Introduction
The communication dysfunction between tumor cells
and stromal cells is essential for tumorigenic processes, including
cellular proliferation, differentiation, and apoptosis (1). Tumor cells must evade and/or suppress
immune responses locally. As chronic inflammation is an important
feature of the tumor microenvironment (1), various immune cells infiltrate the tumor
tissue, including T cells, B cells, macrophages, and dendritic
cells. These immune cells interact with tumor cells, stroma cells
and each other, constituting a complex tumor microenvironment,
however, the immune response remains weak, even in the presence of
chronic inflammation.
Recently, exosomes have emerged as potent
communicators between tumors and their microenvironment, and
potential contributors to tumorigenesis and subsequent metastasis
(2,3).
Exosomes are nanoparticles of 50–100 nm in size, and are reported
to be derived from endosomes or multivesicular bodies. Exosomes are
present in the extracellular microenvironment and in bodily fluids
including blood and urine (4,5).
Accumulating research indicates that exosomes have
diverse functions in an array of biological events, such as
cellular proliferation and migration (6). The contents of exosomes are complex and
may include diverse RNAs, cytokines, growth factors and lipids
(7–10), which are specific to their function
and indicate their biological significance. Exosomes contain
selectively enriched mRNAs and miRNAs that regulate gene expression
in target cells, including antigen-presenting cells; cell-derived
exosomes act to induce an immune response, whereas tumor-derived
exosomes act to suppress it
The essential roles of exosomes have been
demonstrated in a number of studies conducted in various tumor
types, such as prostate cancer, and melanoma (9,11,12). Exosomes derived from different types
of cancer contain different combinations of biological molecules,
and have distinctive effects on tumor behavior. Many miRNAs have
been discovered in exosomes from tumor cells and stroma cells,
which are essential in the regulation of tumor cell migration
(9,10,12–16).
Certain miRNAs may be involved in multiple stages of tumor
development; for example, miR-31, −185, and −34b are involved in
melanoma invasion (10). Therefore,
exosomes have been proposed to be potential markers for the
prognosis and diagnosis of various cancer types, and may be
potential therapeutic targets (9,13,14).
Lymphocyte infiltration and activation in tumor
microenvironments may have a significant effect on tumor prognosis,
growth and metastasis (5,17,18). It
has been hypothesized that well-established cancer cells utilize
multiple pathways to evade and/or suppress the immune response
against tumors (18); manipulating
the underlying mechanisms of this immune response has therefore
been the focus of many therapies for the treatment of various types
of cancer.
Breast cancer is one of the most common cancers in
females, and one of the greatest threats to female health.
Globally, breast cancer was the cause of 522,000 mortalities (15%
of cancer mortalities in females) in 2012 (19). Rapid progress in medical treatment has
markedly improved the prognosis and diagnosis, however, surgery
remains the most effective treatment, which often has a significant
physiological and psychological impact on patients (20,21).
Elucidation of the mechanisms underlying tumorigenesis in breast
cancer would aid in the development of treatments for this
condition. Although certain studies have indicated the involvement
of cancer cell-derived exosomes in tumor growth, few have explored
the role of exosomes in the development of breast cancer (22,23).
The present study explores the immunoregulatory
functions of exosomes derived from two different breast cancer cell
lines (MDA-MB-231 and BT-474). As transforming growth factor-β
(TGF-β) and interleukin-10 (IL-10), the two predominantly
considered cytokines, have been demonstrated to suppress the immune
response (24,25), the presence of these factors in the
exosomes from these cell lines was also investigated.
Materials and methods
Cell culture and transfection
BT-474 and MDA-MB-231 breast cancer cells were
purchased from American Type Culture Collection (Manassas, VA,
USA), and cultured according to the supplier's instructions. The
cells were cultured in Dulbecco's modified Eagle's medium (DMEM;
Invitrogen Life Technologies, Carlsbad, CA, USA) supplemented with
10% fetal bovine serum (FBS). When the cells were ~50% confluent,
the medium was changed to serum-free medium prior to exosome
preparation.
Hypoxic exposure
For hypoxic exposure, cells were cultured in an
O2/CO2 incubator (Sanyo; Osaka, Japan) in a
1% O2, 5% CO2 humidified atmosphere at 37°C.
For certain experiments, cells were treated with
dimethyloxalylglycine, a hypoxia-inducible factor hydroxylase
inhibitor (Enzo Life Sciences; Farmingdale, NY, USA) at a final
concentration of 1 mM. This was added to inhibit the effects of
hypoxia.
Normoxic exposure
For normoxic exposure, cells were cultured in an
O2/CO2 incubator (Sanyo) in a 20%
O2, 5% CO2 humidified atmosphere at 37°C.
Exosome preparation
BT-474 and MDA-MB-231 cell lines were cultured in
serum-free medium for exosome preparation. Exosome preparation was
performed as described by Epple et al (26) with slight modification. After four
days of culture, 100 ml of media were collected. Exosomes were
harvested through serial centrifugation of supernatants (2×10 min,
500 × g; 1×20 min, 2,000 × g; 1×30 min, 10,000 × g), followed by
centrifugation (60 min, 100,000 × g). The resulting pellet was
resuspended in 4 ml phosphate buffered saline (PBS), and harvested
again at 100,000 × g, 60 min. The final exosome-containing pellet
was resuspended in 0.4 ml of PBS and quantified. For
quantification, the resulting pellet was subjected to BCA assay
using BCA protein assay kit (Pierce, Rockford, IL, USA). The CD63
expression was also detected using western blotting, which was
normalized based on the cell number. Exosome preparations were
stored at −20°C until use.
Western blot analysis
Exosomes were collected and lysed with 1% Nonidet
P-40 plus protease inhibitor cocktail (Roche, Indianapolis, IN,
USA). Following brief lysis, 5 µg exosome was subjected to 12%
SDS-PAGE and transferred to nitrocellulose membranes. In this
study, CD63 was used as an indicator for exosomes, as CD63 is a
exosome marker. After blocking with 5% fat-free milk for 60 min,
the membrane was incubated with the appropriate primary antibody
(CD63; Santa Cruz Biotech, Santa Cruz, CA, USA; catalog no.
sc-15363), followed by a horseradish peroxidase-conjugated
secondary antibody (Jackson ImmunoResearch, West Grove, PA, USA;
catalog no. 111-035-003). β-actin was used as the loading control.
The blotting bands were visualized with enhanced chemiluminescence
plus immunoblotting detection reagents (Pierce, Rockford, IL, USA).
The membrane was subsequently scanned and the relative densitometry
of the bands was determined using ImageJ (Windows version 1.48,
NIH, Bethesda, MD, USA).
Splenocyte proliferation assay
Splenocytes were isolated from the spleens of female
C57BL/6 mice (6–8 weeks of age). The mice were purchased from the
Shanghai Laboratory Animal Center of the Chinese Academy of
Sciences (Shanghai, China), and all animal experiments were
approved by the Institutional Review Board of Nanfang Medical
University (Guangzhou, Guangdong, China). Splenocytes were
separated and seeded at a density of 5×103 cells/well in
DMEM supplemented with 10% FBS. Anti-CD3 (Invitrogen Life
Technologies; catalog no. 11452D) and anti-CD28 (Invitrogen Life
Technologies; catalog no. 11452D) linked with goat-anti-mouse
antibody were added for activation. Splenocytes at a density of
5×104 cells/well in 100 µl expansion medium were added
to round-bottom 96-well plates (Nunc; Penfield, NY, USA) and
cultured for five days. Activated splenocytes without addition of
any exosomes were used as the control group. The immunosuppressive
effects of the exosomes were evaluated by incubating 10 µl exosomes
per well with the splenocytes for five days. [3H]
thymidine (1 µCi ml−1; Perkin Elmer, San Jose, CA, USA)
was added 18 h prior to the end of incubation. Cells were
subsequently harvested for measurement of [3H] thymidine
incorporation to assess their rate of proliferation. To harvest the
cells, 1 ml 10% trichloroacetic acid (TCA) was gently added to each
well, and incubated at room temperature for 15 min. Following
aspiration with 10% TCA and washing with a further 1 ml 10% TCA,
the TCA was aspirated and 300 µl 0.2N NaOH was added to each well
to dissolve DNA; this was incubated for 15 min at room temperature
with agitation. To measure the thymidine incorporation,
subsequently, 100 µl of each sample was added to a scintillation
vial, and the counts were measured in a scintillation counter
(Beckman Coulter LS6500; Brea, CA, USA). In order to assess the
effects of TGF-β and IL-10 on splenocyte proliferation, anti-TGF-β
and/or anti-IL-10 antibodies (5 µg/ml) were applied at the time of
exosome application to block the potential effects of these
factors.
ELISA
The concentrations of TGF-b and IL-10 from exosome
lysates were determined by ELISA according to the manufacturer's
instructions (R&D, Minneapolis, MN, USA; DB100B and HS100C,
respectively). Briefly, 100 µl/well exosome lysate samples were
added to 96-well plates and incubated for 2 h at room temperature.
Following three washes with washing buffer (R&D), 100 µl/well
polyclonal horseradish peroxidase-conjugated anti-TGF-b1 or
monoclonal alkaline phosphatase-conjugated anti-IL-10 antibody was
added and the samples were incubated at room temperature for 2 h.
The samples were then washed three times with wash buffer, 50 µl
substrate solution (R&D) was added and incubated for 30 min,
followed by the addition of 50 µl stop solution (R&D).
Absorbance was measured using a microplate reader (Biotek Synergy
2; Biotek, Winooski, VT, USA) at a wavelength of 490 nm. The
concentration of cytokines was quantified, according to a
respective standard curve, which was obtained using recombinant
human TGF-b1 or IL-10 as the control.
Statistical analysis
All statistical analyses were performed using SPSS
software, version 20 (IBM SPSS Armonk, NY, USA). Student's
t-test was used for comparison, and P<0.05 was considered
to indicate a statistically significant difference.
Results
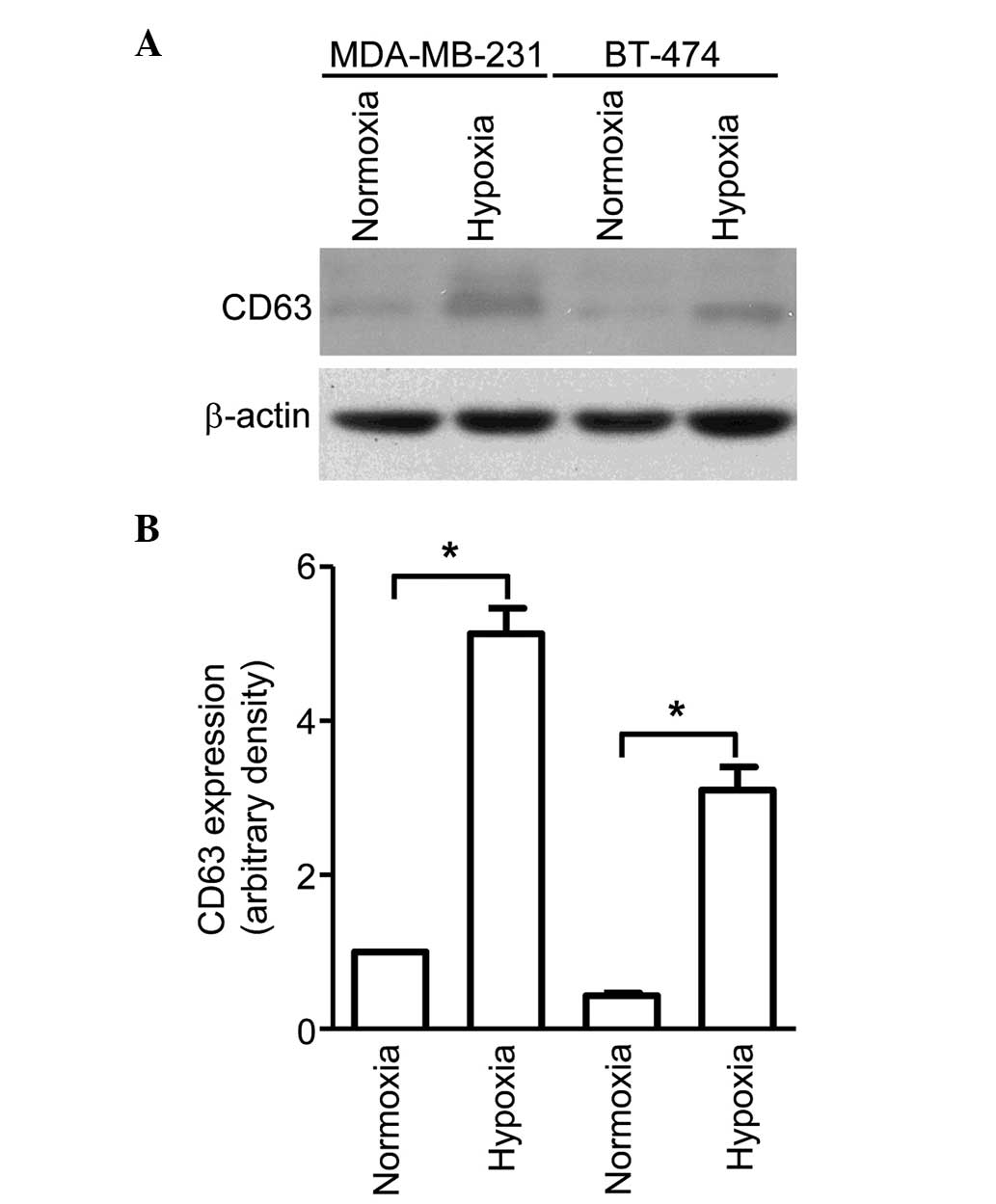
Hypoxia increases exosome secretion
from breast cancer cells
Exosomes were isolated from the conditioned media of
breast cancer cell lines BT-474 and MDA-MB-231, cultured under
normoxic (21% oxygen) or hypoxic (1% oxygen) conditions, using
serial centrifugation. The exosomal secretion of these cells was
evaluated through immunoblotting using CD63, a well-established
exosomal marker. As shown in Fig. 1,
although MDA-MB-231 cells were observed to secrete more exosomes
compared with BT-474 cells, both cell lines exhibited significantly
higher CD63 protein levels for cells cultured in hypoxic conditions
than for those cultured in normoxic conditions. This indicates that
hypoxia is a potent inducer of exosome secretion, which may have a
significant impact on the tumor microenvironment.
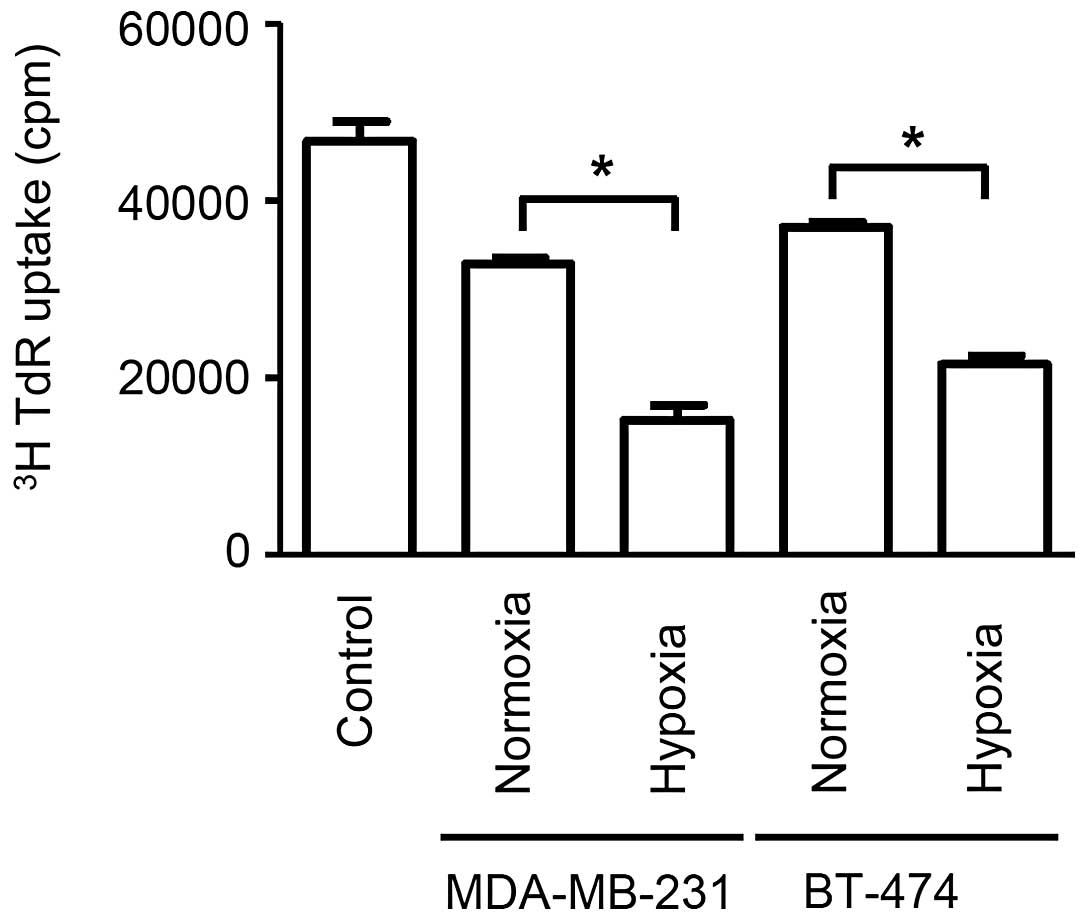
Exosomes derived from cancer cells
suppress splenocyte proliferation
The application of exosomes to proliferating
splenocytes revealed that exosomes secreted from cells cultured
under hypoxic conditions produced significantly stronger
immunosuppressive effects when compared with cells cultured under
normoxic conditions (P<0.05). Additionally, stronger
immunosuppressive effects were observed for exosomes secreted from
cells in hypoxia than that from cells in normoxia (P<0.05).
(Fig. 2). Exosomes derived from
MDA-MB-231 cells under hypoxia exerted the most potent
immunosuppression, followed by exosomes derived from BT-474 cells
under hypoxia, MDA-MB-231 cells under normoxia and BT-474 under
normoxia, which is consistent with the amount of exosome secretion
observed on CD63 immunoblotting. These data demonstrate that
exosomes from breast cancer cells cultured under hypoxic conditions
have potent immunosuppressive effects, which potentially contribute
to rapid tumor growth.
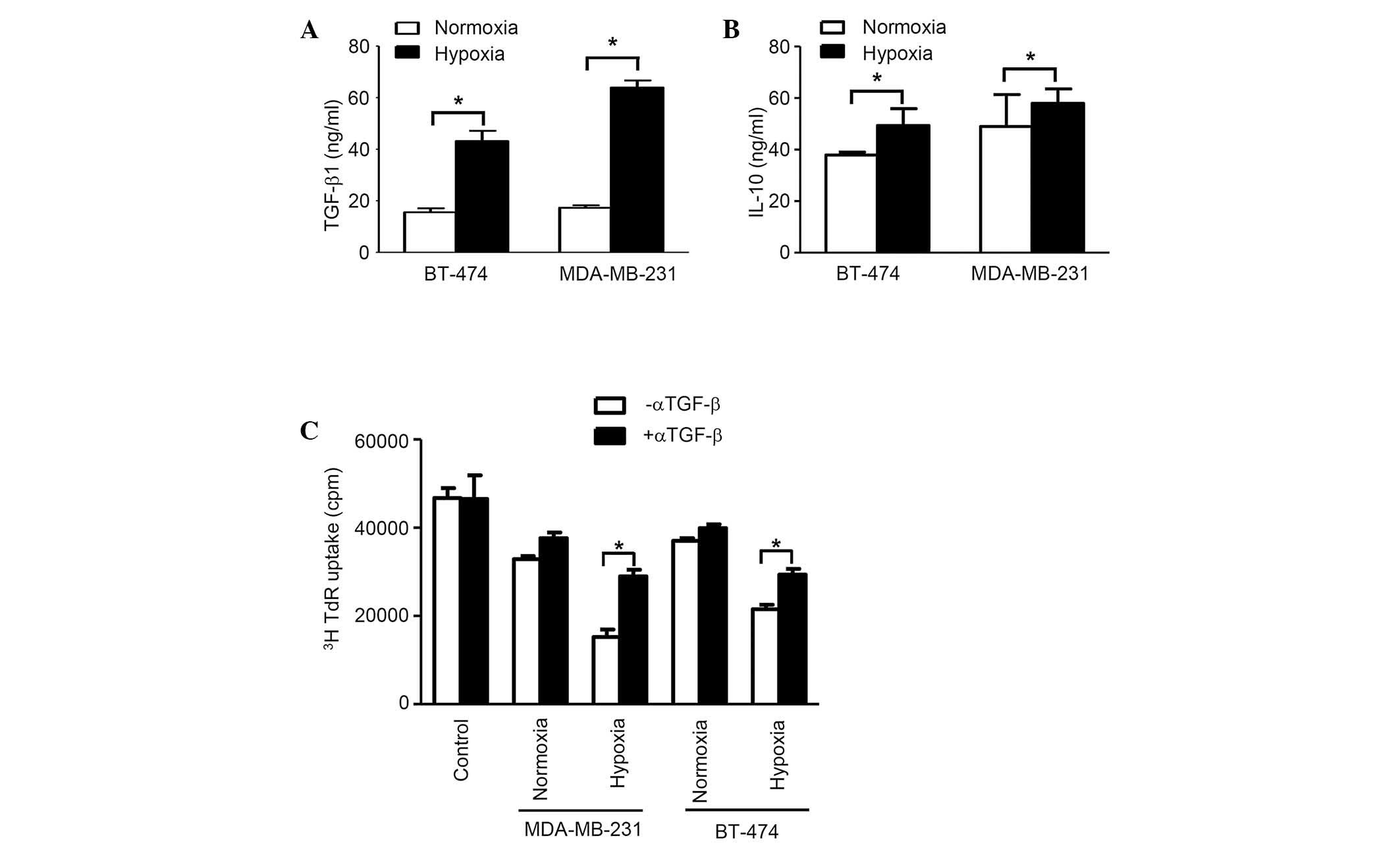
TGF-β in exosomes is responsible for
immunosuppression
The concentration of TGF-β1 in exosomes from
normoxic conditions in both cell lines was considerably higher
compared with that from hypoxic conditions (P<0.05).
Additionally, the concentration of IL-10 in exosomes from normoxic
conditions in cell line BT-474 was significantly higher compared
with hypoxic conditions, however, no considerable difference was
observed for MDA-MB-231 cells. Furthermore, in BT-474 cells, the
difference for TGF-β is much more prominent than for IL-10
(Fig. 3A). The [3H]
thymidine incorporation assay, which was applied to blocking TGF-β1
activity using anti-TGF-β antibodies, revealed that this factor is
essential for immunosuppression, as demonstrated by the evaluation
of splenocyte proliferation, which was restored following TGF-β
neutralization (Fig. 3B).
Collectively, the results demonstrate that exosomes from breast
cancer cells are able to suppress the immune response through
TGF-β, which inhibits splenocyte proliferation.
Discussion
As exosomes contain bulky proteins, proteomic
analysis has allowed for further investigation into the functions
of exosomes. An array of proteins, including vascular endothelial
growth factor, epidermal growth factor and interleukin-4, have been
shown to be present in the exosomes from cancer cells, and are
critical for tumor cell survival, proliferation and migration.
TGF-β1, which is produced by regulatory T cells, has potent
immunosuppressive effects, which are crucial for protecting the
body from excessive immune responses and maintaining immune
homeostasis (27–29). However, these mechanisms may be
manipulated by cancer cells, allowing them to evade and suppress
immune responses. This study demonstrated that exosomes derived
from breast cancer cells contain abundant TGF-β, which facilitates
tumor growth. As hypoxia is a feature of the microenvironment of
solid tumors, it is not surprising that the immunosuppressive
effects of exosomes derived from cancer cells cultured under
hypoxia are much greater than those of exosomes produced under
normoxic conditions. This may explain the rapid growth of solid
cancers in vivo in a hypoxic microenvironment. In the
present study, it was demonstrated that hypoxia promoted the
secretion of exosomes in cancer cells, which contain the
immunosuppressive cytokines, TGF-β and IL-10. It is hypothesized
that exosomes are important for communication among cancer cells,
and are able to fuse with adjacent cancer cells, subsequently
increasing their immunosuppressive effects. Thus, we hypothesize
that preventing communication among cancer cells may provide a
novel method for the inhibition of cancer growth. However, as
breast cancer cell lines were used in the present study the results
may not be applicable in vivo. Therefore, further in
vivo studies using animal models and clinical data are
required. Additionally, the detailed mechanisms of exosome
secretion and cancer cell communication also require further
investigation.
In conclusion, these results revealed a novel aspect
of exosomes derived from breast cancer cells in immunosuppression,
suggesting the essential role of exosomes in tumor microenvironment
constitution. These findings could be potentially significant in
the development of novel therapeutic strategies.
References
|
1
|
Hanahan D and Weinberg RA: Hallmarks of
cancer: the next generation. Cell. 144:646–674. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Corrado C, Raimondo S, Chiesi A, Ciccia F,
De Leo G and Alessandro R: Exosomes as intercellular signaling
organelles involved in health and disease: basic science and
clinical applications. Int J Mol Sci. 14:5338–5366. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Ahmed KA and Xiang J: Mechanisms of
cellular communication through intercellular protein transfer. J
Cell Mol Med. 15:1458–1473. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Denzer K, Kleijmeer MJ, Heijnen HF,
Stoorvogel W and Geuze HJ: Exosome: from internal vesicle of the
multivesicular body to intercellular signaling device. J Cell Sci.
113:3365–3374. 2000.PubMed/NCBI
|
|
5
|
Schorey JS and Bhatnagar S: Exosome
function: from tumor immunology to pathogen biology. Traffic.
9:871–881. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
van Niel G, Porto-Carreiro I, Simoes S and
Raposo G: Exosomes: a common pathway for a specialized function. J
Biochem. 140:13–21. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Bard MP, Hegmans JP, Hemmes A, et al:
Proteomic analysis of exosomes isolated from human malignant
pleural effusions. Am J Respir Cell Mol Biol. 31:114–121. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Street JM, Barran PE, Mackay CL, et al:
Identification and proteomic profiling of exosomes in human
cerebrospinal fluid. J Transl Med. 10:52012. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Hessvik NP, Sandvig K and Llorente A:
Exosomal miRNAs as Biomarkers for Prostate Cancer. Front Genet.
4:362013. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Xiao D, Ohlendorf J, Chen Y, et al:
Identifying mRNA, microRNA and protein profiles of melanoma
exosomes. PloS One. 7:e468742012. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Díaz-Valdés N, Basagoiti M, Dotor J, et
al: Induction of monocyte chemoattractant protein-1 and
interleukin-10 by TGFbeta1 in melanoma enhances tumor infiltration
and immunosuppression. Cancer Res. 71:812–821. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Mizoguchi M, Guan Y, Yoshimoto K, et al:
Clinical implications of microRNAs in human glioblastoma. Front
Oncol. 3:192013. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Gailhouste L, Gomez-Santos L and Ochiya T:
Potential applications of miRNAs as diagnostic and prognostic
markers in liver cancer. Front Biosci (Landmark Ed). 18:199–223.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Kahlert C and Kalluri R: Exosomes in tumor
microenvironment influence cancer progression and metastasis. J Mol
Med (Berl). 91:431–437. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Katakowski M, Buller B, Zheng X, et al:
Exosomes from marrow stromal cells expressing miR-146b inhibit
glioma growth. Cancer Lett. 335:201–204. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Chiba M, Kimura M and Asari S: Exosomes
secreted from human colorectal cancer cell lines contain mRNAs,
microRNAs and natural antisense RNAs, that can transfer into the
human hepatoma HepG2 and lung cancer A549 cell lines. Oncol Rep.
28:1551–1558. 2012.PubMed/NCBI
|
|
17
|
Borghesi L and Milcarek C: Innate versus
adaptive immunity: a paradigm past its prime? Cancer Res.
67:3989–3993. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Lizée G, Overwijk WW, Radvanyi L, Gao J,
Sharma P and Hwu P: Harnessing the power of the immune system to
target cancer. Annu Rev Med. 64:71–90. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Tao Z, Shi A, Lu C, Song T, Zhang Z and
Zhao J: Breast cancer: Epidemiology and etiology. Cell Biochem
Biophys. Dec 28–2014.(Epub ahead of print). PubMed/NCBI
|
|
20
|
Murawa P, Murawa D, Adamczyk B and Połom
K: Breast cancer: Actual methods of treatment and future trends.
Rep Pract Oncol Radiother. 19:165–172. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Edge SB: Quality measurement in breast
cancer. J Surg Oncol. 110:509–517. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Al-Nedawi K: The yin-yang of microvesicles
(exosomes) in cancer biology. Front Oncol. 4:1722014. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Tickner JA, Urquhart AJ, Stephenson SA,
Richard DJ and O'Byrne KJ: Functions and therapeutic roles of
exosomes in cancer. Front Oncol. 4:1272014. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Jui HY, Lin CH, Hsu WT, et al: Autologous
mesenchymal stem cells prevent transplant arteriosclerosis by
enhancing local expression of interleukin-10, interferon-γ, and
indoleamine 2,3-dioxygenase. Cell Transplant. 21:971–984. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Sun L, Akiyama K, Zhang H, et al:
Mesenchymal stem cell transplantation reverses multiorgan
dysfunction in systemic lupus erythematosus mice and humans. Stem
Cells. 27:1421–1432. 2009. View
Article : Google Scholar : PubMed/NCBI
|
|
26
|
Epple LM, Griffiths SG, Dechkovskaia AM,
et al: Medulloblastoma exosome proteomics yield functional roles
for extracellular vesicles. PloS One. 7:e420642012. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Vignali DA, Collison LW and Workman CJ:
How regulatory T cells work. Nat Rev Immunol. 8:523–532. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Belkaid Y and Tarbell K: Regulatory T
cells in the control of host-microorganism interactions(*). Annu
Rev Immunol. 27:551–589. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Zou W: Regulatory T cells, tumour immunity
and immunotherapy. Nat Rev Immunol. 6:295–307. 2006. View Article : Google Scholar : PubMed/NCBI
|