Introduction
Prostate cancer is the most prevalent type of tumor
of the male urinary system. In the United States, prostate cancer
is the second leading cause of cancer in men, the incidence of
which is rising in China as well as in Western countries (1). In general, prostate cancer is a
slow-growing disease, with the onset of symptoms occurring in
advanced-stage disease, following tumor discovery (2). The initial treatment for prostate cancer
is usually hormone therapy; however, when prostate tumors progress
into an androgen-independent state, treatment becomes difficult
(1).
Na+/H+ exchanger regulatory factor 1 (NHERF1) was
identified to be a multifunctional scaffolding protein that has
various functions in numerous types of cancer through its
interactions with oncogenic or tumor-suppressor proteins (3). In breast cancer, NHERF1 acts as a tumor
suppressor protein, which is involved in influencing signal
transduction and modulating the expression of phosphatase and
tensin homolog (PTEN) to regulate the malignant phenotype (4). However, NHERF1 has been shown to have
oncogenic functions in glioma and other types of cancer (5). While the specific role of NHERF1 in
prostate cancer remains to be elucidated, studies have shown that
the NHERF1 protein was overexpressed in tumor specimens in
situ compared with normal prostate tissues (6,7).
Therefore, further studies are required in order to fully elucidate
the role of NHERF1 in prostate cancer.
The present study aimed to determine the effects of
NHREF1 knockdown using short-hairpin RNA (shRNA) in PC-3M
cells, a prostate cancer cell line exhibiting abnormally high
expression of NHERF1.
Materials and methods
Bacterial strains, cell lines and
plasmids
The bacterial strain Escherichia coli DH5α was
purchased from Beijing CoWin Biotech Co., Ltd. (Beijing, China).
The pSuper.puro NHERF1 shRNA plasmid and negative control
plasmid (pSuper.puro luciferase shRNA) were a gift from Professor
Junfang Zheng (Capital Medical University, Beijing, China),
originally constructed by Dr Margaret J. Wheelock (University of
Nebraska Medical Center, Omaha, NE, USA). PC-3M prostate cancer
cells were purchased from the Cell Resource Center of Beijing Xiehe
(Beijing, China).
Reagents
RPMI 1640 medium, fetal calf serum and Lipofectamine
2000 were purchased from Invitrogen Life Technologies (Carlsbad,
CA, USA). Rabbit anti-human NHERF1 (cat no. ab133599) and
Bcl-2-associated X protein (Bax; cat no. ab32503) monoclonal
antibodies, and rabbit anti-human GADPH (cat no. ab181602)
polyclonal antibody were purchased from Abcam (Cambridge, UK).
Rabbit anti-human B cell lymphoma-2 (Bcl-2; cat no. ZA-0536)
monoclonal antibody was purchased from Beijing Zhongshan
GoldenBridge Biotechnology Co., Ltd. (Beijing, China), and goat
anti-rabbit horseradish peroxidase (HRP)-conjugated secondary
antibody (cat no. CW0103) was purchased from Beijing CoWin Biotech
Co., Ltd. MTT was purchased from Sigma-Aldrich (St. Louis, MO,
USA). The Annexin V-FITC Apoptosis Detection kit and Propidium
Staining Cycle Detection kit were obtained from BestBio
Biotechnology Co., Ltd. (Shanghai, China). All other reagents were
of analytic grade.
Conversion and amplification of
plasmid
The interference plasmid pSuper.puro harboring
NHERF1 shRNA and the negative control plasmid pSuper.puro
harboring luciferase shRNA were transformed into competent E.
coli DH5α cells. Briefly, competent E. coli DH5α (100 µl) and
plasmid (2 µl) were added to Eppendorf tubes, and placed in ice for
30 min. Next, the Eppendorf tubes were incubated at 42°C for 90 sec
and then placed back in ice for 2 min. Lysogeny broth (LB) media
without antibiotic (800 µl; Thermo Fisher Scientific, Waltham, MA,
USA) was added and the transformed DH5α cells were incubated at
37°C for 30 min with agitation. Transformed DH5α (50 µl) were
spread on LB culture plates (Corning Incorporated, Corning, NY,
USA) containing ampicillin and cultured at 37°C for 12–16 h.
Positive colonies were selected following cultivation of the
bacteria on LB plates (Beijing CoWin Biotech Co., Ltd.) containing
100 µg/ml ampicillin (Sigma-Aldrich); the plasmid was then
extracted and purified following overnight cultivation of the
bacteria.
Cell culture and the establishment of
a stable expression system
PC-3M cells were cultured in RPMI 1640 medium
containing 10% fetal calf serum and 1% penicillin-streptomycin
(Sigma-Aldrich) at 37°C in an incubator with 5% CO2. Cells were
transfected with 2.5 µg plasmid using Lipofectamine 2000, according
to the manufacturer's instructions. Stably transfected cells were
selected using 2 µg/ml puromycin (Sigma-Aldrich) for 3–4 weeks at
37°C in an incubator with 5% CO2. Clones were maintained in culture
medium containing 1 µg/ml puromycin. Media were replaced every 3
days.
Detection of cell growth
Cells stably expressing NHERF1 shRNA or
negative control shRNA were plated at a density of 5×103 cells/well
onto 96-well plates at 37°C in an incubator with 5% CO2. Cell
proliferation was then assessed every 24 h for 96 h using MTT
assays according to standard protocols (8). For each sample at each time-point, six
wells were analyzed. The experiment was repeated three times.
Western blotting
Cells were collected and lysed in
radioimmunoprecipitation buffer (Beijing CoWin Biotech Co., Ltd.,
Beijing, China) in the presence of protease inhibitors for 20 min
to extract total protein from cells stably expressing NHERF1
shRNA or control cells, and protein levels were quantified using
bicinchoninic acid assays (Beijing CoWin Biotech Co., Ltd.).
Subsequently, 50 µg protein from each sample was loaded onto 8%
sodium dodecyl sulfate (SDS) polyacrylamide gels and subjected to
SDS-polyacrylamide gel electrophoresis (PAGE; Beijing CoWin Biotech
Co., Ltd.). Protein was then transferred to nitrocellulose
membranes (Sigma-Aldrich), which were then blocked with bovine
serum albumin blocking buffer (Invitrogen Life Technologies) for 1
h. Membranes were then incubated with primary antibodies targeting
NHERF1 (1:1,000 dilution), Bcl-2 (1:1,000 dilution), Bax (1:200
dilution) or GAPDH (1:1,000 dilution) overnight at 4°C, followed by
incubation with secondary antibodies conjugated with HRP (1:3,000
dilution) for 1 h at room temperature. Detection was facilitated
using an enhanced chemiluminescence kit and images were analyzed
using ImageJ software (version 1.62; National Institute of Health,
Bethesda, MD, USA).
Wound healing assay
Cells were plated at a density of 2×105 cells/well
onto six-well plates and grown to 80% confluence. The monolayer of
cells was scratched with a 200-µl pipette tip to create a wound and
an image was captured immediately (0 h). Migration was then
observed every 12 h and images of the closing wound were captured
at each time-point with a digital camera connected to a
phase-contrast microscope (CKX41; 10× objective lens; Olympus,
Tokyo, Japan). The same visual field was used throughout the
experiment. Image-Pro Plus software (version 6.0; Media
Cybernetics, Rockville, MD, USA) was used to facilitate the
calculation of the migration rate as follows: Migration rate =
[(relative distance recorded at 0 h - relative distance recorded at
48 h) / relative distance recorded at 0 h] × 100%. The experiment
was repeated three times.
Analysis of cell apoptosis
Annexin V-fluorescein isothiocyanate
(FITC)/propidium iodide (PI) flow cytometry was used to detect the
rate of apoptosis. Cells were collected, washed with
phosphate-buffer saline (PBS) and resuspended in 400 µl buffer
(BestBio Biotechnology Co., Ltd.). Subsequently, 5 µl Annexin
V-FITC (BestBio Biotechnology Co., Ltd.) was added, cells were
mixed and the solution was incubated for 15 min in the dark at room
temperature. PI (10 µl; BestBio Biotechnology Co., Ltd.) was then
added and the samples were incubated in the dark at room
temperature for 5 min. Cells were then analyzed using flow
cytometry within 1 h (Accuri C6; BD Biosciences, Franklin Lakes,
NJ, USA).
Analysis of cell cycle
Cultured cells were removed with trypsin and fixed
with 70% ethanol at 4°C overnight. Subsequently, they were stained
with PI (20 µg/ml PI, 200 µg/ml DNase-free RNase A and 0.1% Triton
X-100, prepared freshly in PBS). The cellular DNA content was
analyzed using a FACSCalibur machine (Becton Dickinson, San Jose,
CA, USA). Data were analyzed using CellQuest software (version 3.0;
Becton Dickinson.)
Statistical analysis
All experiments were repeated at least three times.
SPSS 11.5 software (SPSS, Inc., Chicago, IL, USA) was used to
analyze the results. Growth curves were analyzed using a
repeated-measures analysis of variance with Fisher's least
significant difference post-hoc tests and apoptosis was analyzed
using independent sample t-tests. P<0.05 was considered to
indicate a statistically significant difference between values.
Results
Analysis of NHERF1 knockdown in stably
transfected PC-3M cells
As shown in Fig. 1,
NHERF1 expression was successfully knocked down in NHERF1
shRNA-transfected cells compared with untransfected cells and cells
transfected with the negative control plasmid. These cells were
used in subsequent experiments as NHERF1 knockdown
cells.
Influence of NHERF1 knockdown on the
proliferation capacity of PC-3M cells
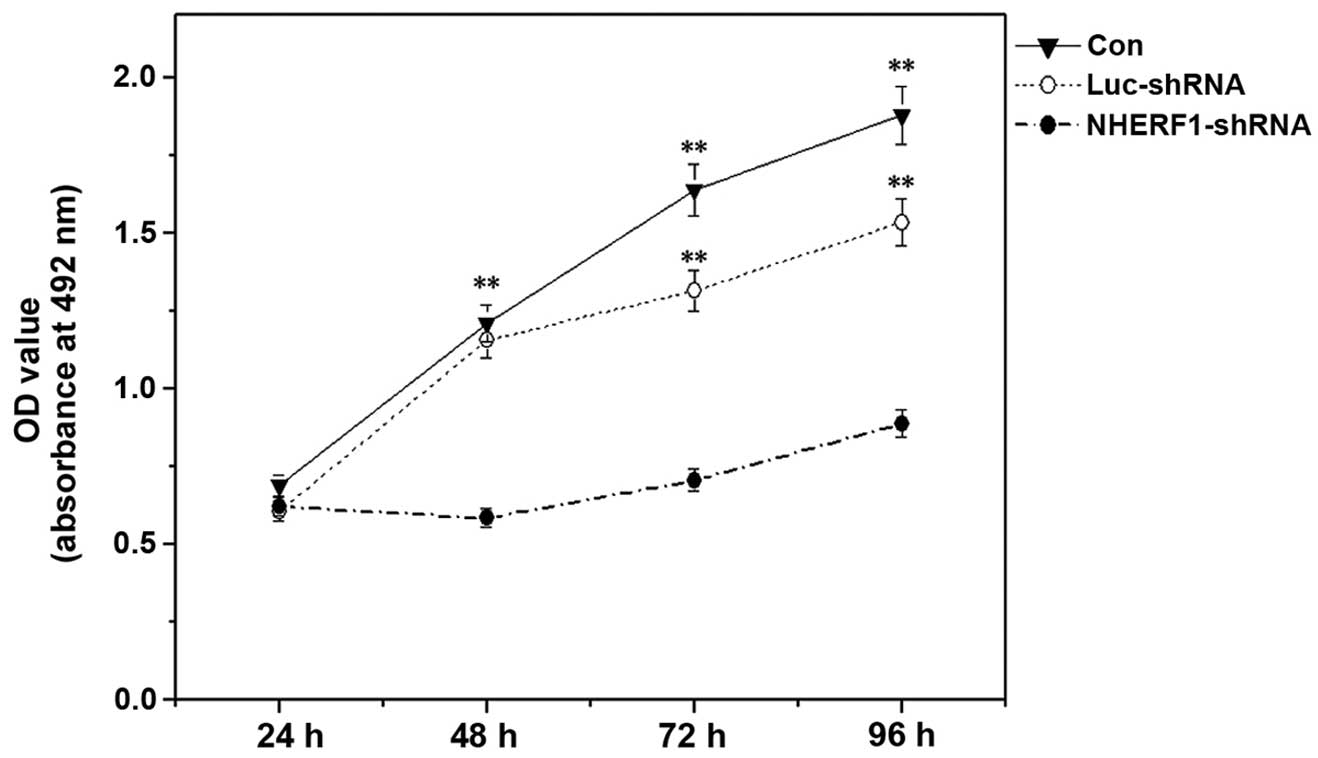
The ability of NHERF1 to modulate the
proliferation of prostate cancer cells was analyzed using an MTT
assay. The results indicated that NHERF1 knockdown
significantly inhibited the proliferation capacity of PC-3M cells
by >50% at all time-points following 24 h of incubation, as
compared with control cells (P<0.01) (Fig. 2). No significant differences were
observed between untransfected cells and luciferase
shRNA-transfected cells.
Influence of NHERF1 knockdown on the
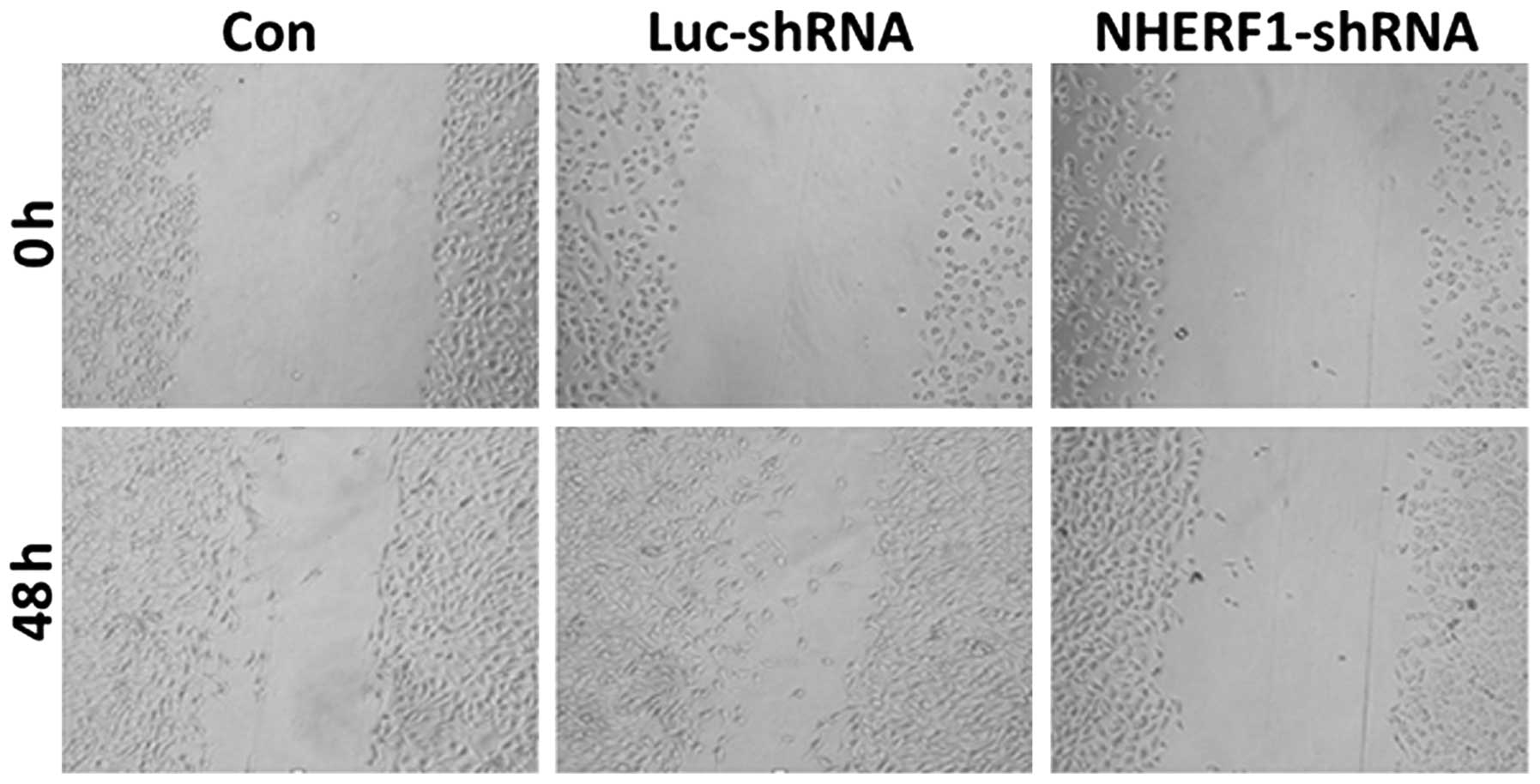
migration capacity of PC-3M cells
At 48 h following wounding, there were no
significant differences in wound healing rates between
untransfected cells and cells transfected with luciferase shRNA
(Fig. 3). However, cells transfected
with NHERF1 shRNA exhibited a significant reduction in
migration capacity (P<0.05), with the wound size measuring
94–98% of the wound size at 0 h (Fig.
3). Thus, these data demonstrated that knockdown of
NHERF1 significantly inhibited the migration capacity of
PC-3M prostate cancer cells.
Influence of NHERF1 knockdown on
apoptosis in PC-3M cells
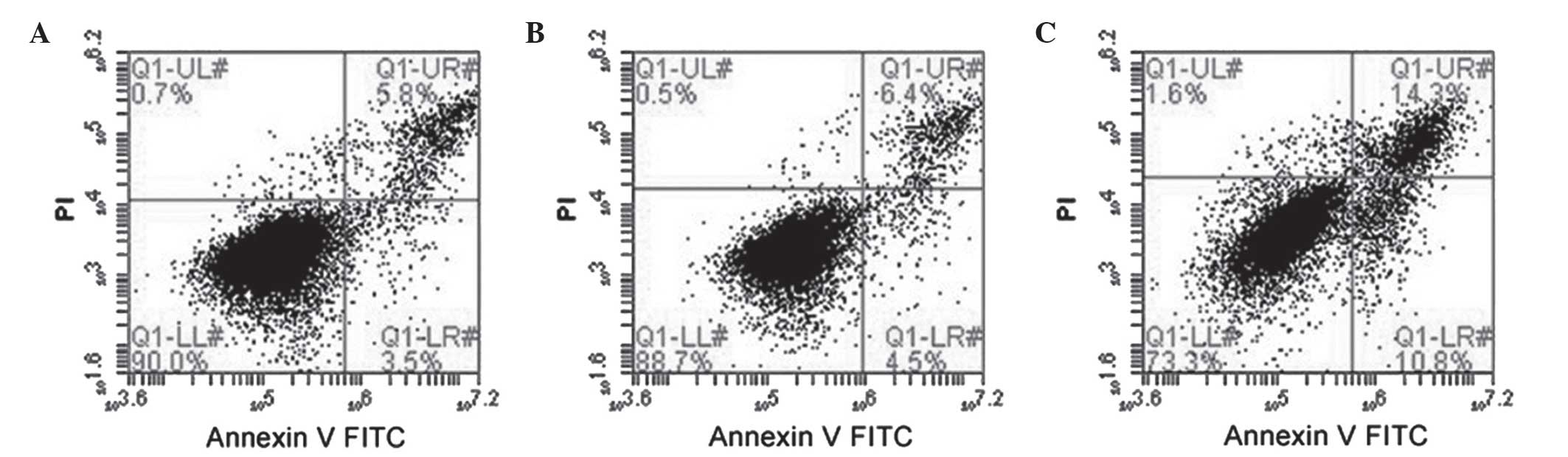
Representative results of flow cytometric analysis
are shown in Fig. 4. In general, the
rates of cell death were low (<5%). However, cells stably
expressing NHREF1 shRNA exhibited a significant increase in
apoptosis by ~4-fold compared with cells transfected with
luciferase shRNA (11.98±3.28 vs. 2.55±0.92%, respectively;
P<0.001); untransfected cells exhibited 3.5±0.96% apoptosis
compared with the luciferase shRNA-transfected cells (P>0.05).
These data indicated that reduced NHERF1 expression substantially
increased apoptosis in PC-3M cells.
Influence of NHERF1 knockdown on cell
cycle in PC-3M cells
Next, the effect of NHERF1 knockdown on the
cell cycle was analyzed. Transfection with NHERF1 shRNA
significantly increased the sub-G1 fraction (the fraction of
apoptotic cells) compared with the luciferase shRNA-transfected and
parental control cells (22, 10 and 8% in NHERF1
shRNA-transfected, luciferase shRNA-transfected and parental cells,
respectively; data not shown). Collectively, these results suggest
that NHERF1 is important in cell growth, as well as cell
cycle progression, in a subset of prostate cancer cell lines.
Influence of NHERF1 knockdown on the
expression of apoptosis-associated proteins in PC-3M cells
As shown in Fig. 5,
the expression of Bax, a pro-apoptotic protein, was unchanged
following transfection with NHERF1 shRNA, while the
expression of Bcl-2, an anti-apoptotic protein, was markedly
decreased (P<0.05). These data supported the results of flow
cytometry analysis indicating that apoptosis was increased
following knockdown of NHERF1.
Discussion
The present study examined the effects of
NHERF1 knockdown on the malignant phenotype in PC-3M
prostate cancer cells. The results demonstrated that NHERF1
knockdown inhibited proliferation and migration, while increasing
apoptosis. Therefore, NHERF1 may represent a novel oncogene
in prostate cancer.
NHERF1 has been reported to be involved in
the development and occurrence of cancer. Of note, abnormalities in
NHERF1 expression have been demonstrated to be associated
with the occurrence, development and metastasis of certain types of
cancer (4,9,10). While
the details of these mechanisms remain to be fully elucidated,
NHERF1 was reported to form a protein complex with epidermal growth
factor receptor (EGFR) and neurofibromatosis type 2 (NF2) at
intercellular adherens junctions (11), thereby mediating the internalization
and signal transduction of EGFR in order to regulate various
oncogenic processes. In addition, NHERF1 was reported to interact
with platelet-derived growth factor receptor (PDGFR) to form a
transduction complex with NF2 (12);
it was suggested that this complex functions to block the signaling
associated with PDGFR, thereby affecting apoptosis during the
epithelial-to-mesenchymal transition and promoting cancer
development and metastasis (13). In
addition, NHERF1 was reported to interact with several other
specific growth factor receptors that have important functions in
proliferation, invasion and angiogenesis (14,15).
Therefore, NHERF1 was thought to directly or indirectly affect the
development and progression of cancer (5).
The expression level of NHERF1 in prostate cancer
varies depending on the pathological pattern and cancer phase
(6). This may be explained by the
observation that different cell systems and tissues have different
genetic backgrounds. In addition, in samples where NHERF1 is
expressed at similar levels, NHERF1 may combine with different
proteins and thus exhibit different functions in samples with
varying genetic backgrounds. In the prostate cancer cell system
adopted in the present study (PC-3M cells), NHERF1 exhibited high
endogenous expression, which was reduced through stable
transfection with a NHERF1-targeting shRNA. The results
indicated that inhibition of NHERF1 expression attenuated the
proliferation and migration rates of PC-3M cells. Of note, major
changes in cell cycle distribution were not observed (data not
shown), although changes were detected in the height of the sub-G1
peak in flow cytometry analysis, which prompted the investigation
of apoptosis in NHERF1-knockdown cells. It was then
demonstrated that knockdown of NHERF1 increased apoptosis,
thereby contributing to the observed inhibition of cell
proliferation. Cell apoptosis, proliferation and differentiation
are all basic, vital functions of cells, which are closely
associated; imbalances in these functions have been associated with
tumor development (16,17). In general, apoptosis has a negative
regulatory role in cancer cells and may inhibit tumor growth
(18). Consistent with the increase
in apoptosis observed in the present study, inhibition of Bcl-2
expression in NHERF1 knockdown cells was detected. Bcl-2 is
an anti-apoptotic protein; thus, inhibition of Bcl-2 expression may
promote apoptosis. Therefore, in the present study, data from
western blot analysis of apoptosis-associated proteins supported
the flow cytometry results, demonstrating the role of NHERF1
in modulating the cell proliferation and apoptosis of prostate
cancer cells.
In conclusion, the results of the present study
demonstrated that NHERF1 was involved in regulating malignant
biological behaviors, including proliferation and migration, in
PC-3M cells. In addition, NHERF1 knockdown was demonstrated
to promote apoptosis through inhibition of Bcl-2 expression.
Therefore, these results indicated that NHERF1 acts as an oncogenic
protein in prostate cancer and may be a potential therapeutic
target for the treatment of prostate cancer. Although these data
support the role of NHERF1 in complexes with growth factor
receptors and ligands or in influencing the expression and
stability of β-catenin (19), further
studies with other associated transmembrane ligands are
required.
Acknowledgements
The present study was supported by the Basic
Clinical Research Cooperation Project of Capital Medical University
(Beijing, China; grant nos. 13JL20 and 13JL69), the Project of
Beijing Obstetrics and Gynecology Hospital, Capital Medical
University (grant no. 201208) the National Natural Science
Foundation of the People's Republic of China (grant no. 81372739)
and the Importation and Development of High-Caliber Talents Project
of Beijing Municipal Institutions (grant nos. CIT&TCD201304187
and CIT&TCD201304190).
References
|
1
|
Jácome-Pita F, Sánchez-Salas R, Barret E,
Amaruch N, Gonzalez-Enguita C and Cathelineau X: Focal therapy in
prostate cancer, The current situation. Ecancermedicalscience.
8:4352014.PubMed/NCBI
|
|
2
|
Roudier MP, True LD, Higano CS, Vesselle
H, Ellis W, Lange P and Vessella RL: Phenotypic heterogeneity of
end-stage prostate carcinoma metastatic to bone. Hum Pathol.
34:646–653. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Georgescu MM, Cote G, Agarwal NK and White
CL III: NHERF1/EBP50 controls morphogenesis of 3D colonic glands by
stabilizing PTEN and ezrin-radixin-moesin proteins at the apical
membrane. Neoplasia. 16:e1–e2. 2014. View Article : Google Scholar
|
|
4
|
Takahashi Y, Morales FC, Kreimann EL and
Georgescu MM: PTEN tumor suppressor associates with NHERF proteins
to attenuate PDGF receptor signaling. EMBO J. 25:910–920. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Molina JR, Agarwal NK, Morales FC, Hayashi
Y, Aldape KD, Cote G and Georgescu MM: PTEN, NHERF1 and PHLPP form
a tumor suppressor network that is disabled in glioblastoma.
Oncogene. 31:1264–1274. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Bartholow TL, Becich MJ, Chandran UR and
Parwani AV: Immunohistochemical analysis of
ezrin-radixin-moesin-binding phosphoprotein 50 in prostatic
adenocarcinoma. BMC Urol. 11:122011. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Prajapati A and Gupta S, Mistry B and
Gupta S: Prostate stem cells in the development of benign prostate
hyperplasia and prostate cancer, Emerging role and concepts. Biomed
Res Int. 2013:1079542013. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
International Organization for
Standardization (ISO): ISO 10993-5: Biological evaluation of
medical devices - Part 5: Tests for in vitro cytotoxicity. ISO.
Geneva: 2009.
|
|
9
|
Malfettone A, Saponaro C, Paradiso A,
Simone G and Mangia A: Peritumoral vascular invasion and NHERF1
expression define an immunophenotype of grade 2 invasive breast
cancer associated with poor prognosis. BMC Cancer. 12:1062012.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Bellizzi A, Malfettone A, Cardone RA and
Mangia A: NHERF1/EBP50 in Breast Cancer, Clinical Perspectives.
Breast Care (Basel). 5:86–90. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Curto M, Cole BK, Lallemand D, Liu CH and
McClatchey AI: Contact-dependent inhibition of EGFR signaling by
Nf2/Merlin. J Cell Biol. 177:893–903. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
James MF, Beauchamp RL, Manchanda N,
Kazlauskas A and Ramesh V: NHERF binding site links the betaPDGFR
to the cytoskeleton and regulates cell spreading and migration. J
Cell Sci. 117:2951–2961. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Malavaki CJ, Roussidis AE, Gialeli C,
Kletsas D Tsegenidis, et al: Imatinib as a key inhibitor of the
platelet-derived growth factor receptor mediated expression of cell
surface heparan sulfate proteoglycans and functional properties of
breast cancer cells. FEBS J. 280:2477–2489. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Yao W, Feng D, Bian W, Yang L, Li Y, Yang
Z, Xiong Y, Zheng J, Zhai R and He J: EBP50 inhibits EGF-induced
breast cancer cell proliferation by blocking EGFR phosphorylation.
Amino Acids. 43:2027–2035. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Jiang Y, Wang S, Holcomb J, Trescott L,
Guan X, Hou Y, Brunzelle J, Sirinupong N, Li C and Yang Z:
Crystallographic analysis of NHERF1-PLCβ3 interaction provides
structural basis for CXCR2 signaling in pancreatic cancer. Biochem
Biophys Res Commun. 446:638–643. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Mamlouk S, Kalucka J, Singh RP, Franke K,
Muschter A, Langer A, Jakob C, Gassmann M, Baretton GB and Wielockx
B: Loss of prolyl hydroxylase-2 in myeloid cells and T-lymphocytes
impairs tumor development. Int J Cancer. 134:849–858. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Chen Q, Lu HS, Gan MF, Chen LX, He K, Fan
GM and Cao XQ: Expression and prognostic role of MEKK3 and pERK in
patients with renal clear cell carcinoma. Asian Pac J Cancer Prev.
16:2495–2499. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Tatsuta T, Sugawara S, Takahashi K, Ogawa
Y, Hosono M and Nitta K: Cancer-selective induction of apoptosis by
leczyme. Front Oncol. 4:1392014. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Kreimann EL, Morales FC, de Orbeta-Cruz J,
Takahashi Y, Adams H, et al: Cortical stabilization of beta-catenin
contributes to NHERF1/EBP50 tumor suppressor function. Oncogene.
26:5290–5299. 2007. View Article : Google Scholar : PubMed/NCBI
|