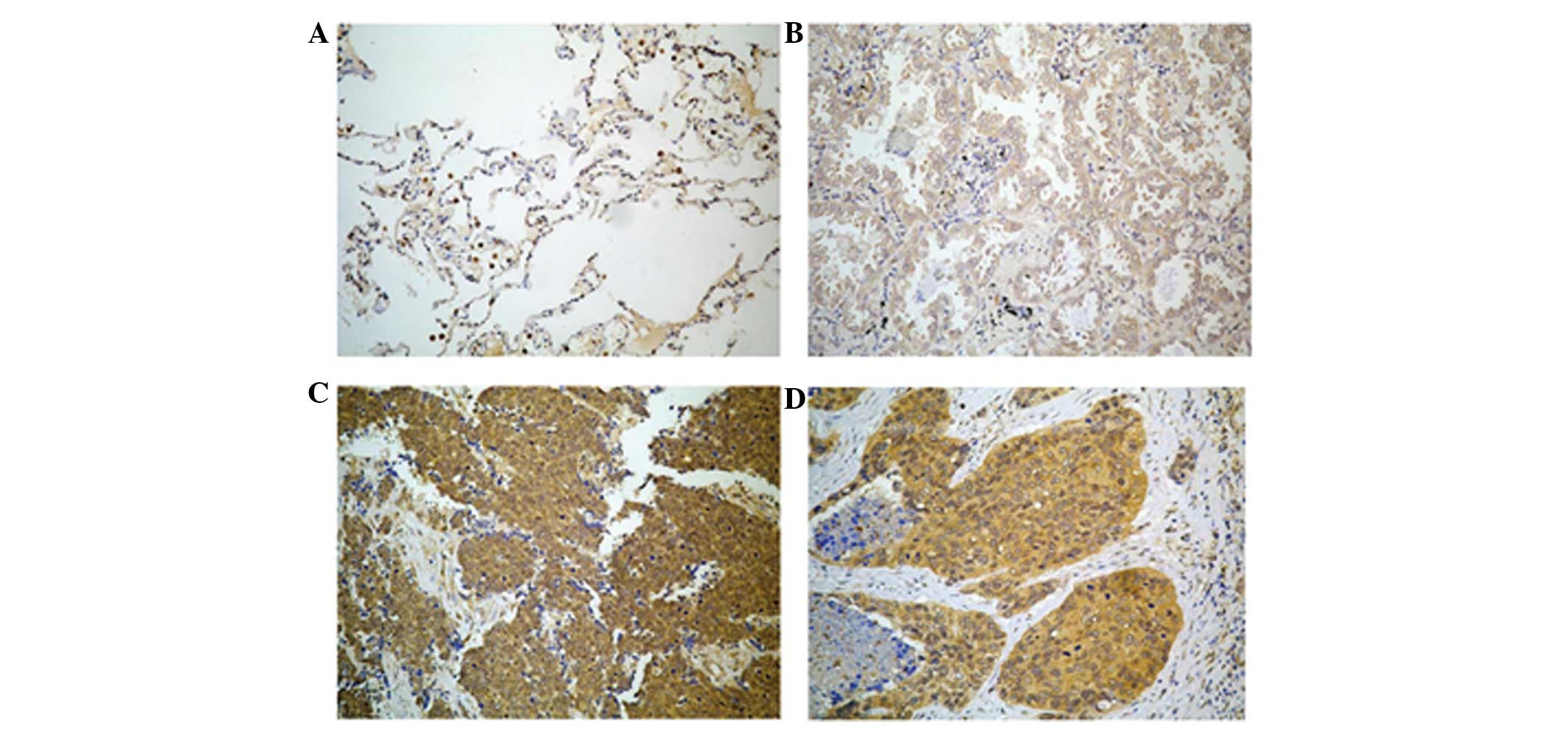
SALL4 is expressed in lung cancer
tissues
The expression of the SALL4 protein was observed
mainly in the cytoplasm and cell membrane of lung cancer cells, but
SALL4 was not expressed in normal or inflammatory lung tissues, or
in normal squamous epithelium (Fig.
1). Mild to weak SALL4 expression was observed in small cell
lung cancer tissues. Strong to mild expression was observed in lung
adenocarcinoma tissues, and strong to weak expression was observed
in squamous cell carcinoma tissues (Fig.
1). In the present study, SALL4 expression was observed in 119
out of 135 tissue samples and the rate of SALL4 expression was 88%
(Table II).
 | Table II.The expression of SALL4 and LGR5 in
each group. |
Table II.
The expression of SALL4 and LGR5 in
each group.
| Markers | Expression in normal
and non-cancerous tissues, n (%) | Expression in lung
cancer tissues, n (%) | χ2 | P-value |
|---|
| Total | 10
(100) | 135
(100) |
|
|
| SALL4 | 0 (0) | 119 (88) | 3.530 | 0.003 |
| LGR5 | 10
(100) | 131 (97) | 0.004 | 0.948 |
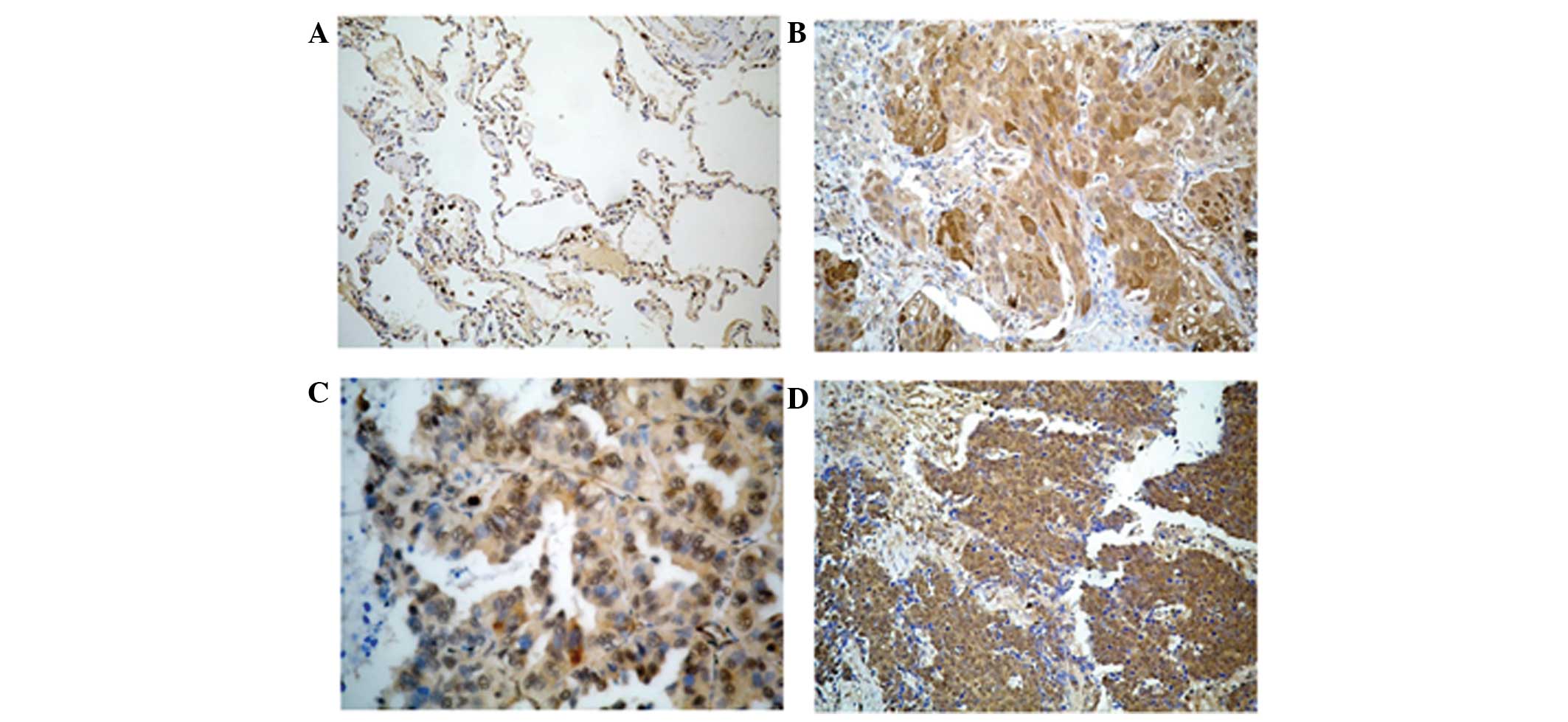
LGR5 is expressed in non-cancerous
cells, consisting of normal lung cells and benign lung lesions, and
lung cancer cells
Medium expression of the LGR5 protein was identified
in normal and inflammatory lung tissues, but LGR5 was not expressed
in normal squamous epithelium, and was only weakly expressed in
macrophages. Strong to mild LGR5 expression was observed in small
cell lung cancer tissues. Mild to weak LGR5 expression was observed
in lung adenocarcinoma and squamous cell lung carcinoma tissues
(Fig. 2). In the present study, LGR5
expression in lung cancer cells was observed in 131 out of 135
tissue samples. The rate of LGR5 expression was 97% (Table II).
Association between the present of
lung cancer cell-associated markers and clinical features
In order to identify the association between these
markers and the clinical features of patients with lung cancer, a
χ2 test was performed. Since the numbers of analyzed
cases of metastatic cancer, carcinoid tumor and differential
carcinoma were too small, these conditions were not included when
analyzing the association. The expression of SALL4 was not
significantly associated with the patient gender or age, lymph node
metastasis, pathological type of cancer or lung cancer stage
(P>0.05). This association is reported in detail in Table III. The expression of LGR5 was
significantly associated with patient gender (P<0.05), but was
not significantly associated with the patient age, presence of
lymph node metastasis, pathological type of cancer or lung cancer
stage (P>0.05). This association is reported in detail in
Table IV.
 | Table III.Association between SALL4 expression
and the clinical features of patients with lung cancer. |
Table III.
Association between SALL4 expression
and the clinical features of patients with lung cancer.
| Feature | Total | SALL4 expression, n
(%) | χ2 | P-value |
|---|
| Age |
|
|
|
|
| <60
years | 80 | 67 (83.75) | 2.386 | 0.122 |
| ≥60
years | 55 | 51 (92.73) |
|
|
| Gender |
|
|
|
|
|
Male | 102 | 91 (89.22) | 1.240 | 0.266 |
|
Female | 33 | 27 (81.82) |
|
|
| Histology |
|
|
|
|
|
Adenocarcinoma | 35 | 32 (91.43) | 2.703 | 0.609 |
|
Squamous cell carcinoma | 64 | 58 (90.63) |
|
|
|
Adenosquamous carcinoma | 9 | 7
(77.78) |
|
|
|
Papillary adenocarcinoma | 9 | 7
(77.78) |
|
|
| Lymph node
metastasis |
|
|
|
|
|
Yes | 41 | 36 (81.80) | 0.004 | 0.95 |
| No | 93 | 82 (88.17) |
|
|
| Stage |
|
|
|
|
|
I–II | 48 | 45 (93.75) | 0.003 | 0.959 |
|
III | 50 | 47 (94.00) |
|
|
 | Table IV.Correlation between LGR5 protein
expression and the clinical features of patients with lung
cancer. |
Table IV.
Correlation between LGR5 protein
expression and the clinical features of patients with lung
cancer.
| Feature | Total | LGR5 expression, n
(%) | χ2 | P-value |
|---|
| Age |
|
|
|
|
| <60
years | 80 | 76
(95.00) | 2.870 | 0.90 |
| ≥60
years | 55 |
55 (100.00) |
|
|
| Gender |
|
|
|
|
|
Male | 102 | 101 (99.02) | 5.704 | 0.017 |
|
Female | 33 | 30
(90.91) |
|
|
| Histology |
|
|
|
|
|
Adenocarcinoma | 35 | 34
(97.14) | 2.343 | 0.673 |
|
Squamous cell carcinoma | 64 | 62
(96.88) |
|
|
|
Adenosquamous carcinoma | 9 |
9
(100.00) |
|
|
|
Papillary adenocarcinoma | 9 |
8 (88.89) |
|
|
| Lymph node
metastasis |
|
|
|
|
|
Yes | 41 | 40
(97.56) | 0.061 | 0.805 |
| No | 93 | 90
(96.77) |
|
|
Correlation between SALL4 and
LGR5
Spearman's rank correlation analysis revealed that
the expression of SALL4 was not significantly correlated with the
expression of LGR5 in lung carcinoma tissues [correlation
coefficient (R/P), 1/0.170; P>0.05; Table V].
 | Table V.Correlation between SALL4 and LGR5
expression. |
Table V.
Correlation between SALL4 and LGR5
expression.
| Protein | Correlation | SALL4 R/P | LGR5 R/P |
|---|
| SALL4 | R/P | 1.000 |
0.170* |
| LGR5 | R/P |
0.170* | 1.000 |
Sensitivity and specificity of SALL4
and LGR5 expression in lung cancer cells
SALL4 was found to be expressed in 88% of lung
cancer tissue specimens and LGR5 was expressed in 97% of lung
cancer tissue specimens. High specificity of SALL4 was identified,
but specificity of LGR5 was not observed (Table VI).
 | Table VI.Comparison of the sensitivity and
specificity of SALL4 and LGR5 expression in lung cancer
tissues. |
Table VI.
Comparison of the sensitivity and
specificity of SALL4 and LGR5 expression in lung cancer
tissues.
| Protein | Sensitivity, % | Specificity, % |
|---|
| SALL4 | 88 | 100 |
| LGR5 | 97 | 0 |
Discussion
The present study investigated the expression and
clinical significance of SALL4 and LGR5 expression in lung cancer
by immunohistochemistry, and also explored the possible use of the
SALL4 and LGR5 proteins as diagnostic markers for lung cancer. The
present results demonstrated that SALL4 and LGR5 are each highly
expressed in lung cancer. SALL4 was found to be expressed in 88% of
lung cancer samples and LGR5 in 97% of lung cancer samples. High
specificity of SALL4 was also identified, but specificity of LGR5
was not observed, indicating that SALL4 may be used as an important
diagnostic marker for lung cancer due to the specific and high
expression in lung cancer cells.
Similar to numerous other studies (3,5,20), the present study supports the
hypothesis that cancer cells exhibit CSC markers, and certain CSC
markers may demonstrate considerable clinical importance in lung
cancer.
Expression analysis of SALL4 in non-cancerous and
cancerous lung tissues revealed that SALL4 is overexpressed in 88%
of lung cancer tissue samples (119 out of 135 cases). Similar
studies have also reported the overexpression of SALL4 in various
malignancies (21–23). In breast cancer, it has been
demonstrated that the SALL4 mRNA level is elevated in 86.1% of
tumor samples (21). In addition, the
increase was observed even in the early stages of tumors (24). However, there was no significant
correlation between the clinicopathological features of the
patients and SALL4 mRNA expression in breast cancer (22). Analysis of SALL4 expression in normal
and tumor colorectal tissues revealed that SALL4 is overexpressed
in almost 90% of CRC samples, in which SALL4 expression was
significantly associated with tumor cell metastasis to lymph nodes
and the grade of tumor cell differentiation (23). It has been revealed that the
expression of SALL4 in all types of testicular GCTs is associated
with the degree of tumor differentiation, and it has been suggested
that SALL4 is essential for the maintenance of the poorly
differentiated status of testicular GCTs (15). In addition, the expression of SALL4
was detected in >90% of the tumor cells of metastatic seminomas,
dysgerminomas and embryonal carcinomas, indicating that SALL4 plays
a role in the development of GCTs. Based on these results, SALL4
has been suggested as a novel sensitive and specific marker for
metastatic GCTs and as a novel diagnostic marker for metastatic
yolk sac tumors from the testis, ovary and extra gonadal sites
(6). A previous study detected the
expression of SALL4 at the mRNA level in cancerous and
non-cancerous lung cancer tissues using reverse
transcription-polymerase chain reaction (RT-PCR) (24). This study found that SALL4 mRNA
expression was present in 80.9% of the cancerous tissues (38 of
47), and revealed that the sensitivity and specificity of SALL4
mRNA were 85.1 and 92.9%, respectively (24).
In the present study, analysis of LGR5 protein
expression in non-cancerous and cancerous lung tissues demonstrated
that LGR5 is overexpressed in almost 97% of lung tissue samples.
The current study found that LGR5 was significantly overexpressed
in 131 out of 135 lung cancer tissue samples. However, the
expression of LGR5 was also detected in non-cancerous tissue
samples, consisting of 5 normal lung tissues, 4 tissue samples of
lung infections and one tubercle nodule.
LGR5 acts as a CSC marker in CRC, and is likely to
play an important role in maintaining the undifferentiated state of
CSCs. In a previous study, LGR5 was found to be expressed in 35 out
of 41 (85%) esophageal adenocarcinoma (EAC) tissues with Barrett's
esophagus (BE) and in 16 out of 19 (81%) EAC tissues without BE. By
contrast, LGR5 was not found to be expressed in esophageal squamous
cell carcinoma tissues (25). In
another study, RT-quantitative PCR analysis was performed on a
panel of representative cancer cell lines derived from various
organs, consisting of 11 colon cancer, 5 human hepatocellular
carcinoma, 10 ovarian cancer and 11 lung cancer cell lines
(26). LGR5 expression in each of the
cell lines was normalized in the 37 cell lines on average. LGR5
mRNA was overexpressed in 5 out of 11 colon cancer cell lines,
whereas LGR5 overexpression was rare in the cell lines derived from
other cancers (27). Several studies
have already focused on the effects of LGR5 expression in the
context of tumor development and progression. LGR5 has been
demonstrated to be involved in the pathogenesis of various human
cancers, including hepatocellular carcinoma (26), basal cell carcinoma (28), endometrial cancer (29), colon cancer and ovarian cancer
(7). Numerous studies have revealed
that LGR5 is also expressed in adenoma and CRC cells (18,26,27). These
findings suggest that LGR5 may be a marker for intestinal stem
cells, as well as for colorectal adenoma and carcinoma in
humans.
All the aforementioned studies, including the
present study, indicate the notable association between CSCs and
cancer, in addition to the importance of various tumor markers in
the diagnosis of various cancers. Overall, these findings and the
present data indicate that the SALL4 expression may be a novel
diagnostic marker for lung cancer. LGR5 expression is not an
effective diagnostic marker for lung cancer due to its poor
specificity for lung cancer. Additional investigation of the
diagnostic potential of SALL4 in the early stages of lung cancer
may have a profound clinical impact.
Acknowledgements
This study was supported by the Divisions of
Pathology and Respiratory Disease of the Affiliated Hospital of
Guilin Medical University. This study was also supported by the Key
Research Project Grant of Guangxi Health Department (grant no.
2012003).
Glossary
Abbreviations
Abbreviations:
|
CSC
|
cancer stem cell
|
|
CRC
|
colorectal carcinoma
|
|
ESCC
|
esophageal squamous cell carcinoma
|
|
EAC
|
esophageal adenocarcinoma
|
|
BE
|
Barrett's esophagus
|
|
RT-PCR
|
reverse transcription polymerase chain
reaction
|
|
GCT
|
germ cell tumor
|
References
|
1
|
Jemal A, Bray F, Center MM, et al: Global
cancer statistics. CA Cancer J Clin. 61:69–90. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Reya T, Morrison SJ, Clarke MF and
Weissman IL: Stem cells, cancer and cancer stem cells. Nature.
414:105–111. 2001. View
Article : Google Scholar : PubMed/NCBI
|
|
3
|
Visvader JE and Lindeman GJ: Cancer stem
cells in solid tumours: Accumulating evidence and unresolved
questions. Nat Rev Cancer. 8:755–768. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Hassan KA, Chen G, Kalemkerian GP, Wicha
MS and Beer DG: An embryonic stem cell-like signature identifies
poorly differentiated lung adenocarcinoma but not squamous cell
carcinoma. Clin Cancer Res. 15:6386–6390. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Cui W, Kong NR, Ma Y, et al: Differential
expression of the novel oncogene, SALL4, in lymphoma, plasma cell
myeloma and acute lymphoblastic leukemia. Mod Pathol. 19:1585–1592.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Cao D, Humphrey PA and Allan RW: SALL4 is
a novel sensitive and specific marker for metastatic germ cell
tumors, with particular utility in detection of metastatic yolk sac
tumors. Cancer. 115:2640–2651. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
McClanahan T, Koseoglu S, Smith K, et al:
Identification of overexpression of orphan G protein-coupled
receptor GPR49 in human colon and ovarian primary tumors. Cancer
Biol Ther. 5:419–426. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Sakaki-Yumoto M, Kobayashi C, Sato A, et
al: The murine homolog of SALL4, a causative gene in Okihiro
syndrome, is essential for embryonic stem cell proliferation and
cooperates with Sall1 in anorectal, heart, brain and kidney
development. Development. 133:3005–3013. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Elling U, Klasen C, Eisenberger T, Anlag K
and Treier M: Murine inner cell mass-derived lineages depend on
Sall4 function. Proc Natl Acad Sci USA. 103:16319–16324. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Zhang J, Tam WL, Tong GQ, et al: Sall4
modulates embryonic stem cell pluripotency and early embryonic
development by the transcriptional regulation of Pou5f1. Nat Cell
Biol. 8:1114–1123. 2006. View
Article : Google Scholar : PubMed/NCBI
|
|
11
|
Lim CY, Tam WL, Zhang J, et al: Sall4
regulates distinct transcription circuitries in different
blastocyst-derived stem cell lineages. Cell Stem Cell. 3:543–554.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Yang J, Gao C, Chai L and Ma Y: A novel
SALL4/OCT4 transcriptional feedback network for pluripotency of
embryonic stem cells. PLoS One. 5:e107662010. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Yang J, Chai L, Gao C, et al: SALL4 is a
key regulator of survival and apoptosis in human leukemic cells.
Blood. 112:805–813. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Yang J, Chai L, Liu F, et al: Bmi-1 is a
target gene for SALL4 in hematopoietic and leukemic cells. Proc
Natl Acad Sci USA. 104:10494–10499. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Cao D, Li J, Guo CC, et al: SALL4 is a
novel diagnostic marker for testicular germ cell tumors. Am J Surg
Pathol. 33:1065–1077. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
McDonald T, Wang R, Bailey W, et al:
Identification and cloning of an orphan G protein-coupled receptor
of the glycoprotein hormone receptor subfamily. Biochem Biophys Res
Commun. 247:266–270. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
McClanahan T, Koseoglu S, Smith K, et al:
Identification of overexpression of orphan G protein-coupled
receptor GPR49 in human colon and ovarian primary tumors. Cancer
Biol Ther. 5:419–426. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Hsu SY, Liang SG and Hsueh AJ:
Characterization of two LGR genes homologous to gonadotropin and
thyrotropin receptors with extracellular leucine-rich repeats and a
G-protein-coupled, seven-transmembrane region. Mol Endocrinol.
12:1830–1845. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Vermeulen L, Todaro M, de Sousa Mello F,
et al: Single-cell cloning of colon cancer stem cells reveals a
multi-lineage differentiation capacity. Proc Natl Acad Sci USA.
105:13427–13432. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Barker N, Ridgway RA, van Es JH, et al:
Crypt stem cells as the cells of-origin of intestinal cancer.
Nature. 457:608–611. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Takashima S, Kadowaki M, Aoyama K, et al:
The Wnt agonist R-spondin1 regulates systemic graft-versus-host
disease by protecting intestinal stem cells. J Exp Med.
208:285–294. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Kobayashi D, Kuribayshi K, Tanaka M and
Watanabe N: SALL4 is essential for cancer cell proliferation and is
overexpressed at early clinical stages in breast cancer. Int J
Oncol. 38:933–939. 2011.PubMed/NCBI
|
|
23
|
Forghanifard MM, Moghbeli M, Raeisossadati
R, et al: Role of SALL4 in the progression and metastasis of
colorectal cancer. J Biomed Sci. 20:62013. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Kobayashi D, Kuribayashi K, Tanaka M and
Watanabe N: Overexpression of SALL4 in lung cancer and its
importance in cell proliferation. Oncol Rep. 26:965–970.
2011.PubMed/NCBI
|
|
25
|
von Rahden BH, Kircher S, Lazariotou M, et
al: LgR5 expression and cancer stem cell hypothesis: Clue to define
the true origin of esophageal adenocarcinomas with and without
Barrett's Esophagus? J Exp Clin Cancer Res. 30:232011. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Yamamoto Y, Sakamoto M, Fujii G, et al:
Overexpression of orphan G-protein-coupled receptor, Gpr49, in
human hepatocellular carcinomas with beta-catenin mutations.
Hepatology. 37:528–533. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Uchida H, Yamazaki K, Fukuma M, et al:
Overexpression of leucine-rich repeat-containing Gprotein-coupled
receptor 5 in colorectal cancer. Cancer Sci. 101:1731–1737. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Tanese K, Fukuma M, Yamada T, et al:
G-protein-coupled receptor GPR49 is up-regulated in basal cell
carcinoma and promotes cell proliferation and tumor formation. Am J
Pathol. 173:835–843. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Sun X, Jackson L, Dey SK and Daikoku T: In
Pursuit of leucine-rich repeat-containing G protein-coupled
receptor-5 regulation and function in the uterus. Endocrinology.
150:5065–5073. 2009. View Article : Google Scholar : PubMed/NCBI
|