Introduction
Gastric carcinoma (GC) is the second most common
tumor worldwide (1). The highest
mortality rates for GC have been reported in East Asia, including
Japan, Korea and China (2).
Currently, typical treatments for GC comprise surgery and
chemotherapy, however recurrence frequently occurs, particularly
with advanced stage GC (3).
Currently, the majority of chemotherapy regimens are only able to
achieve a low clinical complete response rate, and are not capable
of improving overall survival rates (4). Therefore, the development of novel
therapies for the treatment of GC is urgently required.
Epidermal growth factor receptor (EGFR/ErbB1) is a
member of the ErbB family of receptor tyrosine kinases. The EGFR
gene is located on the short arm of human chromosome 7 and produces
a 170 kDa transmembrane glycoprotein (5). When EGFR binds certain ligands,
including epidermal growth factor or transforming growth factor-α,
it is capable of activating a number of intracellular signaling
cascades, for example, the RAS/mitogen activated protein kinase,
phosphatidylinositol-3-kinase and signal transducer and activator
of transcription-3 signal transduction pathways (6,7). These
pathways regulate cell proliferation, migration, adhesion,
differentiation and survival (7,8).
Overexpression and/or increased activity of EGFR may
be detected in a number of human tumors and is frequently
associated with aggressive tumor behaviors and poor prognosis
(7,9).
Therefore, EGFR is considered to be a significant therapeutic
target for the treatment of human cancer. EGFR-targeting drugs have
been developed and approved for use in the treatment of patients
exhibiting EGFR-expressing non-small cell lung cancer (NSCLC) and
colorectal carcinoma (CRC) (9,10).
Cetuximab and panitumumab are EGFR-binding monoclonal antibodies
(mAbs), which are currently approved for use in the treatment of
CRC (11). Gefitinib and erlotinib
are EGFR tyrosine kinase inhibitors that are approved for use in
the treatment of NSCLC (12).
In patients exhibiting advanced NSCLC, a positive
response following treatment with tyrosine kinase inhibitor
gefitinib was correlated with increased EGFR gene copy number and
protein expression (13). Certain
studies have identified overexpression of EGFR as a potential
prognostic indicator for GC (14,15). A
small number of phase II and III clinical trials, in which GC was
treated with cetuximab, have been performed, however ambiguous
results were obtained (16,17). It has been reported that the
alteration of EGFR expression in GC may affect the sensitivity of
EGFR-targeted therapies (18).
The incidence of EGFR overexpression and
abnormalities in the EGFR gene may vary markedly across ethnicities
(19). A small number of studies
concerning EGFR status in Chinese GC patients have been published.
The present study systemically evaluated EGFR protein expression
and gene copy number in 150 samples of GC from Chinese patients.
The associations between EGFR status, clinicopathological
parameters and treatment outcomes were retrospectively analyzed.
The present study may aid in the investigation of the viability of
EGFR-targeting therapies as a potential treatment for GC in Chinese
patients.
Patients and methods
Case selection and clinicopathological
features
Patients pathologically diagnosed with gastric
adenocarcinoma between April 2005 and June 2007 at the Second
Affiliated Hospital of Dalian Medical University (Dalian, China)
were selected for the current study. The current study was approved
by the Institutional Review Board of Dalian Medical University. All
participants signed a consent form prior to the commencement of
surgical procedures and initiation of the study. Pathological
specimens collected from the primary surgery were routinely fixed
in formalin (Kan Nai Xin Zhongshan Biotechnology Co., Ltd.,
Zhongshan, China) and embedded in paraffin (Shanghai Hualing Health
Machinery Plant, Shanghai, China). Each slide was re-evaluated by a
pathologist, with no knowledge of the patient's pathological
diagnosis, prior to the performance of experiments.
Clinicopathological parameters were noted, including gender, age,
tumor/node/metastasis (TNM) and pathological stages, depth of
invasion, the presence of lymph node or distant metastasis and
tumor location. Patient characteristics and details of each sample
are listed in Table I.
 | Table I.Correlation of EGFR expression with
clinicopathological parameters. |
Table I.
Correlation of EGFR expression with
clinicopathological parameters.
|
|
| EGFR expression |
|
|---|
|
|
|
|
|
|---|
| Clinicopathological
parameter | n | − | 1+ | 2+/3+ | P-value |
|---|
| Gender |
|
|
|
| 0.315 |
|
Male | 122 | 56 | 48 | 18 |
|
|
Female | 28 | 11 | 15 | 2 |
|
| Age, years |
|
|
|
| 0.113 |
|
﹤65 | 81 | 41 | 33 | 7 |
|
|
≥65 | 69 | 26 | 30 | 13 |
|
| Diameter of tumor,
cm |
|
|
|
| 0.786 |
| ﹤5 | 77 | 35 | 32 | 9 |
|
| ≥5 | 73 | 31 | 31 | 11 |
|
| Tumor location |
|
|
|
|
0.013b |
| Cardia
and fundus | 17 | 2 | 10 | 5 |
|
|
Body | 46 | 19 | 23 | 4 |
|
| Pylorus
and antrum | 87 | 63 | 20 | 15 |
|
|
Differentiation |
|
|
|
| 0.367 |
|
Well/moderate | 28 | 9 | 15 | 4 |
|
|
Poor | 110 | 54 | 41 | 15 |
|
|
Mucinousa | 12 | 4 | 7 | 1 |
|
| Invasion depth |
|
|
|
| 0.301 |
|
Mucosa/submucosa | 12 | 4 | 7 | 1 |
|
|
Muscular/serosa | 25 | 16 | 7 | 2 |
|
| Out of
the serosa | 87 | 38 | 35 | 14 |
|
| Other
organs | 26 | 9 | 14 | 3 |
|
| Lymph node
metastases |
|
|
|
| 0.086 |
| 0 | 43 | 14 | 21 | 8 |
|
|
1–6 | 54 | 31 | 20 | 3 |
|
| ≥7 | 53 | 22 | 22 | 9 |
|
| Distant
metastases |
|
|
|
| 0.150 |
| − | 127 | 61 | 50 | 16 |
|
| + | 23 | 6 | 13 | 4 |
|
|
Tumor/Node/Metastasis stage |
|
|
|
| 0.525 |
| I | 22 | 9 | 10 | 3 |
|
| II | 31 | 13 | 14 | 4 |
|
|
III | 67 | 36 | 23 | 8 |
|
| IV | 30 | 9 | 16 | 5 |
|
Survival times were calculated from the initial
surgery, and were considered censored for patients who were alive
at the final follow-up or who succumbed with no evidence of GC
recurrence. Clinical outcome was determined from the date of
surgery until mortality, or 31 November 2013, which resulted in a
follow-up period of 1–104 months (mean, 49 months). A total of 189
GC cases were included at the initiation of the present study,
however 39 cases were lost to follow-up. Patients (150 cases) who
possessed complete prognosis data were included in the
analysis.
Tissue array method
An expert pathologist evaluated the hematoxylin and
eosin-stained (Sinopharm Chemical Reagent Co., Ltd., Shanghai,
China) slides in order to ensure that the tissue-containing tumor
cells were studied. Core tissue biopsy specimens (diameter, 2 mm)
were obtained from individual paraffin-embedded GC samples and
arranged in recipient paraffin blocks. In order to account for
tumor heterogeneity, two separate core samples per tumor were
obtained. Non-neoplastic gastric mucosa specimens, which were
obtained from adjacent normal tissue, were included in each of the
array blocks; in total, 40 specimens were included. The tissue
array blocks contained up to 30 cores, meaning that 12 array blocks
were formed from the 150 cases.
Immunohistochemistry (IHC) and
interpretation of immunohistochemical results
Immunohistochemical staining of samples was
performed using rabbit polyclonal IgG against EGFR (anti-EGFR;
1:50; sc03; Santa Cruz Biotechnology, Inc., Dallas, TX, USA) and
avidin-biotin-peroxidase techniques (VECTASTAIN® Elite ABC kit;
Vector Laboratories, Inc., Burlingame, CA, USA). Paraffin-embedded
tissue sections (4 µm) were deparaffinized and rehydrated.
Endogenous peroxidase activity was ablated using 2%
H2O2-methanol (Tianjin Kermel Chemical
Reagent Co., Ltd., Tianjin, China). Antigen retrieval was performed
by microwave heating of slides in 10 mmol/l citrate buffer
(Zhengzhou Cengfeng Chemical Products Co., Ltd., Zhengzhou, China)
at pH 6.0, and 5% normal sheep serum (Zhongshan Jinqiao
Biotechnology Co., Inc., Beijing, China) was added to suppress
nonspecific protein binding. Tissue sections were incubated at 37°C
for 1 h, and subsequently incubated at 4°C overnight with primary
EGFR antibody. Certain tissue sections were incubated with 5% serum
in phosphate buffered saline (PBS; Zhongshan Jinqiao Biotechnology
Co., Inc.) without antibody as a negative control. EGFR antibody
was diluted 1:50 in 5% serum. Sections were then washed with PBS
and incubated with biotinylated goat anti-rabbit immunoglobulin G
secondary antibody at 37°C for 1 h. Following this initial
incubation, cells were incubated a second time with avidin-biotin
complex from the kit at 37°C for 45 min. Color was developed using
3,3′-diaminobenzidine tetrahydrochloride (Zhongshan Jinqiao
Biotechnology Co., Inc.). Slides were counterstained with
hematoxylin.
Immunohistochemical staining for EGFR was evaluated
using the following criteria: 0, no discernible
staining/background-type staining; 1+, ambiguous discontinuous
membrane staining; 2+, moderate intensity membrane staining; and
3+, strong and complete plasma membrane staining (14,20).
Immunohistochemical staining scores of 2+ and 3+ were considered to
indicate EGFR overexpression.
Fluorescence in situ hybridization
(FISH)
Commercially available probes for the EGFR gene and
centromere 7 (GLP EGFR/CSP 7 Dual Color Probe; Beijing Jinpujia
Medical Technology Co., Ltd, Beijing, China) were utilized in the
present study. Procedures were performed according to standard
protocols (21). Briefly, 3–5 µm
sectioned tissue array slides were deparaffinized, dehydrated, and
incubated in 20% sodium bisulphate/2X standard saline citrate (2X
SSC; Zhongshan Jinqiao Biotechnology Co., Inc.), at 75°C for 20
min. Following washing in 2X SSC, slides were treated with
proteinase K (Amresco LCC, Solon, OH, USA) at 37°C for 20 min,
rinsed in 2X SSC at room temperature for 5 min and dehydrated using
ethanol (Hongming Chemical Reagent Co.) in a series of increasing
concentrations (60, 85, 95 and 100%). EGFR and CEP7 probes were
applied to each slide, covered with a glass coverslip and sealed
using rubber cement (Citotest Labware Manufacturing Co., Ltd.,
Nanjing, China). Slides were denatured for 5 min at 83°C in a
hybrite chamber (Citotest Labware Manufacturing Co., Ltd.) and
hybridized overnight (for ≥8 h) at 37°C. Following
post-hybridization washing, slides were counterstained using 10 µl
DAPI (Beyotime Biotechnology Co., Ltd., Shanghai, China) in
antifade solution (Beijing Jinpujia Medical Technology Co., Ltd.),
cover-slipped and examined under a fluorescence microscope (BX41;
Olympus, Tokyo, Japan).
At least 60 tumor cell nuclei were counted per
sample. Numbers of red (EGFR) and green (chromosome 7 centromere)
signals were counted manually by Dr Xiaotang Yu and a technique
assistant (Miss Li Wang of Beijing Jinpujia Medical Technology Co.,
Ltd.). For each FISH probe tested, the status of the chromosome was
defined by the presence of the centromeric probe; CEP7 signals
served as a control. The ratio of gene probe:centromeric probe was
calculated. High levels of polysomy and gene amplification were
regarded as a positive FISH result. Gene amplification was
considered if tight EGFR gene clusters, a ratio of EGFR
gene:chromosome of ≥2 or ≥15 copies of EGFR/cell in ≥10% of
analyzed cells was observed. Polysomy was considered when ≥4 copies
of the EGFR gene were identified in ≥40% of cells, and was termed
inconclusive if the EGFR:CEP7 signal ratio was observed to be ≥2 in
≤10 % of the analyzed cells (15,22).
Statistical analysis
Statistical analysis was performed using SPSS
version 19.0 (IBM SPSS, Armonk, NY, USA). Associations between EGFR
expression and clinicopathological characteristics were assessed by
Kruskal-Wallis and Mann-Whitney U test analysis. Groups were
compared using the Pearson χ2 test. Survival curves were
constructed using Kaplan-Meier analysis, and the significance of
differences between survival curves was determined using the
log-rank test. Multivariate analysis was performed using Cox
proportional hazards regression. All statistical tests were
two-sided. P<0.05 was considered to indicate a statistically
significant difference.
Results
High EGFR expression is correlated
with the presence of GC at the cardia and fundus
EGFR protein expression was determined using IHC in
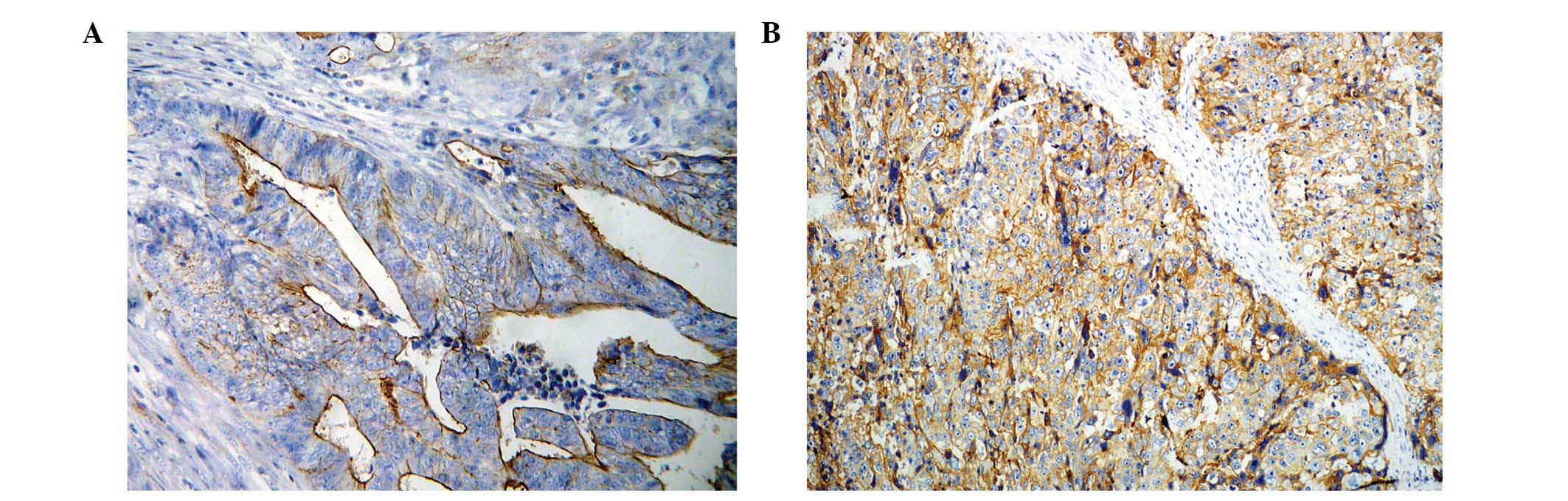
tissue array slides containing 150 samples of GC (Fig. 1). EGFR staining in GC was detected in
the membrane and/or cytoplasm. Out of a total of 150 samples, 67
cases (44.67%) scored 0, 63 (42.00%) scored 1+ and 20 (13.33%)
scored 2+ or 3+. A score of 1+ was considered to demonstrate weak
EGFR expression, while a score of 2+ or 3+ was considered to
demonstrate EGFR overexpression. Associations between EGFR protein
expression and clinicopathological parameters were analyzed and are
summarized in Table I. The results of
the present study revealed that EGFR is highly expressed in GC
located at the cardia and fundus (P=0.012). There were no
significant correlations observed between EGFR protein expression
and any other clinicopathological features (P>0.05).
Increased EGFR gene amplification is
correlated with high levels of EGFR protein expression
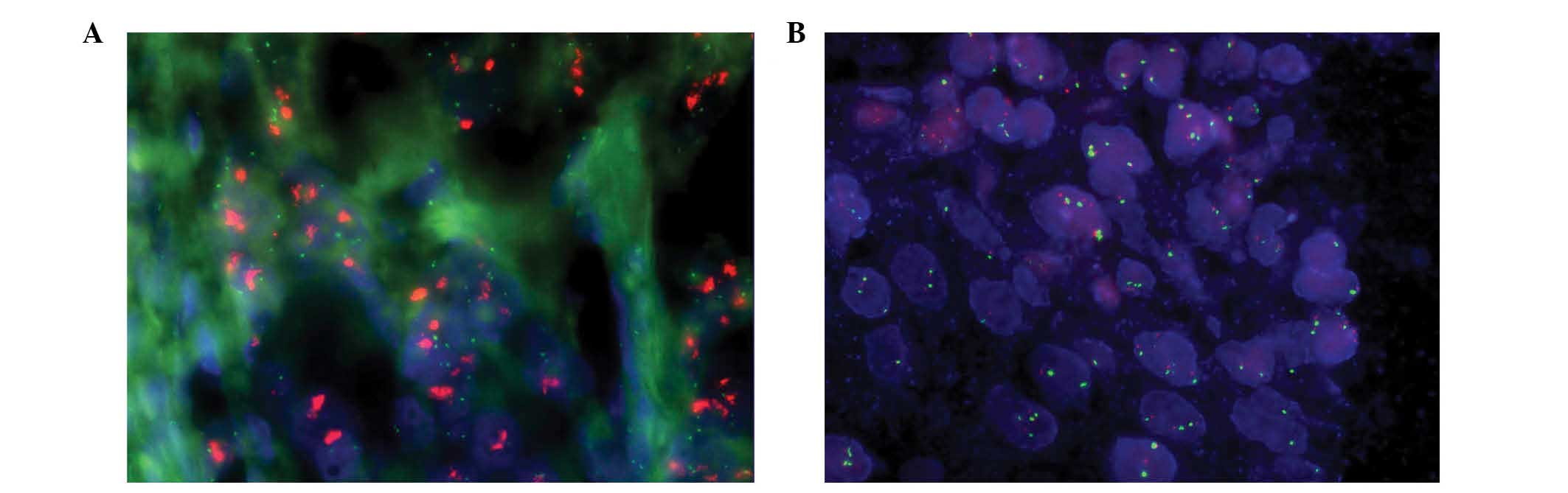
EGFR gene amplification was detected using FISH in
tissue array slides containing 150 GC samples. In a total of 150
cases of GC, 46 failed to produce a clear signal for evaluation,
while 104 cases exhibited a clear signal that was able to be used
for enumerating analysis. EGFR protein expression in these 104
cases of GC was as follows: 41 cases scored 0, 45 cases scored 1+
and 18 scored 2+ or 3+. All EGFR signals were compared with signals
for centromeric probes for chromosome 7. EGFR amplification was
detected in 5.77% (6/104) of the cases, which exhibited red cluster
signals for EGFR (Fig. 2A). Four
cases demonstrated EGFR protein overexpression (2+/3+) and two
cases exhibited weak EGFR expression (1+). An increased gene copy
number due to polysomy was detected in 4.81% (5/104) of the cases
(Fig. 2B); three of these cases
demonstrated EGFR protein overexpression and two exhibited weak
expression. The other 93 cases that produced clear signals for
analysis possessed balanced EGFR and CEP7 copy numbers. In the EGFR
overexpression and EGFR weak expression groups, the frequencies of
gene copy number abnormalities were 38.89 (7/18) and 8.89% (4/45),
respectively. None of the 41 cases demonstrating negative EGFR
expression exhibited EGFR gene amplification or high polysomy.
Mann-Whitney U test analysis revealed that the correlation between
IHC and FISH results was statistically significant (P<0.001). It
was concluded that increased copy number of the EGFR gene was
associated with GC cases with high EGFR protein expression
levels.
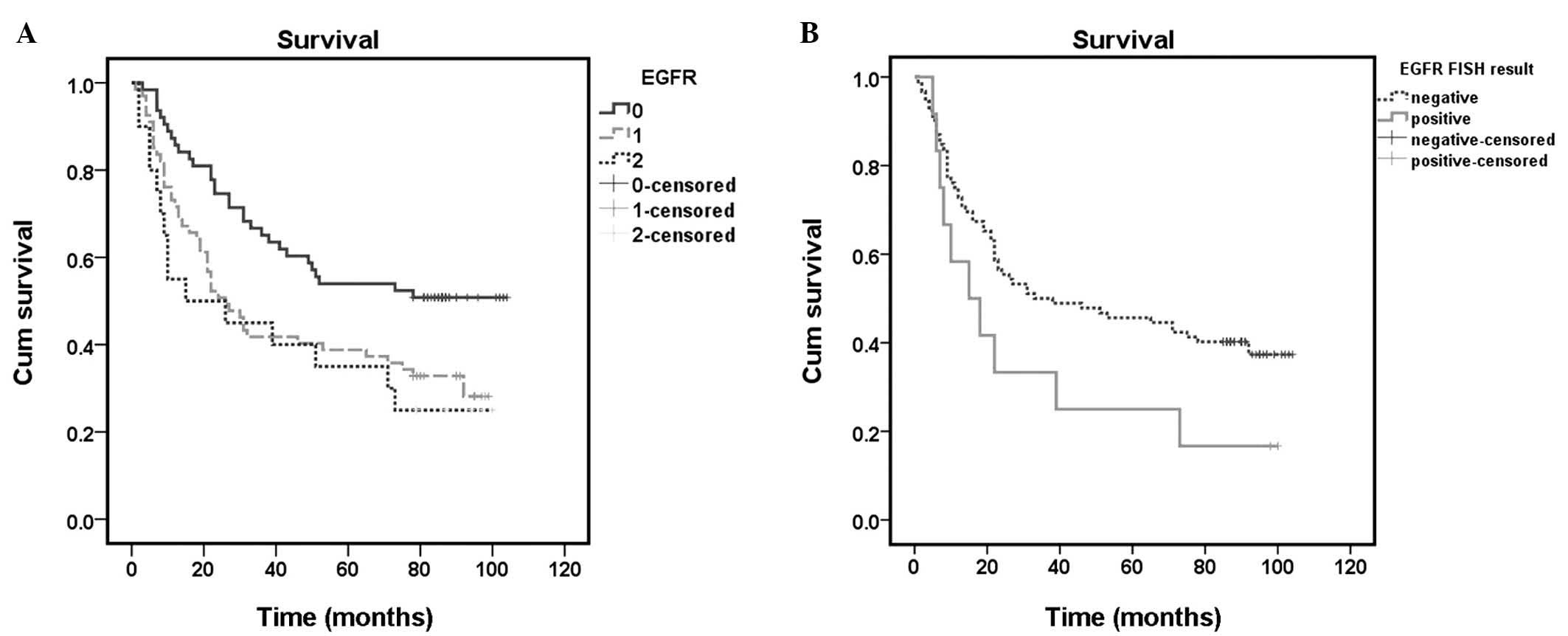
Survival analysis
The duration of follow-up was 1–104 months (mean,
48.9 months) subsequent to surgery; 92/150 patients (61.33%)
succumbed during this period. Univariate analysis revealed that
tumor diameter, site, depth of invasion, presence of lymph node or
distant metastases, TNM stage, EGFR expression and EGFR gene
amplification were associated with prognosis (Table II). Additional factors, including
gender, age and differentiation of GC, were not associated with
prognosis (P>0.05). The overall survival rate of patients
exhibiting negative EGFR expression, as determined using the
log-rank test, was significantly increased compared with the
survival rate of those patients demonstrating EGFR expression
(P=0.001; Fig. 3A). However, there
was no significant difference in survival rate between patients
exhibiting weak EGFR expression and EGFR overexpression. Patients
exhibiting EGFR FISH(+) GC possessed a less favorable prognosis
compared with those exhibiting EGFR FISH(−) GC (P=0.036; Fig. 3B).
 | Table II.Univariate analysis of EGFR status,
clinicopathological parameters and overall cancer survival in
subjects with gastric carcinoma. |
Table II.
Univariate analysis of EGFR status,
clinicopathological parameters and overall cancer survival in
subjects with gastric carcinoma.
|
|
|
| Overall
survival |
|---|
|
|
|
|
|
|---|
| Clinicopathological
parameter | n | Mean overall
survival, months | 95% CI | P-value |
|---|
| Gender |
|
|
|
0.747 |
|
Male | 122 | 52.643 |
37.978–67.308 |
|
|
Female | 28 | 54.869 |
47.441–62.297 |
|
| Age, years |
|
|
|
0.169 |
|
﹤65 | 81 | 58.508 |
49.011–68.001 |
|
|
≥65 | 69 | 50.886 |
41.558–60.213 |
|
| Diameter, cm |
|
|
|
0.005b |
| ﹤5 | 77 | 64.602 |
55.414–73.786 |
|
| ≥5 | 73 | 44.562 |
35.338–53.785 |
|
| Tumor location |
|
|
|
0.022b |
| Cardia
and fundus | 17 | 31.882 |
16.042–47.693 |
|
|
Body | 46 | 56.043 |
44.558–67.652 |
|
| Pylorus
and antrum | 87 | 58.437 |
49.453–67.420 |
|
|
Differentiation |
|
|
|
0.682 |
|
Well/moderate | 28 | 56.357 |
41.111–71.604 |
|
|
Poor | 110 | 55.138 |
47.481–62.795 |
|
|
Mucinousa | 12 | 43.583 |
18.995–68.172 |
|
| Invasion depth |
|
|
|
<0.000b |
|
Mucosa/submucosa | 12 | 80.583 |
65.241–95.925 |
|
|
Muscular/serosa | 25 | 84.880 |
71.800–97.960 |
|
| Out of
the serosa | 87 | 52.243 |
43.781–60.704 |
|
| Other
organs | 26 | 19.385 |
10.732–28.037 |
|
| Lymph node
metastases |
|
|
|
<0.000b |
| 0 | 43 | 72.519 |
60.360–84.679 |
|
|
1–6 | 54 | 57.630 |
47.716–67.543 |
|
| ≥7 | 53 | 35.679 |
25.672–45.687 |
|
| Distant
metastases |
|
|
|
<0.000b |
| − | 127 | 61.643 |
54.455–68.830 |
|
| + | 23 | 18.000 |
9.222–26.778 |
|
|
Tumor/Node/Metastasis stage |
|
|
|
<0.000b |
| I | 22 | 94.000 | 84.029–103.971 |
|
| II | 31 | 64.323 |
50.520–78.117 |
|
|
III | 67 | 53.836 |
44.441–63.261 |
|
| IV | 30 | 16.567 |
9.463–23.671 |
|
| EGFR |
|
|
|
0.011b |
| − | 67 | 66.635 |
56.692–76.578 |
|
| 1+ | 63 | 46.507 |
37.066–55.949 |
|
|
2+/3+ | 20 | 41.650 |
24.353–58.947 |
|
| EGFR gene
amplification and polysomy |
|
|
|
0.040b |
| − | 93 | 54.222 |
45.419–63.024 |
|
| + | 11 | 32.182 |
13.421–50.942 |
|
Clinicopathological parameters, including gender,
age, tumor diameter, location and differentiation, TNM stage and
EGFR expression were included in a multivariate analysis. The
results of this analysis revealed that EGFR expression, tumor
location and TNM stage were independent prognostic indicators of GC
(Table III). However, there were no
significant differences between subjects exhibiting EGFR FISH(+) GC
and those demonstrating EGFR FISH(−) GC (P=0.682) detected in the
multivariate analysis.
 | Table III.Multivariate analysis of predictive
indicators for survival of patients exhibiting gastric
carcinoma. |
Table III.
Multivariate analysis of predictive
indicators for survival of patients exhibiting gastric
carcinoma.
| Clinicopathological
parameter | B | Standard error | Walda | Mean RR | 95% CI of RR | P-value |
|---|
| Gender |
0.403 | 0.288 |
1.961 | 1.497 | 0.851–2.632 | 0.161 |
| Age |
0.191 | 0.224 |
0.730 | 1.211 | 0.781–1.878 | 0.393 |
| Diameter | −0.156 | 0.235 |
0.441 | 0.855 | 0.539–1.357 | 0.507 |
| Site | −0.442 | 0.168 |
6.930 | 0.643 | 0.462–0.893 | 0.008b |
|
Differentiation |
0.102 | 0.243 |
0.178 | 1.108 | 0.689–1.782 | 0.673 |
| Invasive depth |
0.341 | 0.239 |
2.033 | 1.407 | 0.880–2.248 | 0.154 |
| TNM stage |
0.806 | 0.202 | 16.005 | 2.240 | 1.509–3.325 | 0.000b |
| EGFR
expression |
0.387 | 0.150 |
6.640 | 1.473 | 1.097–1.976 | 0.010b |
Discussion
EGFR inhibitors are utilized in the management of a
number of solid malignant tumors, including CRC and metastatic
NSCLC (23,24). Due to the development of EGFR-targeted
treatments, the EGFR gene has been identified in a variety of
studies investigating numerous malignancies (25,26). In
the present study, EGFR protein expression and gene amplification
were systemically evaluated in GC samples from Chinese
patients.
IHC is the typical tool for the determination of
EGFR expression levels and for the identification of patients
likely to benefit from EGFR-targeted therapies in CRC and NSCLC
(27). In GC, EGFR protein expression
has been analyzed in several previous studies. Kim et al
(15) evaluated EGFR status in 511
Korean GC cases; 27.4% of these cases demonstrated EGFR
overexpression. Takehana et al (28) identified negative EGFR protein
expression in 89.6%, low levels in 8.2% and high levels of
expression in 2.2% of 413 GC specimens from Japanese patients.
Gamboa-Dominguez et al (14)
investigated EGFR status in 87 cases of GC from Mexican patients;
18.0% demonstrated moderate EGFR expression and 10.1% exhibited
strong EGFR expression. The results of the present study revealed
that EGFR expression was observed in 83/150 (55.33%) GC cases, and
20 (13.33%) cases demonstrated EGFR overexpression. In the present
and previous studies, the frequency of EGFR overexpression, as
revealed by IHC, ranged between 2–30%. Potential reasons for this
wide variation may include differences in fixation techniques,
antibodies and scoring systems used in IHC (29).
EGFR overexpression may occur as a result of the
presence of an increased gene copy number. The present study
investigated EGFR gene copy number using FISH analysis in tissue
array slides. A total of 104 cases of GC exhibited a positive
signal; while the remaining 46 cases failed to produce any signal.
This low rate of success in the FISH analysis may be a result of
the extended storage time of samples, as well as the tissue array
slides used in FISH analysis. A number of the wax tissue blocks had
been stored for >10 years prior to being utilized for the
present study. A longer storage time may lead to fewer positive
results, as the following factors may exhibit a considerable impact
on the preservation of DNA/mRNA: Oxidation, hydrolysis, sun or
light exposure, fixation time and type of fixative (30). This problem may be resolved by
punching multiple small cores from different regions to capture the
heterogeneity of the tumors more effectively. In addition, the
detection of oncogene amplification by fluorescent in situ
hybridization on tissue microarray may be a reliable tool for large
retrospective studies. Various tissues arranged in one tissue array
slide may require alternative experimental conditions to achieve a
positive result (31). Furthermore,
due to the heterogeneity of various areas of GC tumors, the rate of
increased gene copy number that was detected in the tissue array
slides may be lower than indicated according to the results of the
present study.
In these 104 cases of GC, EGFR gene amplification
was detected in six cases and polysomy in five. A number of EGFR
overexpression cases (7/18; 38.89%) demonstrated EGFR gene
amplification or high polysomy as revealed by FISH, whereas these
features were observed in only 4/45 cases (8.89%) exhibiting weak
EGFR expression. The present study confirmed that EGFR IHC scores
were significantly correlated with EGFR gene expression levels.
This observation also suggested that not all GC cases exhibiting
increased EGFR gene copy number demonstrated EGFR protein
overexpression; and that a number of cases exhibited weak
expression. Therefore, if patients possessing increased EGFR gene
copy number are sensitive to EGFR-targeted drugs, certain patients
demonstrating weak EGFR expression may also potentially benefit
from treatment with these drugs. Univariate analysis revealed that
EGFR overexpression and gene copy number were associated with
unfavorable prognoses. Multivariate analysis revealed that EGFR
expression was an independent prognostic indicator. The potential
value of the results of the present study is that they may
facilitate the identification of a subset of patients exhibiting
tumors that may be sensitive to EGFR-targeted therapy. Patients
exhibiting EGFR overexpression possess a poor prognosis, however
these patients may benefit from EGFR-targeted therapy.
The EGFR gene status of GC has been analyzed in
previous studies. Kim et al (15) evaluated the EGFR gene copy number in
GC tissues from 511 Korean patients; 13/21 (61.9%) cases
demonstrating EGFR overexpression also exhibited EGFR gene
amplification or increased polysomy, while only 14/119 (11.8%)
cases possessing weak EGFR expression exhibited EGFR gene
amplification or high polysomy. Liang et al (20) detected the EGFR gene copy number in
100 cases of GC; 16% of these GC specimens demonstrated positive
FISH results (20). These results
were consistent with those of the present study. The frequency of
increased EGFR gene copy number in GC is reduced compared with
certain other malignancies, including NSCLC, CRC and high-grade
gliomas (13,32,33).
Clinical trials have been undertaken to investigate
the effect of EGFR-targeting mAbs in GC. In a multicenter phase II
Japanese study, 13/75 metastatic GC patients receiving gefitinib
treatment achieved disease control (34). By contrast, a phase II trial of
erlotinib conducted in two groups of patients with gastroesophageal
junction (GEJ) or cardia and distal gastric adenocarcinomas
demonstrated that erlotinib is an effective treatment for patients
with GEJ adenocarcinomas, however, it appears ineffective for the
treatment of distal GC (35). An
additional phase III clinical trial (EXPAND) revealed a small
subset of patients that responded to treatment with cetuximab
(16). The results of these previous
studies emphasize the requirement for the identification of latent
responders.
In NSCLC and CRC, patients exhibiting EGFR
overexpression and/or an increased gene copy number have been
demonstrated to possess a positive response to EGFR-targeted
therapies for carcinoma (29,32,36). A
number of previous in vitro studies have suggested that GC
patients exhibiting EGFR overexpression or gene amplification may
benefit from EGFR-targeted therapy. Fukuda et al (37) identified that the combination of
5-fluorouracil and cetuximab synergistically inhibited cell
proliferation and exhibited an enhanced pro-apoptotic effect in GC
cells demonstrating EGFR overexpression. A preclinical trial
identified that GC patient-derived xenografts responded to
cetuximab, and efficacy was dependent on EGFR overexpression and
gene amplification (17). IHC
analysis of EGFR expression, including FISH analysis of the EGFR
gene, may be a favorable option for the identification of latent
patients, who may respond to EGFR-targeted therapies.
In conclusion, the results of the present study
provide evidence that EGFR expression may be significantly
associated with an unfavorable prognosis in GC. The present study
additionally identified that gene amplification and polysomy were
low frequency events in GC, although were associated with poor
prognosis. An increased copy number of the EGFR gene was
significantly correlated with protein overexpression. The results
of the present study therefore suggest that there is a potential
group of GC patients that may benefit from treatment with
EGFR-targeted agents.
Acknowledgements
This present study was supported by the Department
of Education, Liaoning Province (grant no. L2011157).
References
|
1
|
Amedei A, Benagiano M, della Bella C,
Niccolai E and D'Elios MM: Novel immunotherapeutic strategies of
gastric cancer treatment. J Biomed Biotechnol. 2011:4373482011.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Ferlay J, Shin HR, Bray F, Forman D,
Mathers C and Parkin DM: Estimates of worldwide burden of cancer in
2008: GLOBOCAN 2008. Int J Cancer. 127:2893–2917. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
De Vita F, Di Martino N, Fabozzi A,
Laterza MM, Ventriglia J, Savastano B, Petrillo A, Gambardella V,
Sforza V, Maranwo L, et al: Clinical management of advanced gastric
cancer: the role of new molecular drugs. World J Gastroenterol.
20:14537–14558. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Roviello F, Caruso S, Neri A and Marrelli
D: Treatment and prevention of peritoneal carcinomatosis from
gastric cancer by cytoreductive surgery and hyperthermic
intraperitoneal chemotherapy: Overview and rationale. Eur J Surg
Oncol. 39:1309–1316. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Yk W, Cf G, T Y, Z C, Xw Z, Xx L, Nl M and
Wz Z: Assessment of ERBB2 and EGFR gene amplification and protein
expression in gastric carcinoma by immunohistochemistry and
fluorescence in situ hybridization. Mol Cytogenet. 4:142011.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Ghosh MK, Sharma P, Harbor PC, Rahaman SO
and Haque SJ: PI3K-AKT pathway negatively controls EGFR-dependent
DNA-binding activity of Stat3 in glioblastoma multiforme cells.
Oncogene. 24:7290–7300. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Yu XT, Zhu SN, Xu ZD, Hu XQ, Zhu TF, Chen
JQ and Lu SL: Roles of EGFR-Stat3 signal pathway in carcinogenesis
of experimental hepatoma in rats. J Cancer Res Clin Oncol.
133:145–152. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Mok TS, Lee K and Leung L: Targeting
epidermal growth factor receptor in the management of lung cancer.
Semin Oncol. 41:101–109. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Ruzzo A, Graziano F, Canestrari E and
Magnani M: Molecular predictors of efficacy to anti-EGFR agents in
colorectal cancer patients. Curr Cancer Drug Targets. 10:68–79.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Liang JL, Ren XC and Lin Q: Treating
advanced non-small-cell lung cancer in Chinese patients: Focus on
icotinib. Onco Targets Ther. 7:761–770. 2014.PubMed/NCBI
|
|
11
|
Diasio RB and Fourie J: Targeting the
epidermal growth factor receptor in the treatment of colorectal
cancer: State of the art. Drugs. 66:1441–1463. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Lynch TJ, Bell DW, Sordella R,
Gurubhagavatula S, Okimoto RA, Brannigan BW, Harris PL, Haserlat
SM, Supko JG, Haluska FG, et al: Activating mutations in the
epidermal growth factor receptor underlying responsiveness of
non-small-cell lung cancer to gefitinib. N Engl J Med.
350:2129–2139. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Cappuzzo F, Hirsch FR, Rossi E, Bartolini
S, Ceresoli GL, Bemis L, Haney J, Witta S, Danenberg K, Domenichini
I, et al: Epidermal growth factor receptor gene and protein and
gefitinib sensitivity in non-small-cell lung cancer. J Natl Cancer
Inst. 97:643–655. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Gamboa-Dominguez A, Dominguez-Fonseca C,
Quintanilla-Martinez L, Reyes-Gutierrez E, Green D, Angeles-Angeles
A, Busch R, Hermannstädter C, Nährig J, et al: Epidermal growth
factor receptor expression correlates with poor survival in gastric
adenocarcinoma from Mexican patients: A multivariate analysis using
a standardized immunohistochemical detection system. Mod Pathol.
17:579–587. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Kim MA, Lee HS, Lee HE, Jeon YK, Yang HK
and Kim WH: EGFR in gastric carcinomas: Prognostic significance of
protein overexpression and high gene copy number. Histopathology.
52:738–746. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Lordick F, Kang YK, Chung HC, Salman P, Oh
SC, Bodoky G, Kurteva G, Volovat C, Moiseyenko VM, Gorbunova V, et
al: Arbeitsgemeinschaft Internistische Onkologie and EXPAND
Investigators: Capecitabine and cisplatin with or without cetuximab
for patients with previously untreated advanced gastric cancer
(EXPAND): A randomised, open-label phase 3 trial. Lancet Oncol.
14:490–499. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Zhang L, Yang J, Cai J, Song X, Deng J,
Huang X, Chen D, Yang M, Wery JP, Li S, et al: A subset of gastric
cancers with EGFR amplification and overexpression respond to
cetuximab therapy. Sci Rep. 3:29922013.PubMed/NCBI
|
|
18
|
Kim JG: Molecular targeted therapy for
advanced gastric cancer. Korean J Intern Med. 28:149–155. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Metzger B, Chambeau L, Begon DY, Faber C,
Kayser J, Berchem G, Pauly M, Boniver J, Delvenne P, Dicato M and
Wenner T: The human epidermal growth factor receptor (EGFR) gene in
European patients with advanced colorectal cancer harbors
infrequent mutations in its tyrosine kinase domain. BMC Med Genet.
12:1442011. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Liang Z, Zeng X, Gao J, Wu S, Wang P, Shi
X, Zhang J and Liu T: Analysis of EGFR, HER2, and TOP2A gene status
and chromosomal polysomy in gastric adenocarcinoma from Chinese
patients. BMC Cancer. 8:3632008. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Wang Y, Jiang CQ, Guan J, Yang GF, Yue JQ,
Chen HL, Xue JL, Xu ZG, Qian Q and Fan LF: Molecular alterations of
EGFR in small intestinal adenocarcinoma. Int J Colorectal Dis.
28:1329–1335. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Ciardiello F and Tortora G: Epidermal
growth factor receptor (EGFR) as a target in cancer therapy:
Understanding the role of receptor expression and other molecular
determinants that could influence the response to anti-EGFR drugs.
Eur J Cancer. 39:1348–1354. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Castañón E, Martín P, Rolfo C, Fusco JP,
Ceniceros L, Legaspi J, Santisteban M and Gil-Bazo I: Epidermal
Growth Factor Receptor targeting in non-small cell lung cancer:
revisiting different strategies against the same target. Curr Drug
Targets. 15:1273–1283. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Yarom N and Jonker DJ: The role of the
epidermal growth factor receptor in the mechanism and treatment of
colorectal cancer. Discov Med. 11:95–105. 2011.PubMed/NCBI
|
|
25
|
Lozano MD, Zulueta JJ, Echeveste JI,
Gúrpide A, Seijo LM, Martín-Algarra S, Del Barrio A, Pio R, Idoate
MA, Labiano T and Perez-Gracia JL: Assessment of epidermal growth
factor receptor and K-ras mutation status in cytological stained
smears of non-small cell lung cancer patients: Correlation with
clinical outcomes. Oncologist. 16:877–885. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Scartozzi M, Bearzi I, Mandolesi A,
Pierantoni C, Loupakis F, Zaniboni A, Negri F, Quadri A, Zorzi F,
Galizia E, et al: Epidermal Growth Factor Receptor (EGFR) gene copy
number (GCN) correlates with clinical activity of
irinotecan-cetuximab in K-RAS wild-type colorectal cancer: a
fluorescence in situ (FISH) and chromogenic in situ hybridization
(CISH) analysis. BMC Cancer. 9:303 View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Takehana T, Kunitomo K, Suzuki S, Kono K,
Fujii H, Matsumoto Y and Ooi A: Expression of epidermal growth
factor receptor in gastric carcinomas. Clin Gastroenterol Hepatol.
1:438–445. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Terashima M, Kitada K, Ochiai A, Ichikawa
W, Kurahashi I, Sakuramoto S, Katai H, Sano T, Imamura H and Sasako
M: ACTS-GC Group: Impact of expression of human epidermal growth
factor receptors EGFR and ERBB2 on survival in stage II/III gastric
cancer. Clin Cancer Res. 18:5992–6000. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Cappuzzo F, Finocchiaro G, Rossi E, Jänne
PA, Carnaghi C, Calandri C, Bencardino K, Ligorio C, Ciardiello F,
Pressiani T, et al: EGFR FISH assay predicts for response to
cetuximab in chemotherapy refractory colorectal cancer patients.
Ann Oncol. 19:717–723. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Economou M, Schöni L, Hammer C, Galván JA,
Mueller DE and Zlobec I: Proper paraffin slide storage is crucial
for translational research projects involving immunohistochemistry
stains. Clin Transl Med. 3:42014. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Brown LA and Huntsman D: Fluorescent in
situ hybridization on tissue microarrays: challenges and solutions.
J Mol Histol. 38:151–157. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Viana-Pereira M, Lopes JM, Little S,
Milanezi F, Basto D, Pardal F, Jones C and Reis RM: Analysis of
EGFR overexpression, EGFR gene amplification and the EGFRvIII
mutation in Portuguese high-grade gliomas. Anticancer Res.
28:913–920. 2008.PubMed/NCBI
|
|
33
|
Rojo F, Tabernero J, Albanell J, Van
Cutsem E, Ohtsu A, Doi T, Koizumi W, Shirao K, Takiuchi H, Cajal
Ramon YS and Baselga J: Pharmacodynamic studies of gefitinib in
tumor biopsy specimens from patients with advanced gastric
carcinoma. J Clin Oncol. 24:4309–4316. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Dragovich T, McCoy S, Fenoglio-Preiser CM,
Wang J, Benedetti JK, Baker AF, Hackett CB, Urba SG, Zaner KS,
Blanke CD and Abbruzzese JL: Phase II trial of erlotinib in
gastroesophageal junction and gastric adenocarcinomas: SWOG 0127. J
Clin Oncol. 24:4922–4927. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Gancberg D, Di Leo A, Rouas G, Järvinen T,
Verhest A, Isola J, Piccart MJ and Larsimont D: Reliability of the
tissue microarray based FISH for evaluation of the HER-2 oncogene
in breast carcinoma. J Clin Pathol. 55:315–317. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Lièvre A, Bachet JB, Le Corre D, Boige V,
Landi B, Emile JF, Côté JF, Tomasic G, Penna C, Ducreux M, et al:
KRAS mutation status is predictive of response to cetuximab therapy
in colorectal cancer. Cancer Res. 66:3992–3995. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Fukuda K, Saikawa Y, Takahashi M,
Takahashi T, Wada N, Kawakubo H, Takeuchi H and Kitagawa Y:
Antitumor effect of cetuximab in combination with S-1 in
EGFR-amplified gastric cancer cells. Int J Oncol. 40:975–982.
2012.PubMed/NCBI
|