Introduction
Colorectal cancer (CRC) is a type of malignant tumor
developed in the rectum or colon, and it is estimated that there
are currently >1 million people who are living with CRC in the
USA (1). It is the second leading
cause of cancer-associated mortalities among all cancer types that
affect both females and males (2). An
association has been identified between CRC and a relatively high
5-year survival rate at early stages of the disease, while the
survival is poor when distant and nodal metastasis is exhibited
(3,4).
The number of patients with metastatic status is up to 30%, and
these patients present a 5-year survival rate of <10% (5,6). In China,
the mortality associated with CRC increased by 17.9% between
1973–1975 and 2004–2005 (7). Thus, it
is of great importance to identify the genetic changes and
potential underlying mechanisms in CRC.
Genes involved in the metastasis process in CRC have
been extensively studied. Kirsten rat sarcoma viral oncogene
homolog (KRAS) mutations have been reported to be a
predictor of resistance to the monoclonal antibodies targeting the
epidermal growth factor receptor (EGFR) in metastatic CRC (8). In addition, B-Raf proto-oncogene,
serine/threonine kinase (BRAF), and phosphatase and tensin
homolog (PTEN) mutations are considered to be implications
for the targeted treatment in metastatic CRC (9). Furthermore, a previous study revealed
that the forced expression of chemokine (C-X-C motif) receptor 3
(CXCR3) promoted preferential metastasis to draining lymph
nodes in CRC, while CXCR3 knockdown significantly decreased
dissemination of cancer cells to the lungs and liver; thus,
CXCR3 may be a potential therapy against metastatic CRC
(10). The receptor of advanced
glycation end products is a prognostic biomarker of CRC metastasis
(11).
Furthermore, numerous metastasis-associated genes
have been screened by gene expression profiling. For instance,
Stange et al (12) used
microarray data to identify genes associated with CRC metastasis to
the liver. As a result, 163 unique genes were identified to be
significantly overexpressed, whereas 15 genes were significantly
downregulated (12). These genes,
including CYP2E1, CYP4A11, CRP, ORM1,
SAA1, APOA1, FGA and FGB, may be
associated with metabolism and inflammatory response. Del Rio et
al (13) identified the 33-gene
signature to classify the hepatic metastases, primary tumors,
normal colon mucosa and normal liver tissues, and indicated that
these genes may influence the CRC metastasis to the liver by
involving the extracellular matrix remodeling. However, the genes
identified in a single dataset may be limited if they have not been
confirmed in other datasets.
The present study aimed to use the data of Stange
et al (12) and Del Rio et
al (13) together, to further
detect the candidate metastasis-associated genes in CRC. The metaDE
package in R language, which implements 12 major meta-analyses in
differential expression screening (14), was used to screen the differentially
expressed genes (DEGs) between primary and metastatic cancer
samples in the two datasets. Functional enrichment was also
conducted for the significantly associated functions and pathways.
By calculating the standard deviations of common DEGs in another
dataset with clinical data, candidate metastasis-associated genes
were collected, followed by survival analysis.
Materials and methods
Microarray data
The Gene Expression Omnibus (GEO; http://www.ncbi.nlm.nih.gov/geo/) database was
retrieved for obtaining the microarray data with the following
accession numbers: GSE14297 (12),
GSE49355 (13,15) and GSE29621 (16). A total of 18 primary CRC and 18
matched liver metastasis samples were available in the GSE14297
dataset, based on the GPL6370 Illumina human-6 v2.0 expression
beadchip (extended). Similarly, the expression data of 20 primary
CRC and 19 matched liver metastasis samples in the GSE49335
dataset, based on GPL10430 Rosenstiel Fundulus heteroclitus
7K array, were downloaded. In addition, the GSE29621 dataset
included 46 primary CRC samples, 18 metastatic samples and 1 sample
with unknown metastatic status, based on the platform of GPL570
(HG_U133_Plus_2) Affymetrix Human Genome U133 Plus 2.0 Array.
Data processing
The gene expression profiles of GSE14297 were
prenormalized, while the raw CEL data (media.affymetrix.com/support/developer/powertools/changelog/gcos-agcc/cel.html)
in GSE49335 and GSE29621 were initially normalized using the robust
multi-array average method in R software, version 2.6.0 (R
Foundation for Statistical Computing, Vienna, Austria; www.R-project.org/). The median value of multiple
probes corresponding to a same gene was used as the expression
value.
Microarray meta-analysis for DEGs
Initially, the expression values obtained from the
GSE14297 and GSE49335 datasets were used to screen DEGs between
primary and metastatic CRC samples. To eliminate discrepancies, the
metaDE package in R language (14) in
R was used as a supplement for Fishers exact test. Next, clustering
analysis was performed to detect the distinguishing effect of
metaDE on differential expression in different sample groups. The
threshold for DEGs was a false discovery rate (FDR) of <0.05 in
metaDE, combined with a P value of <0.05 in Fisher's exact test.
P<0.05 and FDR<0.05 were considered to indicate a
statistically significant difference.
DEG enrichment analysis
In order to identify the significantly altered
functions and pathways during the metastasis of CRC, the online
tools in Database for Annotation, Visualization, and Integrated
Discovery (DAVID version 6.7; http://david.abcc.ncifcrf.gov/) were used for the
enrichment analysis. FDR<0.01 was set as the cut-off value for
the enrichment process.
Metastasis-associated genes in
CRC
Fold-changes (increase or decreased) in the
expression of the selected genes were investigated, and DEGs with a
fold-change of >2 were considered to be the
metastasis-associated genes in CRC. Common metastasis-associated
genes were obtained by comparing data in the profiles of GSE14297
and GSE49335. Next, the expression values of these common
metastasis-associated genes recorded in GSE29621 were obtained, in
order to calculate the standard deviation of common genes in all
the 65 samples. Genes with a high standard deviation in their
expression were defined as the candidate metastatic genes in
CRC.
Survival analysis
The clinical data of CRC patients were also obtained
from the GSE29621 dataset, and were subjected to survival analysis.
Using the log-rank test, the effects of metastasis and adjuvant
chemotherapy on survival were detected. Additionally, all the 65
samples were divided into the high expression and low expression
groups, according to the expression levels of the candidate
metastatic genes. Subsequently, the log-rank test was conducted to
identify the effect of these candidate genes on the survival of CRC
patients.
Results
Data processing and DEG screening
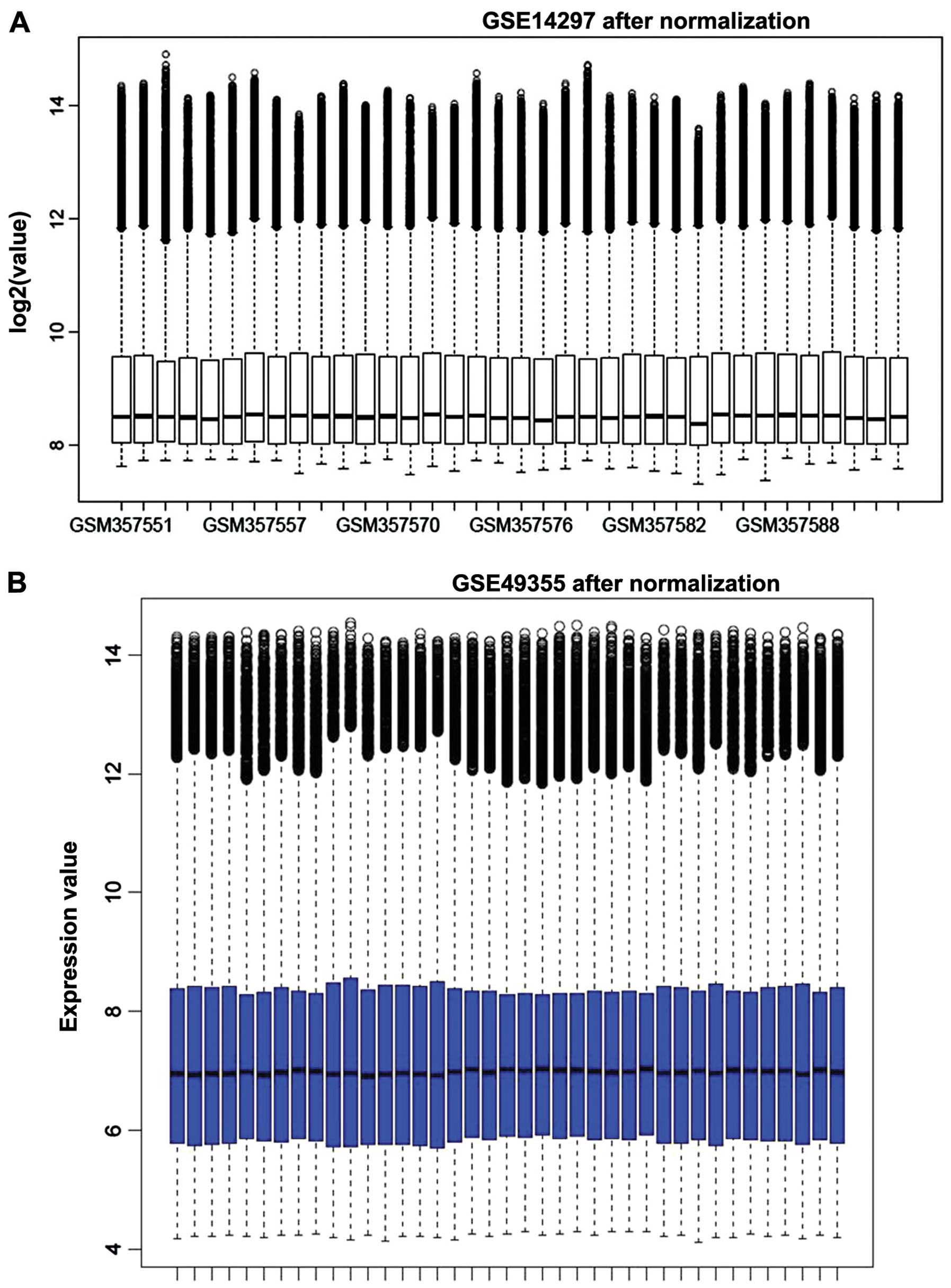
Initially, the expression profiles were normalized,
and the results are shown in Fig. 1.
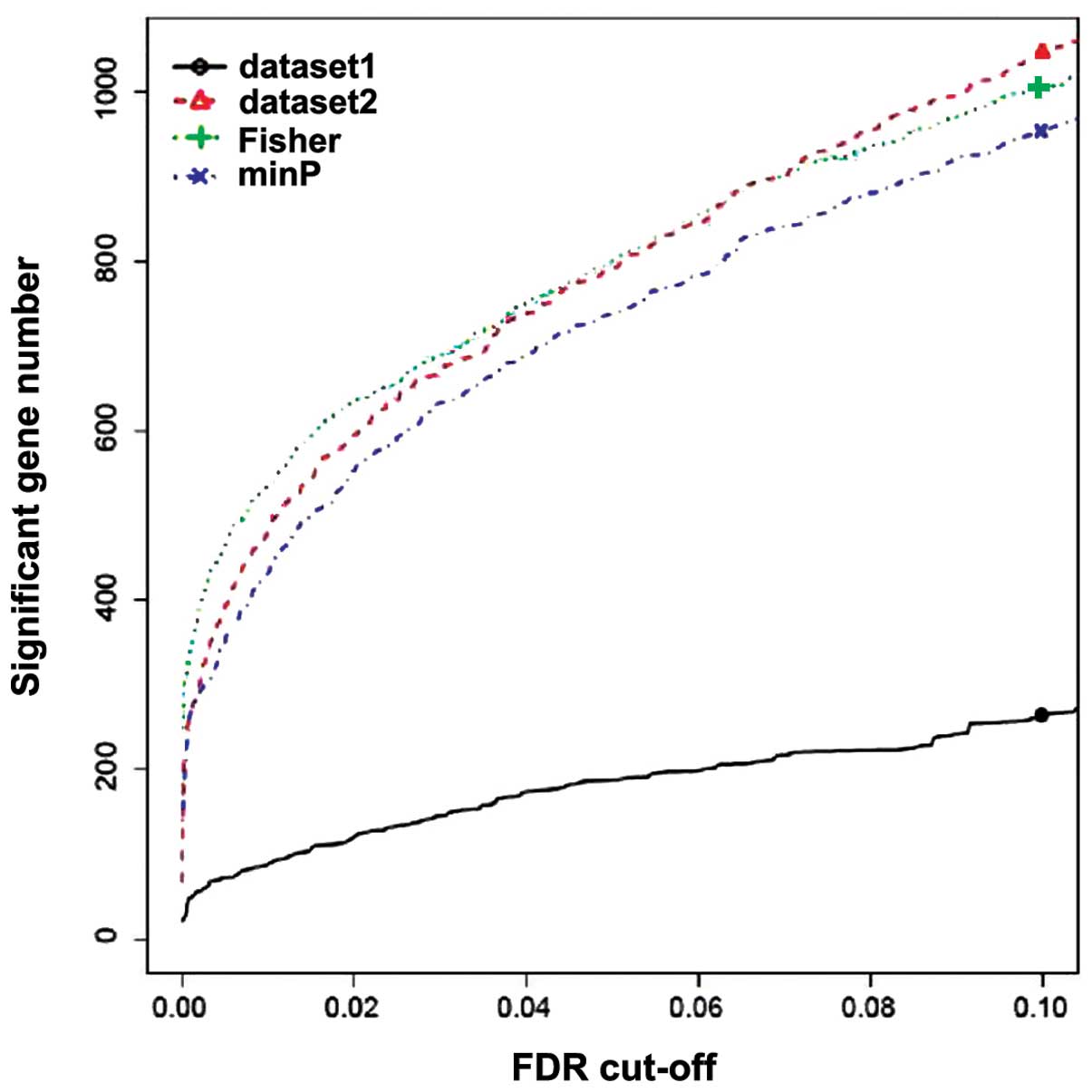
Using the threshold of FDR<0.05 in metaDE and P<0.05 in
Fisher's exact test, a total of 370 genes were identified to be
differentially expressed between the primary and metastatic CRC
samples in GSE14297 and GSE49335, among which 153 upregulated and
217 downregulated genes were detected. Fig. 2 shows the number of genes that are
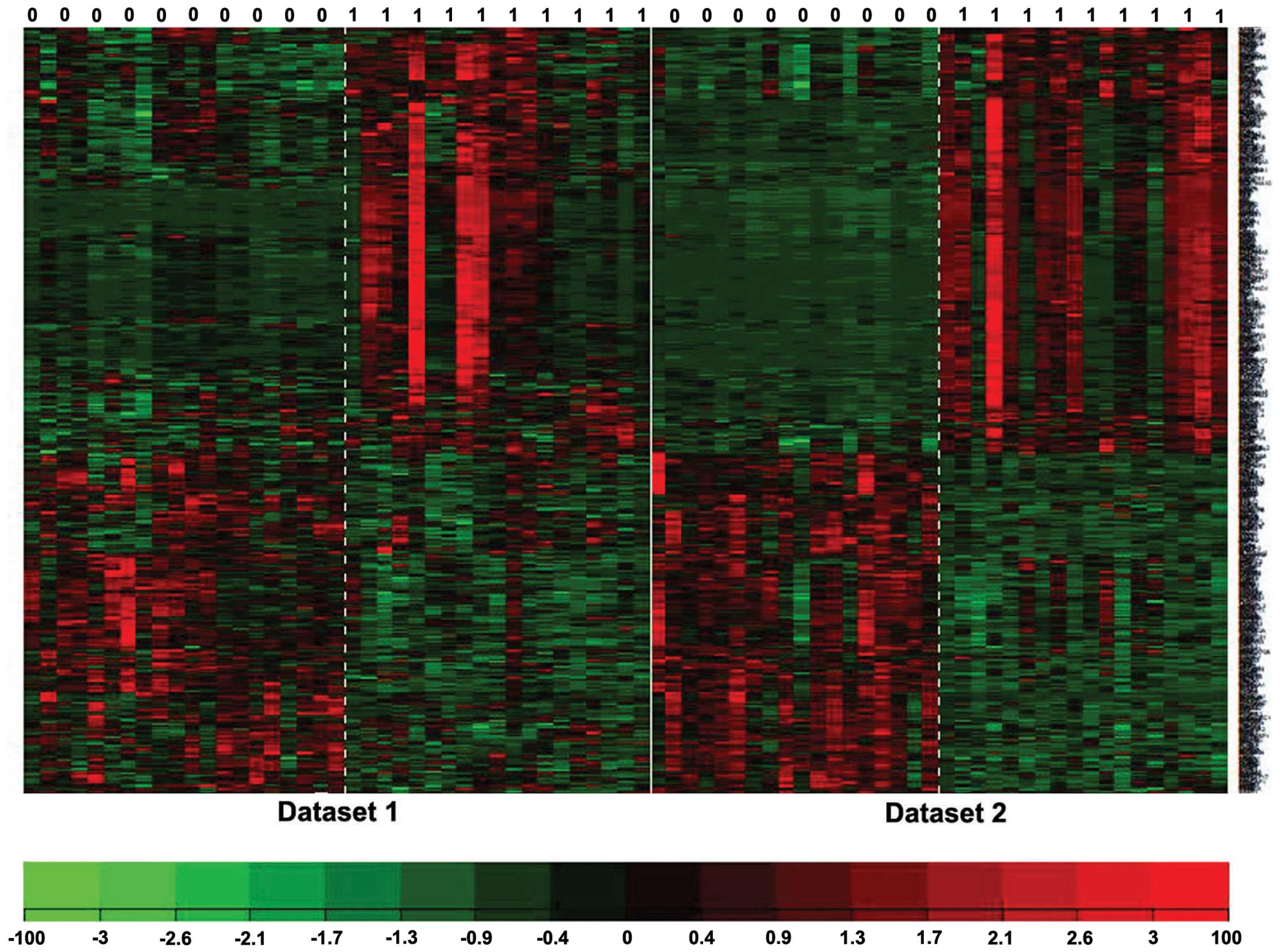
differentially expressed against various FDR values. The clustering
analysis results indicated that the DEGs selected using metaDE
analysis may be able to distinguish metastatic cancer samples from
non-metastatic samples (Fig. 3).
DEG enrichment analysis
DAVID was applied to investigate the function and
pathway enrichment. Upregulated genes were mainly enriched in
immune-associated functions, such as repose to wounding and
inflammatory response, as well as in metabolic process-associated
functions, including peptidase inhibitor activity and carbohydrate
binding. Complement and coagulation cascades, and drug metabolism
were among the significantly enriched pathways (Table I). Downregulated genes were
significantly enriched in the functions of vasculature development,
cell adhesion, biological adhesion and extracellular matrix
structure; however, no pathways were enriched (Table II).
 | Table I.Enriched functions and pathways of
upregulated genes in the GSE14927 and GSE40355 datasets. |
Table I.
Enriched functions and pathways of
upregulated genes in the GSE14927 and GSE40355 datasets.
| Category | Term | Count | FDR |
|---|
| GOTERM_BP_FAT | GO:0009611 -
response to wounding | 62 |
6.60×10−35 |
| GOTERM_BP_FAT | GO:0002526 - acute
inflammatory response | 31 |
1.14×10−28 |
| GOTERM_BP_FAT | GO:0006954 -
inflammatory response | 40 |
1.52×10−21 |
| GOTERM_BP_FAT | GO:0006952 -
defense response | 47 |
2.73×10−17 |
| GOTERM_BP_FAT | GO:0006956 -
complement activation | 17 |
5.18×10−16 |
| GOTERM_CC_FAT | GO:0005615 -
extracellular space | 72 |
3.85×10−38 |
| GOTERM_CC_FAT | GO:0005576 -
extracellular region | 106 |
1.02×10−32 |
| GOTERM_CC_FAT | GO:0044421 -
extracellular region part | 73 |
2.28×10−29 |
| GOTERM_CC_FAT | GO:0034364 -
high-density lipoprotein particle | 13 |
5.44×10−13 |
| GOTERM_CC_FAT | GO:0032994 -
protein-lipid complex | 14 |
1.92×10−12 |
| GOTERM_MF_FAT | GO:0004866 -
endopeptidase inhibitor activity | 25 |
1.14×10−15 |
| GOTERM_MF_FAT | GO:0030414 -
peptidase inhibitor activity | 25 |
4.15×10−15 |
| GOTERM_MF_FAT | GO:0004857 - enzyme
inhibitor activity | 30 |
4.14×10−14 |
| GOTERM_MF_FAT | GO:0004867 -
serine-type endopeptidase inhibitor activity | 18 |
2.68×10−11 |
| GOTERM_MF_FAT | GO:0030246 -
carbohydrate binding | 26 |
1.05×10−7 |
| KEGG_PATHWAY | hsa04610:
Complement and coagulation cascades | 31 |
1.22×10−28 |
| KEGG_PATHWAY | hsa00982: Drug
metabolism | 15 |
1.31×10−7 |
| KEGG_PATHWAY | hsa00980:
Metabolism of xenobiotics by cytochrome P450 | 13 |
1.32×10−5 |
| KEGG_PATHWAY | hsa00830: Retinol
metabolism | 11 |
4.99×10−4 |
 | Table II.Enriched functions of downregulated
genes in datasets of GSE14927 and GSE40355. |
Table II.
Enriched functions of downregulated
genes in datasets of GSE14927 and GSE40355.
| Category | Term | Count | FDR |
|---|
| GOTERM_BP_FAT | GO:0001944 -
vasculature development | 17 |
3.30×10−6 |
| GOTERM_BP_FAT | GO:0007155 - cell
adhesion | 26 |
1.42×10−5 |
| GOTERM_BP_FAT | GO:0022610 -
biological adhesion | 26 |
1.46×10−5 |
| GOTERM_BP_FAT | GO:0001568 - blood
vessel development | 15 |
1.42×10−4 |
| GOTERM_BP_FAT | GO:0048514 - blood
vessel morphogenesis | 13 |
1.31×10−3 |
| GOTERM_BP_FAT | GO:0042127 -
regulation of cell proliferation | 23 |
7.84×10−3 |
| GOTERM_CC_FAT | GO:0044421 -
extracellular region part | 53 |
2.54×10−22 |
| GOTERM_CC_FAT | GO:0005576 -
extracellular region | 72 |
1.22×10−21 |
| GOTERM_CC_FAT | GO:0031012 -
extracellular matrix | 29 |
8.38×10−15 |
| GOTERM_CC_FAT | GO:0005578 -
proteinaceous extracellular matrix | 28 |
1.31×10−14 |
| GOTERM_CC_FAT | GO:0005615 -
extracellular space | 31 |
8.12×10−9 |
| GOTERM_CC_FAT | GO:0044420 -
extracellular matrix part | 11 |
3.75×10−4 |
| GOTERM_CC_FAT | GO:0030934 -
anchoring collagen | 5 |
1.56×10−3 |
| GOTERM_CC_FAT | GO:0005581 -
collagen | 7 |
1.59×10−3 |
| GOTERM_MF_FAT | GO:0005201 -
extracellular matrix structural constituent | 11 |
1.11×10−5 |
Screening of candidate
metastasis-associated genes
Following the intersection of DEGs in the GSE49355
and GSE14927 datasets, a total of 77 common DEGs were obtained,
consisting of 17 downregulated and 59 upregulated genes. The
expression values of these 77 common DEGs in the GSE29621 dataset
were used to calculate the standard deviation, and 12 genes with a
standard deviation >1 were identified: carbonic anhydrase II
(CA2); carcinoembryonic antigen-related cell adhesion
molecule 7 (CEACAM7); chloride channel accessory 4
(CLCA4); CLCA1; CXC ligand 14 (CXCL14); Fc
fragment of immunoglobulin G binding protein (FCGBP);
immunoglobulin J polypeptide, linker protein for immunoglobulin α
and µ polypeptides (IGJ); lipocalin 2 (LCN2); matrix
metallopeptidase 1 (MMP1); MMP3; peptidase inhibitor
3, skin-derived (PI3); and placenta-specific 8
(PLAC8). A larger deviation value indicated great changes in
the samples; thus, these 12 genes were regarded as candidate
metastasis-associated genes, and may exert an intensive effect on
the survival of patients.
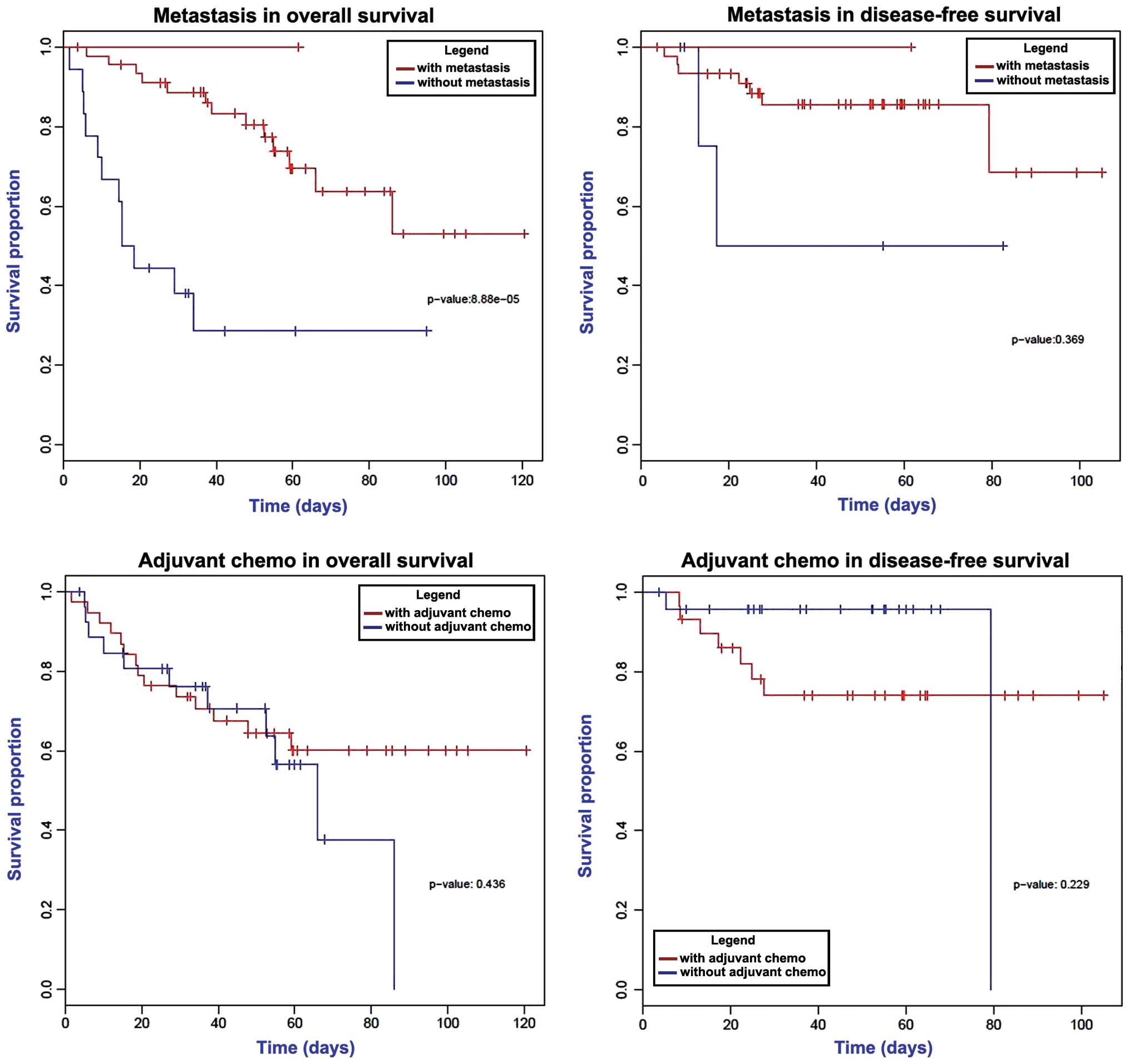
Survival analysis
Univariate survival analysis was conducted via the
log-rank test. As shown in Fig. 4, a
significantly decreased overall survival time was observed in
patients with metastasis (P<0.05), while the disease-free
survival times were not changed according to the metastatic status
of the cancer. Furthermore, adjuvant chemotherapy demonstrated no
statistically significant effect on the overall and disease-free
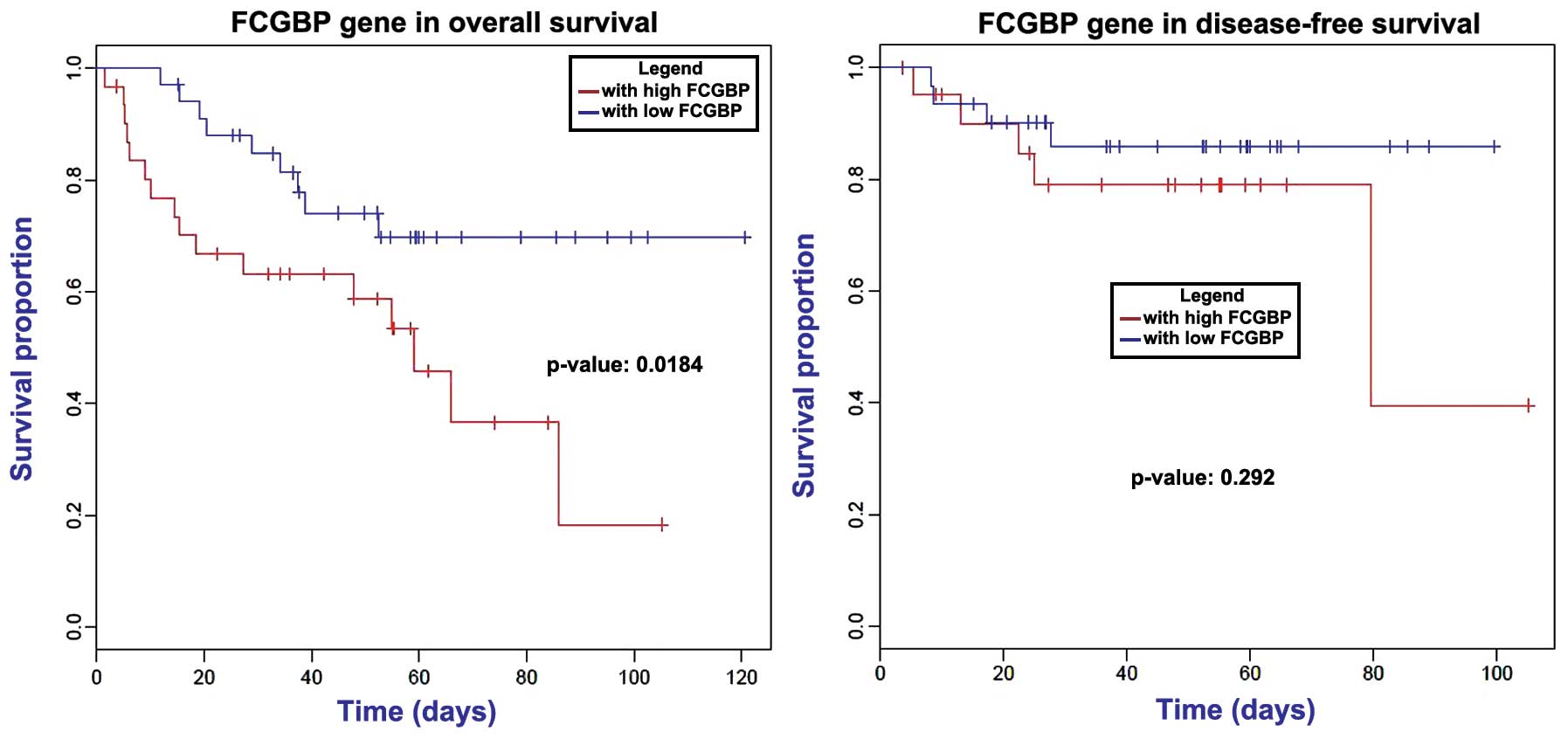
survival times (P>0.05). Analysis of the 12 candidate
metastasis-associated genes revealed that downregulation of
FCGBP markedly decreased the patients' overall survival time
(P=0.184); however, no effect was observed on the disease-free
survival time (Fig. 5).
Discussion
At present, metastasis remains an important factor
contributing towards the majority of cancer-associated mortalities,
as metastatic cancer is often resistant to conventional therapies
(17,18). However, reliable and sensitive methods
for detecting early metastasis in CRC are not currently available.
In the present study, gene expression data were initially analyzed
using the metaDE package in R language, in order to identify the
DEGs in CRC samples, and a total of 370 genes were identified as
DEGs between primary and metastatic cancer samples. In addition,
clustering analysis demonstrated the reliability of the identified
DEGs. By comparing the expression levels in different datasets, 12
genes were identified among the DEGs as the metastasis-associated
genes in CRC, including CA2, CEACAM7, CLCA4,
CLCA1, CXCL14, FCGBP, IGJ, LCN2,
MMP1, MMP3, PI3 and PLAC8.
Subsequently, FCGBP was demonstrated to influence the
patients' survival.
The carbonic anhydrase family is a set of proteins
that are important to the pH regulatory system. Carbonic anhydrase
IX (CA9) is overexpressed in numerous types of cancer cells,
and various agents targeting CA9 are currently in
preclinical or clinical development for cancer therapy (19,20).
CA9 is also a therapeutic target for metastasis (21). CA2 and CA9 were
demonstrated to be overexpressed in endometrial adenocarcinoma,
regulating the pH in the tumor microenvironment (22). Similarly, CA2 was identified as
a metastasis-associated gene in CRC in the present study.
Proteomics analysis identified CA2 as a potential biomarker,
which exerted significant inhibitory effects on cell growth in CRC
(23). CA2 expression was also
associated with lymph node metastasis in gastric cancer, and a
previous study reported that its reduction may contribute to tumor
metastasis (24). Therefore,
CA2 may also be used as a therapeutical target for
metastasis.
CEACAM7 is expressed in normal colon tissues,
but is downregulated in CRC (25). By
contrast, its expression is upregulated in liver metastases
(26). CEACAM6 also belongs to
the CEACAM subgroup, which is mainly associated with the cell
membrane. A previous study suggested that CEACAM6 plays a
role in tumor cell migration, adhesion and invasion, as well as the
formation of distant metastases (27). In addition, the expression of
CEACAM6 in CRC is an independent prognostic factor (28). Furthermore, downregulation of
CEACAM7 in early-stage adenomas represents certain
observable molecular events that may lead to CRC (29). In the present study, CEACAM7
was also identified as a metastasis-associated gene.
FCGBP has been reported to be implicated in
ulcerative colitis, which is a chronic inflammatory disease
predisposing to CRC (30,31). This gene may play a role in
anti-inflammation and cell protection in tissues (32). The mutation of FCGBP has been
previously identified in CRC (33),
while it was also identified to be a DEG in metastatic CRC in the
current study. Notably, in gallbladder adenocarcinoma, FCGBP
functions as an important maker for clinical prognosis (34). Using the univariate Cox model and
Kaplan-Meier survival curve, a correlation between FCGBP
expression and overall survival in ovarian adenocarcinoma was
observed (35). The survival curve of
CRC patients with liver metastasis in the present study also
detected an association of FCGBP with prognosis.
Furthermore, FCGBP was significantly enriched in the
function of cell adhesion. Metastasis is facilitated by the
cell-cell interactions between the endothelium and tumor cells in
distant tissues. Cell adhesion occurring in vasculature of specific
organs is an essential step in cancer metastasis (36). Thus, we infer that FCGBP may
exert an important role during metastasis in CRC by participating
in cell adhesion.
In conclusion, using the metaDE package in R
language, DEGs were screened in two sets of gene expression
profiles, which may be able to distinguish the metastasis samples
from the primary cancer samples. Candidate metastasis-associated
genes identified by comparing with a third expression dataset were
further subjected to survival analysis. Therefore, CA2 has
the potential to be used as a therapeutic target for metastatic
CRC, while FCGBP may affect patients' survival by
participating in cell adhesion.
References
|
1
|
National Cancer Institute. SEER Cancer
Statistics Factsheets: Colon and Rectum Cancer, National Cancer
Institute (Bethesda, MD, USA). simpleseer.cancer.gov/statfacts/html/colorect.html
|
|
2
|
U.S. Cancer Statistics Working Group:
United States cancer statistics: 1999–2008 incidence and mortality
web-based report. U.S. Department of Health and Human Services.
Centers for Disease Control and Prevention and National Cancer
Institute (Atlanta, GA). 2012.
|
|
3
|
Lansdorp-Vogelaar I, van Ballegooijen M,
Zauber AG, Habbema JDF and Kuipers EJ: Effect of rising
chemotherapy costs on the cost savings of colorectal cancer
screening. J Natl Cancer Inst. 101:1412–1422. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Pickhardt PJ, Hassan C, Halligan S and
Marmo R: Colorectal cancer: CT colonography and colonoscopy for
detection - systematic review and meta-analysis. Radiology.
259:393–405. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Di Nicolantonio F, Martini M, Molinari F,
Sartore-Bianchi A, Arena S, Saletti P, De Dosso S, Mazzucchelli L,
Frattini M, Siena S, et al: Wild-type BRAF is required for response
to panitumumab or cetuximab in metastatic colorectal cancer. J Clin
Oncol. 26:5705–5712. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Hoff PM, Ansari R, Batist G, Cox J, Kocha
W, Kuperminc M, Maroun J, Walde D, Weaver C, Harrison E, et al:
Comparison of oral capecitabine versus intravenous fluorouracil
plus leucovorin as first-line treatment in 605 patients with
metastatic colorectal cancer: Results of a randomized phase III
study. J Clin Oncol. 19:2282–2292. 2001.PubMed/NCBI
|
|
7
|
Zhou M, Wang X, Hu J, et al: Geographical
distribution of cancer mortality in China, 2004–2005. Zhonghua yu
fang yi xue za zhi. Chin J Prev Med. 44:303–308. 2010.(In
Chinese).
|
|
8
|
Zhang W, Winder T, Ning Y, Pohl A, Yang D,
Kahn M, Lurje G, Labonte MJ, Wilson PM, Gordon MA, et al: A let-7
microRNA-binding site polymorphism in 3-untranslated region of KRAS
gene predicts response in wild-type KRAS patients with metastatic
colorectal cancer treated with cetuximab monotherapy. Ann Oncol.
22:104–109. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
De Roock W, De Vriendt V, Normanno N,
Ciardiello F and Tejpar S: KRAS, BRAF, PIK3CA, and PTEN mutations:
Implications for targeted therapies in metastatic colorectal
cancer. Lancet Oncol. 12:594–603. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Murakami T, Kawada K, Iwamoto M, Akagami
M, Hida K, Nakanishi Y, Kanda K, Kawada M, Seno H, Taketo MM, et
al: The role of CXCR3 and CXCR4 in colorectal cancer metastasis.
Int J Cancer. 132:276–287. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Dahlmann M, Okhrimenko A, Marcinkowski P,
Osterland M, Herrmann P, Smith J, Heizmann CW, Schlag PM and Stein
U: RAGE mediates S100A4-induced cell motility via MAPK/ERK and
hypoxia signaling and is a prognostic biomarker for human
colorectal cancer metastasis. Oncotarget. 5:3220–3233. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Stange DE, Engel F, Longerich T, Koo BK,
Koch M, Delhomme N, Aigner M, Toedt G, Schirmacher P, Lichter P, et
al: Expression of an ASCL2 related stem cell signature and IGF2 in
colorectal cancer liver metastases with 11p15.5 gain. Gut.
59:1236–1244. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Del Rio M, Mollevi C, Vezzio-Vie N, Bibeau
F, Ychou M and Martineau P: Specific extracellular matrix
remodeling signature of colon hepatic metastases. PLoS One.
8:e745992013. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Chen DT, Hernandez JM, Shibata D, McCarthy
S, Humphries LA, Clark W, Elahi A, Gruidl M, Coppola D and Yeatman
T: Complementary strand microRNAs mediate acquisition of metastatic
potential in colonic adenocarcinoma. J Gastrointest Surg.
6:905–912. 2012. View Article : Google Scholar
|
|
15
|
Del Rio M, Molina F, Bascoul-Mollevi C,
Copois V, Bibeau F, Chalbos P, Bareil C, Kramar A, Salvetat N,
Fraslon C, et al: Gene expression signature in advanced colorectal
cancer patients select drugs and response for the use of
leucovorin, fluorouracil, and irinotecan. J Clin Oncol. 25:773–780.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Wang X, Kang DD, Shen K, Song C, Lu S,
Chang LC, Liao SG, Huo Z, Tang S, Ding Y, et al: An R package suite
for microarray meta-analysis in quality control, differentially
expressed gene analysis and pathway enrichment detection.
Bioinformatics. 28:2534–2536. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Kroon J, in't Veld LS, Buijs JT, Cheung H,
van der Horst G and van der Pluijm G: Glycogen synthase kinase-3β
inhibition depletes the population of prostate cancer
stem/progenitor-like cells and attenuates metastatic growth.
Oncotarget. 5:8986–8994. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Li Y, Rogoff HA, Keates S, Gao Y,
Murikipudi S, Mikule K, Leggett D, Li W, Pardee AB and Li CJ:
Suppression of cancer relapse and metastasis by inhibiting cancer
stemness. Proc Nat Acad Sci USA. 112:1839–1844. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
McDonald PC, Winum J-Y, Supuran CT and
Dedhar S: Recent developments in targeting carbonic anhydrase IX
for cancer therapeutics. Oncotarget. 3:84–97. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Lou Y, McDonald PC, Oloumi A, Chia S,
Ostlund C, Ahmadi A, Kyle A, dem Keller Auf U, Leung S, Huntsman D,
et al: Targeting tumor hypoxia: Suppression of breast tumor growth
and metastasis by novel carbonic anhydrase IX inhibitors. Cancer
Res. 71:3364–3376. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Tafreshi NK, Lloyd MC, Bui MM, Gillies RJ
and Morse DL: Carbonic anhydrase IX as an imaging and therapeutic
target for tumors and metastases. Carbonic Anhydrase. Mechanism.
Regulation, Links to Disease, and Industrial Applications. Frost SC
and McKenna R: (1st). (New York, NY). Springer. 221–254. 2014.
|
|
22
|
Hynninen P, Parkkila S, Huhtala H,
Pastorekova S, Pastorek J, Waheed A, Sly WS and Tomas E: Carbonic
anhydrase isozymes II, IX, and XII in uterine tumors. APMIS.
120:117–129. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Zhou R, Huang W, Yao Y, Wang Y, Li Z, Shao
B, Zhong J, Tang M, Liang S, Zhao X, et al: CA II, a potential
biomarker by proteomic analysis, exerts significant inhibitory
effect on the growth of colorectal cancer cells. Int J Oncol.
43:611–621. 2013.PubMed/NCBI
|
|
24
|
Li XJ, Xie HL, Lei SJ, Cao HQ, Meng T-Y
and Hu YL: Reduction of CAII expression in gastric cancer:
Correlation with invasion and metastasis. Chin J Cancer Res.
24:196–200. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Thompson J, Seitz M, Chastre E, Ditter M,
Aldrian C, Gespach C and Zimmermann W: Down-regulation of
carcinoembryonic antigen family member 2 expression is an early
event in colorectal tumorigenesis. Cancer Res. 57:1776–1784.
1997.PubMed/NCBI
|
|
26
|
Kleivi K, Lind GE, Diep CB, Meling GI,
Brandal LT, Nesland JM, Myklebost O, Rognum TO, Giercksky KE,
Skotheim RI, et al: Gene expression profiles of primary colorectal
carcinomas, liver metastases, and carcinomatoses. Mol Cancer.
6:22007. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Blumenthal RD, Leon E, Hansen HJ and
Goldenberg DM: Expression patterns of CEACAM5 and CEACAM6 in
primary and metastatic cancers. BMC Cancer. 7:22007. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Jantscheff P, Terracciano L, Lowy A,
Glatz-Krieger K, Grunert F, Micheel B, Brümmer J, Laffer U, Metzger
U, Herrmann R, et al: Expression of CEACAM6 in resectable
colorectal cancer: A factor of independent prognostic significance.
J Clin Oncol. 21:3638–3646. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Schölzel S, Zimmermann W, Schwarzkopf G,
Grunert F, Rogaczewski B and Thompson J: Carcinoembryonic antigen
family members CEACAM6 and CEACAM7 are differentially expressed in
normal tissues and oppositely deregulated in hyperplastic
colorectal polyps and early adenomas. Am J Pathol. 156:595–605.
2000. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Kim M, Lee S, Yang S-K, Song K and Lee I:
Differential expression in histologically normal crypts of
ulcerative colitis suggests primary crypt disorder. Oncol Rep.
16:663–670. 2006.PubMed/NCBI
|
|
31
|
Risques RA, Lai LA, Himmetoglu C, Ebaee A,
Li L, Feng Z, Bronner MP, Al-Lahham B, Kowdley KV, Lindor KD, et
al: Ulcerative colitis-associated colorectal cancer arises in a
field of short telomeres, senescence, and inflammation. Cancer Res.
71:1669–1679. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Selbach M and Mann M: Protein interaction
screening by quantitative immunoprecipitation combined with
knockdown (QUICK). Nat Methods. 3:981–983. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Spisák S, Kalmár A, Galamb O, Wichmann B,
Sipos F, Péterfia B, Csabai I, Kovalszky I, Semsey S, Tulassay Z,
et al: Genome-wide screening of genes regulated by DNA methylation
in colon cancer development. PLoS One. 7:e462152012. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Xiong L, Wen Y, Miao X and Yang Z: NT5E
and FcGBP as key regulators of TGF-1-induced epithelial-mesenchymal
transition (EMT) are associated with tumor progression and survival
of patients with gallbladder cancer. Cell Tissue Res. 355:365–374.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Choi CH, Choi JJ, Park YA, Lee YY, Song
SY, Sung CO, Song T, Kim MK, Kim TJ, Lee JW, et al: Identification
of differentially expressed genes according to chemosensitivity in
advanced ovarian serous adenocarcinomas: Expression of GRIA2
predicts better survival. Br J Cancer. 107:91–99. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Bendas G and Borsig L: Cancer cell
adhesion and metastasis: selectins, integrins, and the inhibitory
potential of heparins. Int J Cell Biol. 2012:6767312012. View Article : Google Scholar : PubMed/NCBI
|