Introduction
Gastrointestinal stromal tumors (GISTs) are the most
frequent mesenchymomas of the gastrointestinal tract with a smooth
muscle origin. The application of immunohistochemistry to the study
of GISTs, provides novel insights into the disorder, revealing the
contribution of the interstitial cells of Cajal, the spindle cells
of the gut wall (1–4). The majority of GISTs are located within
the stomach (50–70%) or the small intestine (20–30%), with as few
as 10% of the tumors developing in the rectum, and only 5%
developing in the large intestine, the retroperitoneal space and a
variety of other locations (i.e., appendix and pancreas) (5,6). These
tumors are even less frequent within the mesentery, omentum and
esophagus (7).
The age of onset for GIST patients is broad, but the
tumors commonly occur at 50–60 years. Precise GIST diagnostics
became possible only after the 1998 discovery of the c-kit
proto-oncogene and cluster of differentiation (CD)117 protein
overexpression in the tumor cells (8). The biological nature of these tumors,
indispensably link the activity of v-kit Hardy-Zuckerman 4 feline
sarcoma viral oncogene homolog (KIT) kinase and platelet-derived
growth factor receptor α (PDGFRA) in cancer progression
(9–11). Notably, 10–15% of tumor cases are not
associated with these genes (designated as KIT/PDGFRA
wild-type), but rather to different carcinogenesis contributors,
such as the succinate dehydrogenase complex and mutations of
neurofibromin 1, B-Raf proto-oncogene, serine/threonine kinase or
Kirsten rat sarcoma viral oncogene homolog kinase (12,13). This
broad genetic heterogeneity of GIST highlights the complexity of
the tumor origin, but more importantly, further affects the varied
responsiveness of GISTs to treatment with the tyrosine kinase
inhibitors (14).
Meckel's diverticulum (MD) is the most frequent
congenital defect of the small intestine and is present in ~2% of
the general population. Neoplasms of MD are diagnosed only in
0.5–3.2% of the population carrying this anatomical defect
(15). To date, GISTs of MD origin
have not been investigated thoroughly at the genomic level.
However, a few case studies have described the tumor tissue
examination, showing positive immunohistochemistry reactions for
vimentin and c-kit, therefore indicating the GIST nature (15–17). A
previous epidemiological study on 163 MD cases has indicated that
MD is a cancer ‘hot-spot’, comprising an attractive location for
tumor development (18). Moreover,
when comparing different types of cancer, an apparent preponderance
of adenocarcinomas versus malignant carcinoids (2:1 ratio) was
observed.
The present study attempted to comprehensively
explore and reveal the genetic nature of the rare cancer tissue
located in MD and investigate its GIST origin.
Patients and methods
Patient
A 71-year-old woman reported to the Obstetrics and
Gynecology Emergency Room (Clinical Hospital, Poznan, Poland) on
December 3, 2011, due to newly occurring postmenopausal bleeding. A
biopsy of the endometrium was performed and the material obtained
was assessed by the Pathology Laboratory. The results of the
histopathological examination indicated endometrioid adenocarcinoma
(G1). Following the diagnosis, the patient was admitted to the
hospital for further treatment. Subsequent to being admitted to the
Surgical Gynecology Clinic of the Gynecological and Obstetrics
Clinical Hospital, the patient underwent a gynecological
examination and a transvaginal ultrasound. Following a cardiology
consultation, the patient qualified for elective surgical
treatment. During the surgery, the entire uterus with adnexa was
removed, and a solid tumor of ~3 cm in size, which was previously
not visible in the ultrasound image, was found in MD. The procedure
included a complete resection of the tumor along with the
diverticulum and an end to end intestinal anastomosis. Tissue
samples were sent for a histopathological examination. No
intraoperative examinations were performed. There were no
complications in the post-operative period or during recovery
following the intestinal anastomosis. The patient was discharged
from the hospital on the 7th day after the surgery in a good
general condition. The patient was advised to await the
histopathological examination results and was informed that a
decision concerning any further course of treatment would be made
based on these results.
Genetic examination
Once the treatment was completed, advanced molecular
testing was employed in order to identify tumor-specific genome
changes. Briefly, the Genome-Wide Human CytoScan HD Array and
CytoScan 750K (Affymetrix, Santa Clara, CA, USA) was used to
analyze genomic alterations in the tumor sample. Genomic DNA was
obtained from formalin-fixed paraffin-embedded (FFPE) sections
using conventional processing (deparaffinization, enzymatic
treatment and DNA extraction) (19).
The 250 ng of genomic DNA from the tumor was subjected to
microarray examination according to the manufacturer's protocols as
follows: i) Digestion with the restriction enzyme NspI; ii)
adapter ligation, polymerase chain reaction (PCR) amplification and
magnetic bead purification; iii) fragmentation and end-labeling
with biotin; iv) washing and staining using a GeneChip® Fluidics
Station 450; and v) scanning using an Affymetrix GeneChip Scanner
3000 7G (Affymetrix). Scanned data files were generated using
Affymetrix GeneChip Command Console Software, version 1.2, and
analyzed with Affymetrix Chromosome Analysis Suite v 2.0.0.195
(Affymetrix). To calculate the copy number of altered regions, the
data were normalized to baseline reference intensities using NA
32.3 FFPE v.2 reference model (Affymetrix). The hidden Markov model
available within the software package was used to determine the
copy number states (CN) and their breakpoints. Thresholds of
log2 ratio ≥0.58 and ≤1 were used to categorize altered
regions as CN variation (CNV) gains (amplifications) and copy
number losses (deletions), respectively. To prevent the detection
of false-positive CNVs arising due to microarray unspecific
signals, only regions that involved at least 50 consecutive probes
were considered in the analysis of gains or losses in this study.
Amplifications and deletions were analyzed separately. To exclude
aberrations representing common normal CNVs, all the identified
CNVs were compared with those reported in the Database of Genomic
Variants (http://projects.tcag.ca/variation/). To identify the
genes involved in the CNVs further, the UCSC database (http://genome.ucsc.edu) and Ensemble (http://www.ensembl.org) were used. Gene annotation and
gene overlap were determined using the human genome build 19 and
NetAffx (http://www.affymetrix.com). In
addition, the identified alterations were compared with COSMIC
database (http://cancer.sanger.ac.uk) (20) to look for overlap with up-to-date,
known genomic cancer regions and single cancer genes. The algorithm
for the detection of copy number aberrations in tumor cell mixtures
(mosaicism and clonality) considers the comprehensive analysis of
adjacent single copy deletions and gains segments. The algorithm is
designed to be most accurate when the normal/expected CN is diploid
and targets the detection of changes in regions of ~5 Mb or more in
size and variation with a minimum of 500 markers (being typical for
segments of 5,000 markers or more). This approach considers only a
discrete number of mosaicism levels, which are set at 30, 50 and
70%. The range of log ratios is broken into a series of bands
according to the detection level (≥30, ≥50% or 70–100% bands) and
log ratios within each band denote a specific copy number change
event. This tool is most efficient in detecting mosaicism between
30–70% of cells and for copy numbers between 1 and 3. Selected
regions were validated in the present study using quantitative PCR
(2−ΔΔCq; KIT gene).
Results
Clinical findings
The results of the histopathological examination
indicated endometrioid adenocarcinoma (G1) and a uterine leiomyoma;
GIST, T2 (immunophenotype: CD117+, CD34+ and
vimentin+). Laboratory tests, a gynecological
examination and transvaginal ultrasound were performed during the
next hospital stay. No indications for chemotherapy were
identified, the radical nature of the surgery was confirmed, as
well as the complete removal of the lesion. After a single day of
in-patient treatment, the patient was discharged in a good general
condition and was recommended to undergo whole-body positron
emission tomography-computed tomography. The scan was performed 12
months after the last hospitalization. No pathological tracer
uptake areas were identified. At present, the patient is under the
care of the Oncology Outpatient Clinic. No episodes of recurrence
had been identified at the time of preparing the manuscript for
this case study.
Genomic studies
The strategy for the reliable detection of
chromosomal rearrangements assumes usage of the CytoScan 750K chip
in the first step. This investigation resulted in detection of ~62
genomic imbalances that were not filtered out and passed the
genomic criteria (>400 kb; 50 markers). To confirm abnormalities
and fine mapping of the borders for genomic segments, a
high-resolution chip Cytoscan HD (2.7 million probes and high
coverage of 522 cancer genes) was applied. The unbiased density of
probes on the chip enabled improvements to the accuracy and
confidence of detected regions. A total of 45 previously detected
regions were confirmed, and 17 other segments were disregarded as
false-positives (signal noise, more rigorous filtering criteria
compared with CytoScan 750K chip and usage of FFPE reference model
for Cytoscan HD that is far more accurate). In total, the cancer
tissue revealed alterations on 6 chromosomes (chromosomes 7, 8, 10,
12, 13 and 19) and 4 entire chromosome aneuploidies (chromosomes 5,
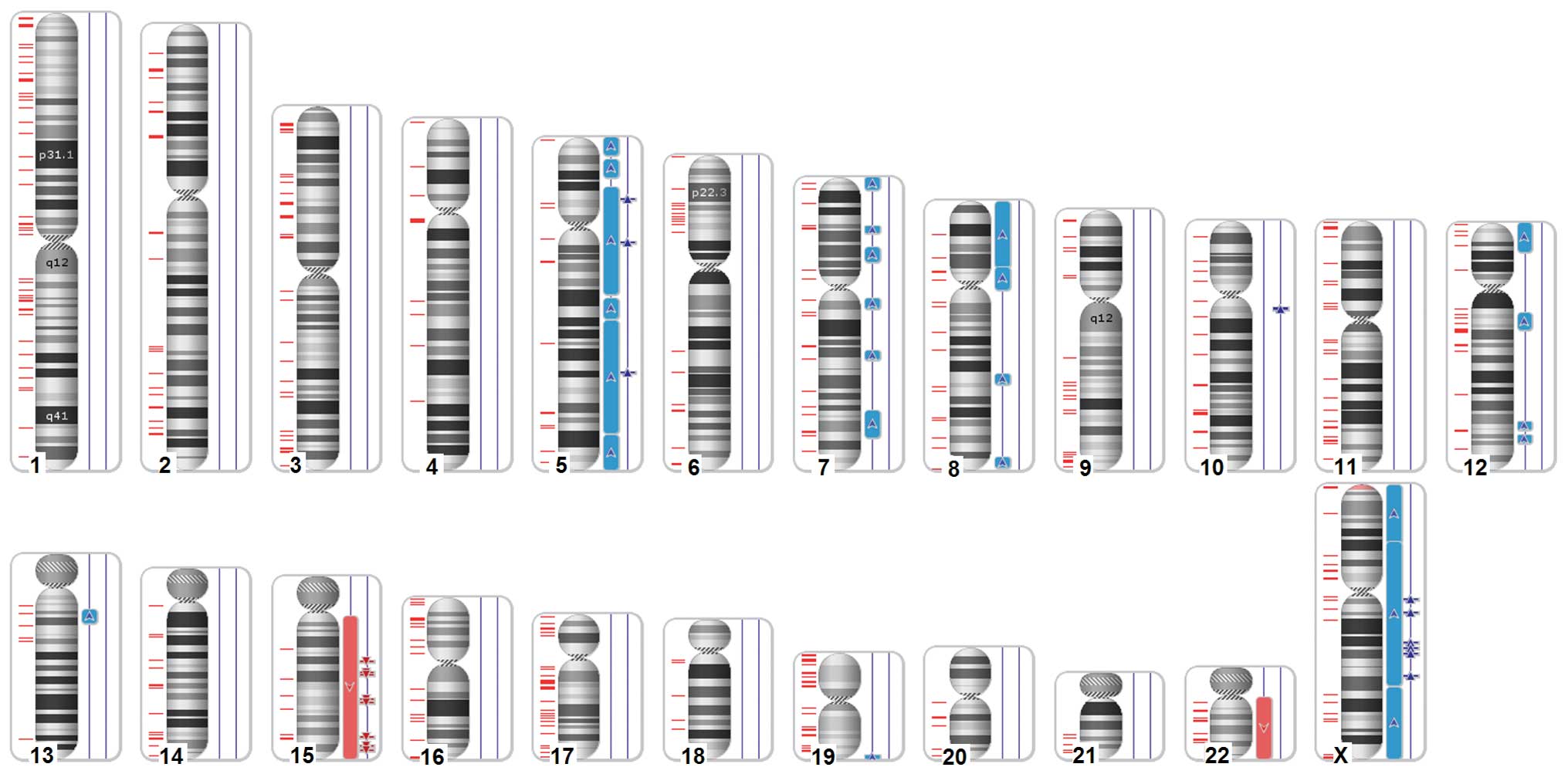
15, 22 and X). A dedicated karyogram illustrating the localization
of each segment is shown in Fig. 1,
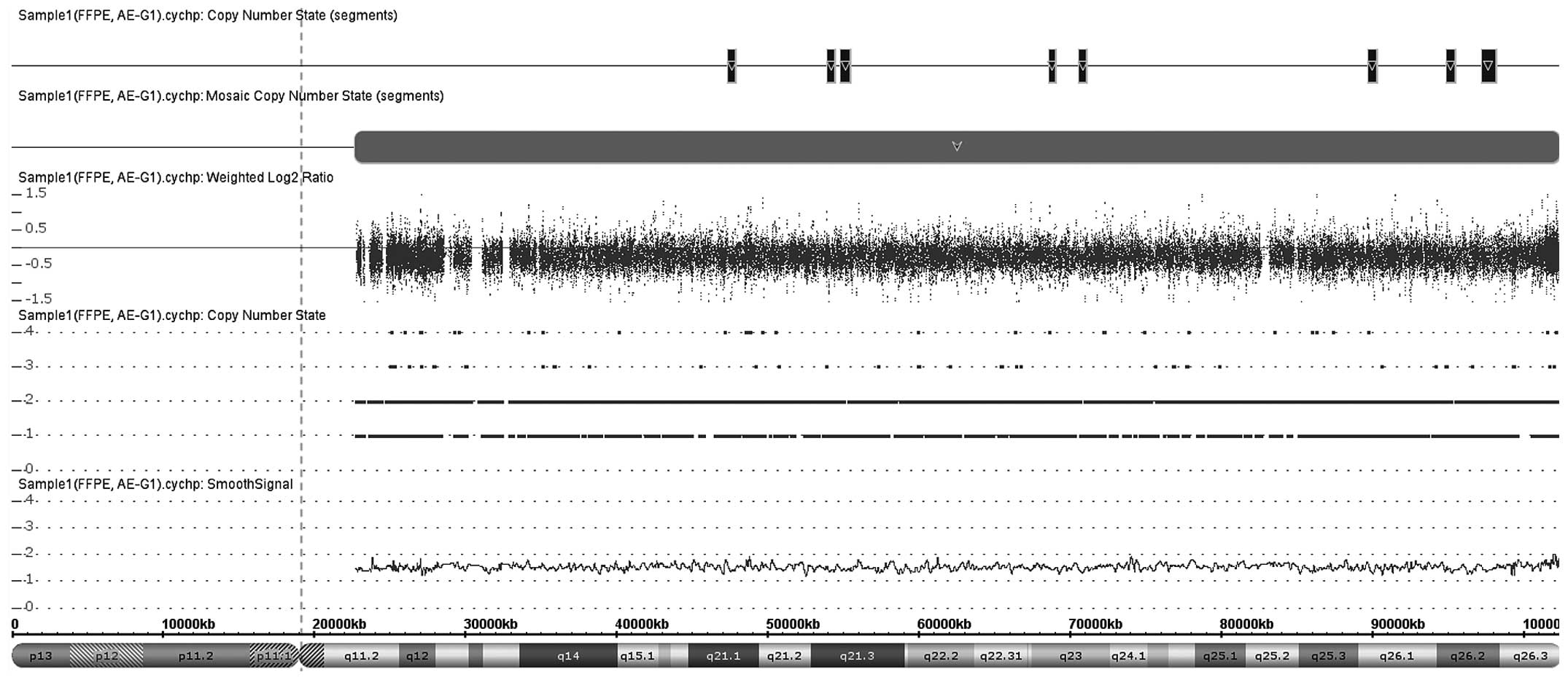
and an example of mosaic aneuploidy (chromosome 15) is shown in
Fig. 2. The genomic regions where
than divided into 4 groups: Low mosaic gains (CN2–3), low mosaic
losses (CN2-1), amplifications (CN>3) and deletions suggestive
for loss of heterozygosity (CN<1). All these data and precise
genomic coordinates were incorporated into Table I. Within detected intervals, 4,803
genes were identified. Having high microarray reproducibility, less
rigorous criteria were applied to narrow down the abnormality size
up to 100 kb (with 50 markers unchanged) to evaluate smaller
rearrangements, expanding the genomic area to 442 segments. A focus
was placed particularly on known cancer genes (522 entities) and
associated pathways that contribute to adenocarcinoma in MD. In
order to divulge putative genes contributing to MD adenocarcinoma,
the regions for COSMIC genes (known also as Census Genes) (21–23) were
assessed and 88 entities were selected, 43 of which were recurrent
(Table II). In order to further
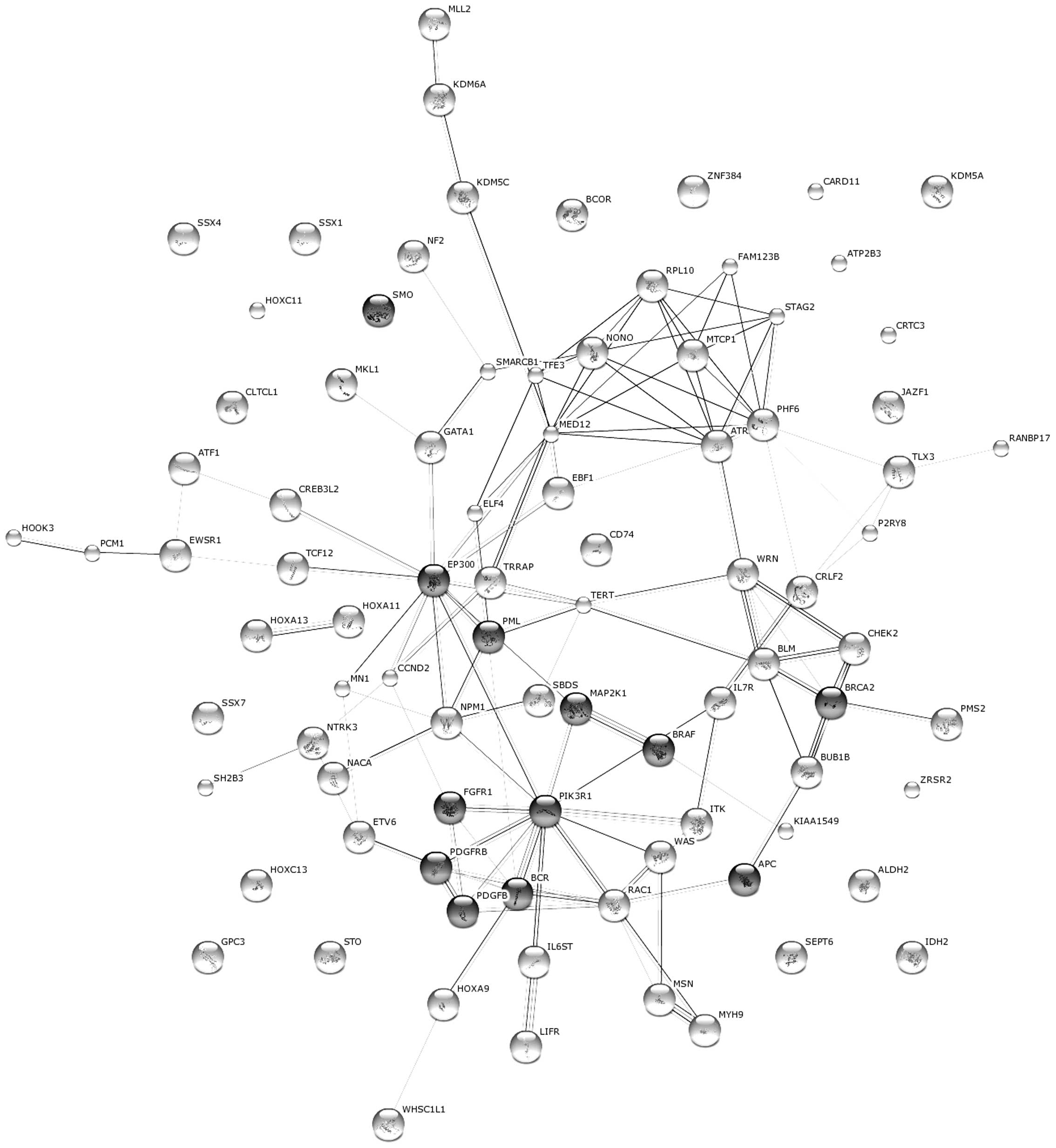
delineate the putative cancer pathways, gene-interactions were
searched for using the Kyoto Encyclopedia of Genes and Genomes
(KEGG) pathway database (http://www.genome.jp/kegg) (24). In total, 16 KEGG pathways were
selected that are associated with the development of different
types of cancer or cellular regulation processes (Table III). Interacting genes were
visualized as a network using String version 9.1 (http://string-db.org) (Fig.
3) (25).
 | Table I.Genomic alterations (>400 kb in
size) found in examined tumor. Any overlapping cancer genes
(COSMIC) are additionally listed using gene ID. |
Table I.
Genomic alterations (>400 kb in
size) found in examined tumor. Any overlapping cancer genes
(COSMIC) are additionally listed using gene ID.
| Case | CN | Type | Chromosome | Cytoband start | Start | End | Size, kbp | Gene count,
total | Census gene
count | Gene name (s) |
|---|
| 1 | 2.29 | Gain | 5 | p15.33 | 113576 | 10070760 | 9957.184 | 63 | 1 | TERT |
| 2 | 2.25 | Gain | 5 | p15.2 | 12452650 | 21850508 | 9397.858 | 19 |
|
|
| 3 | 2.33 | Gain | 5 | p14.1 | 27490784 | 85587655 | 58096.871 | 274 | 4 | IL7R, IL6ST,
PIK3R1, LIFR |
| 4 | >3 | Gain | 5 | p13.3 | 33484586 | 33897522 | 412.936 |
1 |
|
|
| 5 | >3 | Gain | 5 | q11.2 | 56710608 | 57133644 | 423.036 |
1 |
|
|
| 6 | 2.25 | Gain | 5 | q14.3 | 87860680 | 98769659 | 10908.979 | 42 |
|
|
| 7 | 2.33 | Gain | 5 | q21.1 | 99756189 | 160909509 | 61153.32 | 438 | 5 | CD74, ITK, EBF1,
APC, PDGFRB |
| 8 | >3 | Gain | 5 | q23.3 | 127572667 | 128144311 | 571.644 |
1 |
|
|
| 9 | 2.32 | Gain | 5 | q34 | 161866188 | 180719789 | 18853.601 | 159 | 4 | NPM1, RANBP17,
NSD1, TLX3 |
| 10 | 2.23 | Gain | 7 | p22.3 | 43360 | 6483365 | 6440.005 | 75 | 3 | pMS2, CARD11,
RAC1 |
| 11 | 2.21 | Gain | 7 | p15.2 | 26483627 | 30210591 | 3726.964 | 42 | 4 | HOXA11, HOXA9,
HOXA13, JAZF1 |
| 12 | 2.29 | Gain | 7 | p14.1 | 37893184 | 45800150 | 7906.966 | 70 |
|
|
| 13 | 2.22 | Gain | 7 | q11.21 | 66150834 | 71291670 | 5140.836 | 14 |
| SBDS |
| 14 | 2.22 | Gain | 7 | q21.3 | 94402911 | 98900790 | 4497.879 | 32 | 1 | TRRAP |
| 15 | 2.25 | Gain | 7 | q31.33 | 126728205 | 141381467 | 14653 | 127 | 4 | KIAA1549, CREB3L2,
SMO, BRAF |
| 16 | 2.22 | Gain | 8 | p23.3 | 158048 | 35996434 | 35838.386 | 255 |
| pCM1, WRN |
| 17 | 2.24 | Gain | 8 | p12 | 36003284 | 48320037 | 12316.753 | 66 | 3 | HOOK3, FGFR1,
WHSC1L1 |
| 18 | 2.22 | Gain | 8 | q22.1 | 93943335 | 99747954 | 5804.619 | 46 |
|
|
| 19 | 2.22 | Gain | 8 | q24.23 | 139395919 | 145287179 | 5891.26 | 77 |
|
|
| 20 | >3 | Gain | 10 | q11.22 | 46976673 | 48174779 | 1198.106 | 17 |
|
|
| 21 | 2.26 | Gain | 12 | p13.33 | 173786 | 15842149 | 15668.363 | 244 | 4 | ZNF384, KDM5A,
CCND2, ETV6, |
| 22 | 2.22 | Gain | 12 | q13.11 | 49073505 | 57666097 | 8592.592 | 253 | 5 | HOXC11, KMT2D,
HOXC13, NACA, ATF1 |
| 23 | 2.22 | Gain | 12 | q23.3 | 107607681 | 112259343 | 4651.662 | 58 | 2 | SH2B3, ALDH2, |
| 24 | 2.22 | Gain | 12 | q24.21 | 114772266 | 119550739 | 4778.473 | 23 |
|
|
| 25 | 2.24 | Gain | 13 | q12.3 | 31303510 | 38510652 | 7207.142 | 43 | 1 | BRCA2 |
| 26 | 1.47 | Loss | 15 | q11.2 | 22770421 | 102429112 | 79658.691 | 799 | 8 | BLM, PML, NTRK3,
TCF12, IDH2, CRTC3, BUB1B, MAP2K1 |
| 27 | <1 | Loss | 15 | q21.1 | 47407284 | 47903647 | 496.363 |
1 |
|
|
| 28 | <1 | Loss | 15 | q21.3 | 53994492 | 54476263 | 481.771 |
2 |
|
|
| 29 | <1 | Loss | 15 | q21.3 | 54866217 | 55494071 | 627.854 |
2 |
|
|
| 30 | <1 | Loss | 15 | q23 | 68667321 | 69080873 | 413.552 |
3 |
|
|
| 31 | <1 | Loss | 15 | q23 | 70688921 | 71128704 | 439.783 |
2 |
|
|
| 32 | <1 | Loss | 15 | q26.1 | 89819011 | 90281381 |
462.37 | 11 |
|
|
| 33 | <1 | Loss | 15 | q26.2 | 94991252 | 95497814 | 506.562 |
1 |
|
|
| 34 | <1 | Loss | 15 | q26.2 | 97288627 | 98205766 | 917.139 |
1 |
|
|
| 35 | 2.25 | Gain | 19 | q13.43 | 56862882 | 58956888 | 2094.006 | 75 |
|
|
|
| Case | CN | Type | Chromosome | Cytoband start | Start | End | Size, kbp | Gene count,
total | Census gene
count | % of overlap map
item covered by segment |
|
| 36 | 1.7 | Loss | 22 | q11.1 | 16888899 | 50902452 | 34013.553 | 525 | 11 | CHEK2, MKL1,
CLTCL1, NF2, EWSR1, MN1, SMARCB1, PDGFB, EP300, MYH9, BCR |
| 37 | 2.32 | Gain | X | p22.33 | 168546 | 32908751 | 32740.205 | 165 | 3 | CRLF2, ZRSR2,
P2RY8, |
| 38 | 2.46 | Gain | X | p21.1 | 32388483 | 113724212 | 81335.729 | 488 | 14 | SSX1, MSN, KDM5C,
KDM6A, ATRX, BCOR, SSX2, NONO, GATA1, TFE3, SSX4, AMER1, WAS,
MED12 |
| 39 | >3 | Gain | X | q11.2 | 64280772 | 64692636 | 411.864 |
0 |
|
|
| 40 | >3 | Gain | X | q13.2 | 71859817 | 72307913 | 448.096 |
8 |
|
|
| 41 | >3 | Gain | X | q21.31 | 88759463 | 89169371 | 409.908 |
0 |
|
|
| 42 | >3 | Gain | X | q21.32 | 91923926 | 92392650 | 468.724 |
0 |
|
|
| 43 | >3 | Gain | X | q21.33 | 95094694 | 95611952 | 517.258 |
1 |
|
|
| 44 | >3 | Gain | X | q22.3 | 108029759 | 108614975 | 585.216 |
0 |
|
|
| 45 | 2.38 | Gain | X | q23 | 114896478 | 155233731 | 40337.253 | 345 |
8 | ATP2B3, PHF6,
STAG2, SEPT6, GPC3, RPL10, ELF4, MTCP1 |
 | Table II.COSMIC database reporting genes to be
mutated in gastrointestinal tract (site indeterminate and various
tumor types) and small intestine adenocarcinoma.a |
Table II.
COSMIC database reporting genes to be
mutated in gastrointestinal tract (site indeterminate and various
tumor types) and small intestine adenocarcinoma.a
| Case | Gene | Mutated
samples | Sample tested |
|---|
| 1 | NRAS | 79 | 502 |
| 2 | HRAS | 31 | 495 |
|
3 |
TERT | 28 |
97 |
| 4 | KRAS | 19 | 490 |
| 5 | PIK3CA | 14 | 194 |
| 6 | PTEN | 11 | 150 |
|
7 |
BRAF |
8 | 595 |
| 8 | TSHR | 8 | 50 |
| 9 | IDH1 | 6 | 53 |
| 10 | CDKN2A | 4 | 28 |
| 11 | MET | 2 | 41 |
| 12 | TP53 | 2 | 20 |
| 13 | GNAS | 2 | 73 |
| 14 | PAX8 | 0 | 142 |
| 15 | EGFR | 0 | 101 |
| 16 | CCDC6 | 0 | 80 |
| 17 | PDGFRA | 0 | 64 |
| 18 |
KIT |
0 |
64 |
| 19 | NCOA | 0 | 55 |
| 20 | GNA11 | 0 | 46 |
| 21 | GNAQ | 0 | 42 |
| 22 |
PIK3R1 |
0 |
34 |
| 23 |
IDH2 |
0 |
32 |
| 24 | AKT1 | 0 | 23 |
| 25 | ALK | 0 | 22 |
| 26 | RET | 0 | 21 |
| 27 | PRKAR1A | 0 | 16 |
| 28 | MEN1 | 0 | 15 |
| 29 |
APC |
0 |
15 |
| 30 |
MAP2K1 |
0 |
13 |
| 31 | STRN | 0 | 11 |
| 32 | VHL | 0 | 10 |
| 33 |
NPM1 |
0 |
10 |
| 34 | CDH1 | 0 |
8 |
| 35 | TFG | 0 |
6 |
| 36 | TPR | 0 |
6 |
| 37 | JAK2 | 0 |
6 |
| 38 | CTNNB1 | 0 |
4 |
| 39 | SF3B1 | 0 |
4 |
| 40 |
KDM6A |
0 |
4 |
| 41 | SMAD4 | 0 |
4 |
| 42 | PTPN11 | 0 |
3 |
| 43 | TPM3 | 0 |
2 |
 | Table III.Kyoto Encyclopedia of Genes and
Genomes pathway and statistical analysis for involvement of
selected cancer genes present in altered genomic regions and their
contribution to different cancer types or cellular control. |
Table III.
Kyoto Encyclopedia of Genes and
Genomes pathway and statistical analysis for involvement of
selected cancer genes present in altered genomic regions and their
contribution to different cancer types or cellular control.
| Case | Gene ontology
ID | Term | P-value | No. of genes | Genes |
|---|
| 1 | hsa05200 | Pathways in
cancer |
1.29×10−6 | 12 | BCR,
APC, SMO, PDGFRB, PDGFB, EP300,
BRAF, MAP2K1, PIK3R1, BRCA2,
FGFR1, PML |
| 2 | hsa05215 | Prostate
cancer |
1.27×10−7 | 8 | CREB3L2,
PIK3R1, PDGFRB, PDGFB, EP300,
FGFR1, BRAF, MAP2K1 |
| 3 | hsa05218 | Melanoma |
7.8×10−6 | 6 | PIK3R1,
PDGFRB, PDGFB, FGFR1, BRAF,
MAP2K1 |
| 4 | hsa05214 | Glioma |
6.4×10−5 | 5 | PIK3R1,
PDGFRB, PDGFB, BRAF, MAP2K1 |
| 5 | hsa04630 | Jak-STAT signaling
pathway |
8.73×10−5 | 7 | PIK3R1,
IL7R, CCND2, CRLF2, IL6ST,
EP300, LIFR |
| 6 | hsa05211 | Renal cell
carcinoma |
1.08×10−4 | 5 | PIK3R1,
PDGFB, EP300, BRAF, MAP2K1 |
| 7 | hsa05213 | Endometrial
cancer |
4.39×10−4 | 4 | APC,
PIK3R1, BRAF, MAP2K1 |
| 8 | hsa05221 | Acute myeloid
leukemia |
5.87×10−4 | 4 | PIK3R,
PML, BRAF, MAP2K1 |
| 9 | hsa05210 | Colorectal
cancer |
6.73×10−4 | 4 | APC,
PIK3R1, BRAF, MAP2K1 |
| 10 | hsa05212 | Pancreatic
cancer |
1.04×10−3 | 4 | PIK3R1,
BRCA2, BRAF, MAP2K1 |
| 11 | hsa05220 | Chronic myeloid
leukemia |
1.38×10−3 | 4 | PIK3R1,
BRAF, MAP2K1, BCR |
| 12 | hsa04110 | Cell cycle |
1.51×10−3 | 5 | CCND2,
CHEK2, STAG2, EP300, BUB1B |
| 13 | hsa04510 | Focal adhesion |
2.43×10−3 | 6 | PDGFRB,
PDGFB, BRAF, MAP2K1, PIK3R1,
CCND2 |
| 14 | hsa05223 | Non-small cell lung
cancer |
5.86×10−3 | 3 | PIK3R1,
BRAF, MAP2K1 |
| 15 | hsa04062 | Chemokine signaling
pathway |
8.99×10−3 | 5 | PIK3R1,
WAS, BRAF, MAP2K1, ITK |
| 16 | hsa04060 | Cytokine-cytokine
receptor interaction |
9.63×10−3 | 6 | CRLF2,
IL6ST, PDGFRB, PDGFB, LIFR,
IL7R |
Discussion
Current studies are aimed at estimating the
incidence of GIST cases in the population. Swedish studies have
shown that currently, the number of novel GIST cases amounts to
15–16 million/year (26). A tumor may
develop in any section of the gastrointestinal tract, as well as
intraperitoneally and in the retroperitoneal space. The current
state of knowledge and statistical data indicate that when a
radical surgical excision of the tumor is possible, the 5-year
survival rate is 50–60% (27,28). At the same time, literature references
indicate that 80% of patients who undergo surgery experience local
recurrence within 2 years of the procedure, and that liver
metastases additionally appear in 50% of the patients (29,30). In
the case discussed in the present study, the patient was treated
surgically for reasons other than a GIST. This problem is widely
discussed in the literature. Approximately 40% of women with small
intestine GISTs undergo surgery due to genital tract tumors (GIST
mimicking pelvic disorders). The genetic mapping of major molecular
contributors, tyrosine kinases KIT and PDGFRA (ref.
NM_000222 and NM_006206), reveal that they are located very close
to each other on chromosome 4 (31,32) and
are found to be mutated in up to 90% of GIST cases. For each, a
gain of function mutations results in constitutive activation of
the oncogenes, but the clinical consequences may differ
significantly (33,34). The biological consequences related to
abnormal hyperactivity of both proteins observed in cancer cells
for each gene implies the occurrence of a variety of possible
mechanisms, including point mutations and bigger genomic
instabilities (whole gene amplifications or gene fusions). Hence,
the comprehensive molecular portrait of GIST cancers is now
emerging. This is possibly due to usage of high-throughput genomic
approaches that provide valuable data referring to the involvement
of a single gene, but also characterizing entire pathways on
various stages of GIST carcinogenesis (35–37). To
date, high-resolution mapping has been conducted only by two groups
(36,38), with reference to typical GIST
occurrences. Specific chromosomal rearrangements are solid and a
consistent finding accompanying KIT and PDGFRA alterations. Losses
of chromosomes 1, 3, 13, 14, 15 and 22, whereas gains of
chromosomes 4 and 5 may represent clinical utility and prognostic
relevance. In the present case, a 15q loss was identified, which is
regarded as an aggressive course for a GIST. The loss of 1p was
also noted, which is typical for the small intestine localization
of tumors, and the absence of a copy number alteration in the
RB1 locus, which is known as a strong clinical predictor
(39). Application of a
high-resolution microarray allows the identification of CN changes
at a single gene level (for constitutional disorders, even single
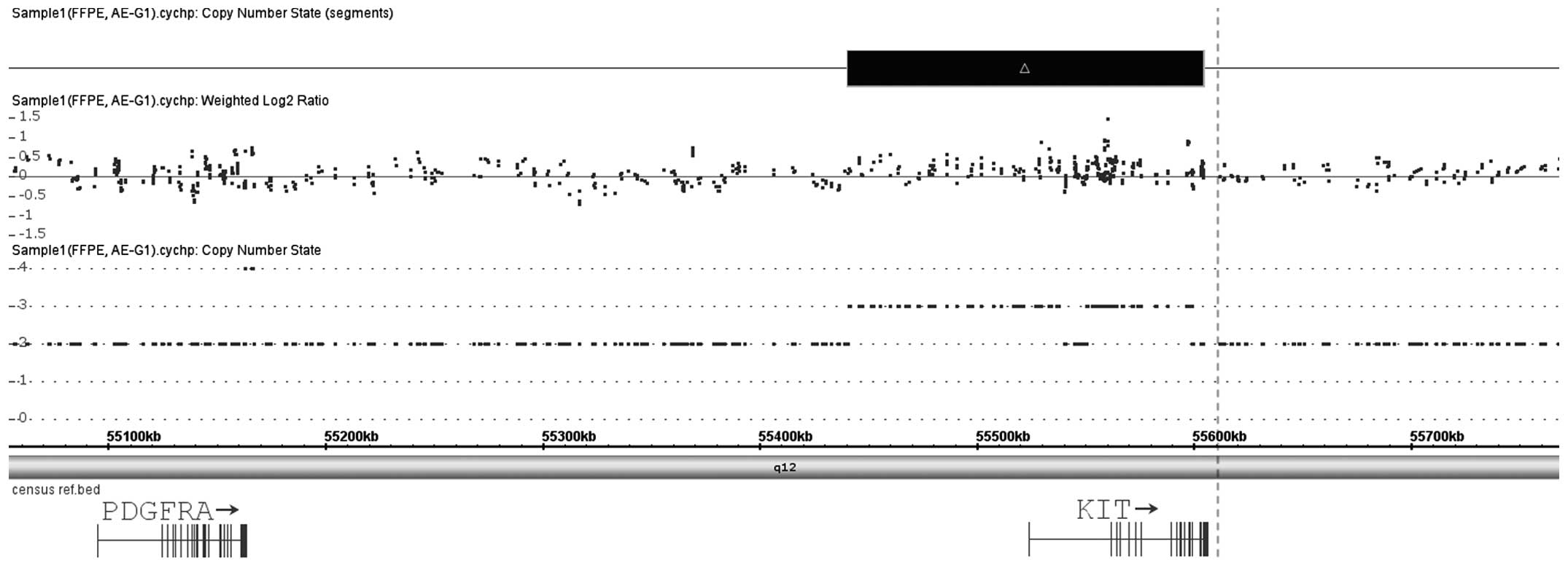
exon rearrangements). In the present study, KIT gene
amplification was detected on chromosome 4, spanning only 163 kb
and comprising the entire genomic sequence of this gene.
Unexpectedly, the closely located PDGFRA gene (400 kb away) was not
altered (Fig. 4). This finding is
reminiscent and indicative of the principal role of KIT in
triggering oncogenesis in this case. However, an activating point
mutation in another gene cannot be excluded. Alteration of two
prominent interacting proteins, PDGFRB and PIK3R1 (40), was also found. Phosphorylation of
phosphatidylinositol 3-kinase (PIK3R1) by KIT leads to the
activation of the v-akt murine thymoma viral oncogene homolog 1
signaling pathway. Activated KIT also transmits signals via growth
factor receptor-bound protein 2 and activation of RAS, Raf-1
proto-oncogene, serine/threonine kinase and the mitogen-activated
protein kinases (MAPKs) MAPK1/ERK2 and/or MAPK3/ERK1.
In conclusion, by employing a high-resolution
microarray, the present study performed a comprehensive genomic
analysis on genes contributing to GISTs, in the rare location of
MD. A principal role of the KIT gene was confirmed in cancer
initiation, which was demonstrated by detailed histopathological
and molecular investigations. The detected chromosomal gains and
losses were consistent with the findings of a GIST and confirm
previous studies. Possible chemokine and cytokine-related signaling
pathways (PIK3R1, BRAF, MAP2K1, PDGFRB and PDGFB) that may
reasonably contribute to cancer progression with GIST
characteristics were also indicated.
Acknowledgements
This study was supported by the grant no.
789/FNiTP/162/2013 Polish Ministry of Science and Education.
References
|
1
|
Sircar K, Hewlett BR, Huizinga JD,
Chorneyko K, Berezin I and Riddell RH: Interstitial cells of Cajal
as precursors of gastrointestinal stromal tumors. Am J Surg Pathol.
23:377–389. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Chan JK: Mesenchymal tumors of the
gastrointestinal tract: A paradise for acronyms (STUMP, GIST GANT,
and now GIPACT), implication of c-kit in genesis, and yet another
of the many emerging roles of the interstitial cell of Cajal in the
pathogenesis of gastrointestinal diseases? Adv Anat Pathol.
6:19–40. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Rutkowski P, Debiec-Rychter M and Ruka W:
Gastrointestinal stromal tumors, Key to diagnosis and choice of
therapy. Mol Diagn Ther. 12:131–143. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Rutkowski P, Wozniak A, Dębiec-Rychter M,
Kąkol M, Dziewirski W, Zdzienicki M, Ptaszynski K, Jurkowska M,
Limon J and Siedlecki JA: Clinical utility of the new American
Joint Committee on Cancer staging system for gastrointestinal
stromal tumors, Current overall survival after primary tumor
resection. Cancer. 117:4916–4924. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Corless CL, Fletcher JA and Heinrich MC:
Biology of gastrointestinal stromal tumors. J Clin Oncol.
22:3813–3825. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Miettinen M, Sarlomo-Rikala M and Lasota
J: Gastrointestinal stromal tumors, Recent advances in
understanding of their biology. Hum Pathol. 30:1213–1220. 1999.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Roberts PJ and Eisenberg B: Clinical
presentation of gastrointestinal stromal tumors and treatment of
operable disease. Eur J Cancer. 38((Suppl 5)): S37–S38. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Neuhaus SJ, Clark MA, Hayes AJ, Thomas JM
and Judson I: Surgery for gastrointestinal stromal tumour in the
post-imatinib era. ANZ J Surg. 75:165–172. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Duensing A, Heinrich MC, Fletcher CD and
Fletcher JA: Biology of gastrointestinal stromal tumors, KIT
mutations and beyond. Cancer Invest. 22:106–116. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Heinrich MC, Rubin BP, Longley BJ and
Fletcher JA: Biology and genetic aspects of gastrointestinal
stromal tumors, KIT activation and cytogenetic alterations. Hum
Pathol. 33:484–495. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Huizinga JD, Thuneberg L, Klüppel M,
Malysz J, Mikkelsen HB and Bernstein A: W/kit gene required for
interstitial cells of Cajal and for intestinal pacemaker activity.
Nature. 373:347–349. 1995. View
Article : Google Scholar : PubMed/NCBI
|
|
12
|
Nannini M, Astolfi A, Urbini M, Indio V,
Santini D, Heinrich MC, Corless CL, Ceccarelli C, Saponara M,
Mandrioli A, et al: Integrated genomic study of quadruple-WT GIST
(KIT/PDGFRA/SDH/RAS pathway wild-type GIST). BMC Cancer.
14:6852014. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Pantaleo MA, Nannini M, Corless CL and
Heinrich MC: Quadruple wild-type (WT) GIST, Defining the subset of
GIST that lacks abnormalities of KIT, PDGFRA, SDH, or RAS signaling
pathways. Cancer Med. 4:101–103. 2014. View
Article : Google Scholar : PubMed/NCBI
|
|
14
|
Heinrich MC, Griffith DJ, Druker BJ, Wait
CL, Ott KA and Zigler AJ: Inhibition of c-kit receptor tyrosine
kinase activity by STI 571, a selective tyrosine kinase inhibitor.
Blood. 96:925–932. 2000.PubMed/NCBI
|
|
15
|
Chandramohan K, Agarwal M, Gurjar G, Gatti
RC, Patel MH, Trivedi P and Kothari KC: Gastrointestinal stromal
tumour in Meckel's diverticulum. World J Surg Oncol. 5:502007.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Khoury MG: 2nd andA ulicino MR:
Gastrointestinal stromal tumor (GIST) presenting in a Meckel's
diverticulum. Abdom Imaging. 32:78–80. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Kosmidis C, Efthimiadis C, Levva S,
Anthimidis G, Baka S, Grigoriou M, Tzeveleki I, Masmanidou M,
Zaramboukas T and Basdanis G: Synchronous colorectal adenocarcinoma
and gastrointestinal stromal tumor in Meckel's diverticulum; an
unusual association. World J Surg Oncol. 7:332009. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Thirunavukarasu P, Sathaiah M, Sukumar S,
Bartels CJ, Zeh H III, Lee KK and Bartlett DL: Meckel's
diverticulum - a high-risk region for malignancy in the ileum.
Insights from a population-based epidemiological study and
implications in surgical management. Ann Surg. 253:223–230. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Gillio-Tos A, De Marco L, Fiano V,
Garcia-Bragado F, Dikshit R, Boffetta P and Merletti F: Efficient
DNA extraction from 25-year-old paraffin-embedded tissues, Study of
365 samples. Pathology. 39:345–348. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Forbes SA, Bindal N, Bamford S, Cole C,
Kok CY, Beare D, Jia M, Shepherd R, Leung K, Menzies A, et al:
COSMIC: Mining complete cancer genomes in the Catalogue of Somatic
Mutations in Cancer. Nucleic Acids Res. 39(Database): D945–D950.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Futreal PA, Coin L, Marshall M, Down T,
Hubbard T, Wooster R, Rahman N and Stratton MR: A census of human
cancer genes. Nat Rev Cancer. 4:177–183. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
de Castro Pérez I, de Cárcer G and
Malumbres M: A census of mitotic cancer genes: New insights into
tumor cell biology and cancer therapy. Carcinogenesis. 28:899–912.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Santarius T, Shipley J, Brewer D, Stratton
MR and Cooper CS: A census of amplified and overexpressed human
cancer genes. Nat Rev Cancer. 10:59–64. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Altermann E and Klaenhammer TR:
PathwayVoyager: Pathway mapping using the Kyoto Encyclopedia of
Genes and Genomes (KEGG) database. BMC Genomics. 6:602005.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Franceschini A, Szklarczyk D, Frankild S,
Kuhn M, Simonovic M, Roth A, Lin J, Minguez P, Bork P, von Mering
C, et al: STRING v9.1: Protein-protein interaction networks, with
increased coverage and integration. Nucleic Acids Res. 41(D1):
D808–D815. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Nilsson B, Bümming P, Meis-Kindblom JM,
Odén A, Dortok A, Gustavsson B, Sablinska K and Kindblom LG:
Gastrointestinal stromal tumors, The incidence, prevalence,
clinical course, and prognostication in the preimatinib mesylate
era - a population-based study in western Sweden. Cancer.
103:821–829. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Dematteo RP, Heinrich MC, El-Rifai WM and
Demetri G: Clinical management of gastrointestinal stromal tumors,
Before and after STI-571. Hum Pathol. 33:466–477. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Tosoni A, Nicolardi L and Brandes AA:
Current clinical management of gastrointestinal stromal tumors.
Expert Rev Anticancer Ther. 4:595–605. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Ruka W, Rutkowski P, Nowecki Z,
Nasierowska-Guttmejer A and Debiec-Rychter M: Other malignant
neoplasms in patients with gastrointestinal stromal tumors (GIST).
Med Sci Monit. 10:LE13–LE14. 2004.PubMed/NCBI
|
|
30
|
Rutkowski P, Nowecki ZI, Michej W,
Debiec-Rychter M, Woźniak A, Limon J, Siedlecki J, Grzesiakowska U,
Kakol M, Osuch C, et al: Risk criteria and prognostic factors for
predicting recurrences after resection of primary gastrointestinal
stromal tumor. Ann Surg Oncol. 14:2018–2027. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Hirota S, Isozaki K, Moriyama Y, Hashimoto
K, Nishida T, Ishiguro S, Kawano K, Hanada M, Kurata A, Takeda M,
et al: Gain-of-function mutations of c-kit in human
gastrointestinal stromal tumors. Science. 279:577–580. 1998.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Chompret A, Kannengiesser C, Barrois M,
Terrier P, Dahan P, Tursz T, Lenoir GM and Bressac-De Paillerets B:
PDGFRA germline mutation in a family with multiple cases of
gastrointestinal stromal tumor. Gastroenterology. 126:318–321.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Kwon JE, Kang HJ, Kim SH, Lee YC, Hyung
WJ, Noh SH, Kim NK and Kim H: Pathological characteristics of
gastrointestinal stromal tumours with PDGFRA mutations. Pathology.
41:544–554. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Schaefer IM, Ströbel P, Cameron S, Beham
A, Otto C, Schildhaus HU and Agaimy A: Rhabdoid morphology in
gastrointestinal stromal tumours (GISTs) is associated with PDGFRA
mutations but does not imply aggressive behaviour. Histopathology.
64:421–430. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Gunawan B, Bergmann F, Höer J, Langer C,
Schumpelick V, Becker H and Füzesi L: Biological and clinical
significance of cytogenetic abnormalities in low-risk and high-risk
gastrointestinal stromal tumors. Hum Pathol. 33:316–321. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Schoppmann SF, Vinatzer U, Popitsch N,
Mittlböck M, Liebmann-Reindl S, Jomrich G, Streubel B and Birner P:
Novel clinically relevant genes in gastrointestinal stromal tumors
identified by exome sequencing. Clin Cancer Res. 19:5329–5339.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Wozniak A, Sciot R, Guillou L, Pauwels P,
Wasag B, Stul M, Vermeesch JR, Vandenberghe P, Limon J and
Debiec-Rychter M: Array CGH analysis in primary gastrointestinal
stromal tumors Cytogenetic profile correlates with anatomic site
and tumor aggressiveness, irrespective of mutational status. Genes
Chromosomes Cancer. 46:261–276. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Astolfi A, Nannini M, Pantaleo MA, Di
Battista M, Heinrich MC, Santini D, Catena F, Corless CL, Maleddu
A, Saponara M, et al: A molecular portrait of gastrointestinal
stromal tumors: An integrative analysis of gene expression
profiling and high-resolution genomic copy number. Lab Invest.
90:1285–1294. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Lagarde P, Pérot G, Kauffmann A, Brulard
C, Dapremont V, Hostein I, Neuville A, Wozniak A, Sciot R,
Schöffski P, et al: Mitotic checkpoints and chromosome instability
are strong predictors of clinical outcome in gastrointestinal
stromal tumors. Clin Cancer Res. 18:826–838. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Liu CH, Chen TC, Chau GY, Jan YH, Chen CH,
Hsu CN, Lin KT, Juang YL, Lu PJ, Cheng HC, et al: Analysis of
protein-protein interactions in cross-talk pathways reveals CRKL
protein as a novel prognostic marker in hepatocellular carcinoma.
Mol Cell Proteomics. 12:1335–1349. 2013. View Article : Google Scholar : PubMed/NCBI
|