Introduction
MicroRNAs (miRNAs/miRs) are ~22-nt, single-stranded,
non-coding RNAs that serve an important role in regulating
carcinogenic pathways (1,2). miRNAs may lead to messenger RNA (mRNA)
degradation or translational repression by binding to the
3′-untranslated regions of target genes, and are involved in a wide
range of biological and pathological processes, including cell
differentiation, migration, growth, proliferation, apoptosis and
metabolism (3–6). miRNA expression is often dysregulated in
various types of cancer, with either an oncogenic function when
overexpressed or a tumor suppressor function when downregulated
(2,7,8). Whether
miRNA functions as an oncogene or tumor suppressor gene is
primarily dependent on the regulation of its target genes. Due to
the presence of complementarity between miRNAs and target gene
mRNAs, each miRNA may regulate multiple mRNAs and each mRNA may be
targeted by multiple miRNAs (7).
miRNAs exert their function in a temporal- and tissue-specific
manner (8). In clinical practice,
miRNAs may prove invaluable, being exploited as biomarkers for
diagnostic, prognostic and monitoring purposes. Additionally,
miRNAs may serve as possible targets for novel therapeutic
approaches, particularly in patients with tumor subtypes that do
not respond to therapies presently available (1).
Kidney cancer is the 14th most prevalent cancer in
the world, with incidence and mortality rates that have recently
plateaued in Europe and North America, but continue to increase in
developing countries (9,10). Renal cell carcinoma (RCC) is the most
frequent kidney cancer that develops in adults, accounting for ~90%
of all renal tumors and 3.9% of all cancers (11,12). The
three most prevalent histological subtypes of RCC include clear
cell RCC (ccRCC), papillary RCC and chromophobe RCC, with a
prevalence of 70, 10 and 5%, respectively (13). RCCs are clinically silent at their
earliest stages, therefore, at the time of the initial diagnosis,
up to 30% of patients have already progressed to a locally advanced
disease state or exhibit metastases (14). If detected promptly, early-stage RCC
is curable in >90% of cases (15).
In 20–40% of patients, a recurring disease will develop following
surgical nephrectomy due to the lack of effective adjuvant therapy,
including chemotherapy or radiotherapy (16).
Therefore, miRNAs have gained increasing attention
as important factors associated with RCC tumorigenesis and
development, serving as biomarkers for early detection and
progression monitoring, and as potential targets for molecular
therapy (16,17). However, to the best of our knowledge,
the expression and function of miR-142-3p in RCC has not yet been
fully investigated. Previous microarray chip studies have
demonstrated that miR-142-3p is overexpressed in RCC tissues
compared with adjacent normal or benign kidney tissues (18–21). It
has also been reported that miR-142-3p is dysregulated in
malignancies of the breast (22),
thyroid (23), liver (24), stomach (25), lung (26), blood (27,28),
colorectum (29), testes (30), esophagus (31), head and neck (32), and bone (33). The present study establishes the
oncogenic role of miR-142-3p in RCC, demonstrating how it regulates
cell migration, proliferation and apoptosis.
Materials and methods
Cell culture and transfection
Human RCC cells (786-O and ACHN) and normal human
embryo kidney cells (293T) from the Guangdong and Shenzhen Key
Laboratory of Male Reproductive Medicine and Genetics (Shenzhen,
China) were seeded and grown in Dulbecco's modified Eagle's medium
(DMEM) (Gibco; Thermo Fisher Scientific, Inc., Waltham, MA, USA)
with 10% fetal bovine serum, and l% glutamine and included 100
µg/ml penicillin and 100 mg/ml streptomycin sulfates (Invitrogen;
Thermo Fisher Scientific, Inc.) at 37°C, in a humidified atmosphere
containing 5% CO2. For the downregulation of miR-142-3p,
synthesized miR-142-3p inhibitor (Shanghai GenePharma, Co., Ltd.,
Shanghai, China) was transfected into cells using Lipofectamine®
2000 (Invitrogen; Thermo Fisher Scientific, Inc.), and then the
cells were mixed in the Opti-MEM® I Reduced Serum Medium (Gibco;
Thermo Fisher Scientific, Inc.) 24 h after plating. Quantitative
polymerase chain reaction (qPCR) was used to verify the efficiency
of transfection. The sequence of the miR-142-3p inhibitor was
5′-UCCAUAAAGUAGGAAACACUACA-3′ and the negative control was
5′-CAGUACUUUUGUGUAGUACAA-3′.
Human patient samples
A total of 53 paired RCC tissues and adjacent
non-cancerous kidney tissues were collected from patients in the
Peking University Shenzhen Hospital (Shenzhen, China), from
September 2012 to November 2014. Written informed consent was
provided by all patients. Histological and pathological diagnostics
for patients with RCC were determined according to the 2009
American Joint Committee on Cancer staging system (34). The patients with RCC had not been
treated with either chemotherapy or radiotherapy prior to tissue
sampling. Tissue samples were snap-frozen and stored in a cryo
freezer at −80°C in RNAlater® RNA Stabilization Agent (Qiagen,
Inc., Valencia, CA, USA) for further research. The
clinicopathological information of all the patients is presented in
Table I.
 | Table I.Clinicopathological features observed
in renal cell carcinoma patients. |
Table I.
Clinicopathological features observed
in renal cell carcinoma patients.
| Characteristic | Value |
|---|
| Mean age (range),
years | 49 (27–71) |
| Gender, n |
|
|
Male | 35 |
|
Female | 18 |
| Histological type,
n |
|
| Clear
cell | 42 |
|
Papillary | 11 |
| pT-stage, n |
|
| T1 | 30 |
| T2 | 20 |
|
T3+T4 | 3 |
| Fuhrman grade,
n |
|
| I | 18 |
| II | 23 |
|
III | 9 |
| IV | 3 |
| AJCC clinical
stage, n |
|
| I | 31 |
| II | 19 |
|
III+IV | 3 |
RNA isolation, cDNA synthesis and
qPCR
Total RNA was extracted from the cell lines and
tissues using TRIzol® Reagent (Invitrogen; Thermo Fisher
Scientific, Inc.), and was purified using the RNeasy® Maxi kit
(Qiagen, Inc.) according to the manufacturer's protocols. The
amount of RNA was measured on a NanoDrop 2000c (Thermo Fisher
Scientific, Inc.) and the RNA samples with 260/280 ratios of
1.8–2.0 were utilized for further experiments. Synthesis of cDNA
with reverse transcriptase was performed with the miScript II RT
kit (Qiagen, Inc.). The sequence of the forward primer for
miR-142-3p was 5′-TGTAGTGTTTCCTACTTTATGGA-3′ and the reverse primer
was provided by the miScript SYBR® Green PCR kit (Qiagen, Inc.).
qPCR analysis was performed using the same kit, according to the
manufacturer's protocols, on the LightCycler® 480 Real-Time PCR
system (Roche Diagnostics, Basel, Switzerland). The thermal cycling
conditions for PCR were as follows: 95°C for 15 min, followed by 40
cycles of 94°C for 15 sec, 55°C for 30 sec and 72°C for 30 sec.
Quantification cycle (Cq) values of miR-142-3p were equilibrated to
U6, which was used as an internal control. The forward primer of U6
was 5′-CTCGCTTCGGCAGCACA-3′ and the reverse primer was
5′-ACGCTTCACGAATTTGCGT-3′. Relative expression was calculated using
the ΔΔCq method.
Wound healing assay
A wound scratch assay was performed to evaluate the
migratory ability of the 786-O and ACHN cells in vitro. A
total of ~3×105 cells were seeded into each well within
a 12-well dish, and each was transfected with either 100 pmol of
chemically synthesized miR-142-3p inhibitor or a negative control,
using Lipofectamine 2000. Following 6 h of transfection, a sterile
200-µl pipette tip was used to scratch a clear line through the
cell layer. The cells were then rinsed with phosphate-buffered
saline (PBS) and cultured in serum-free DMEM at 37°C in a
humidified chamber, containing 5% CO2. Using a digital
camera system, images were captured at 0, 24 and 48 h after the
scratches were made. The experiments were performed in triplicate
and repeated ≥3 times.
Cell proliferation assay using
3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide
(MTT)
MTT assays were conducted to analyze the
proliferation ability of the 786-O or ACHN cells. A total of
~5×103 786-O or ACHN cells were seeded into each well
within 96-well plates, with 5 replicate wells of each condition.
Each well was transfected with either 5 pmol miR-142-3p inhibitor,
or a negative control, and proliferation was measured at 0, 24, 48
or 72 h post-transfection. Prior to measurement, 20 µl MTT (5
mg/ml; Sigma-Aldrich, St. Louis, MO, USA) was added to each well,
with the 96-well plates then incubated at 37°C in a humidified
chamber, containing 5% CO2, for 6h. Subsequently, the
MTT medium mixtures were discarded and 120 µl dimethylsulfoxide
(Sigma-Aldrich, Shanghai, China) was added. Following agitation for
30 min at room temperature, the optical density values were
measured by the iMark Microplate Absorbance Reader (Bio-Rad
Laboratories, Inc., Hercules, CA, USA) at a wave length of 490 nm,
with 630 nm serving as the reference wave length.
Flow cytometry assay
The Coulter® Epics XL® flow cytometer (Beckman
Coulter, Inc., Brea, CA, USA) was used to quantify the early
apoptosis rate of the 786-O and ACHN cells, with Annexin
V-fluorescein isothiocyanate (FITC) and propidium iodide (PI)
staining (Invitrogen; Thermo Fisher Scientific, Inc.). A total of
~3×105 786-O or ACHN cells were seeded into 6-well
plates for the cell apoptosis assay. The cells were transfected
with 200 pmol miR-142-3p inhibitor, or the negative control, for 6
h. Following a 48-h transfection, the cells, including floating
cells, were harvested, washed twice with 4°C PBS and resuspended in
100 µl 1X binding buffer at a concentration of at least
3×106 cells/ml. This suspension (100 µl) was stained
with 5 µl Annexin V-FITC and 5 µl PI for 15 min at room temperature
in the dark. Following the addition of 400 µl binding buffer to
each tube, the cells were analyzed by flow cytometry. Each
experiment was performed at least 3 times.
Statistical analysis
For the comparison of miR-142-3p expression levels
in matched tumor and normal samples, a paired t-test was performed.
The relative expression of miR-142-3p in tissues was presented as
the mean ± standard error. All other data were presented as the
mean ± standard deviation from the three independent experiments.
Statistical analysis was performed using SPSS software, version
19.0 (IBM SPSS, Armonk, NY, USA). Statistical significance was
determined with Student's t-test. P<0.05 was considered to
indicate a statistically significant difference.
Results
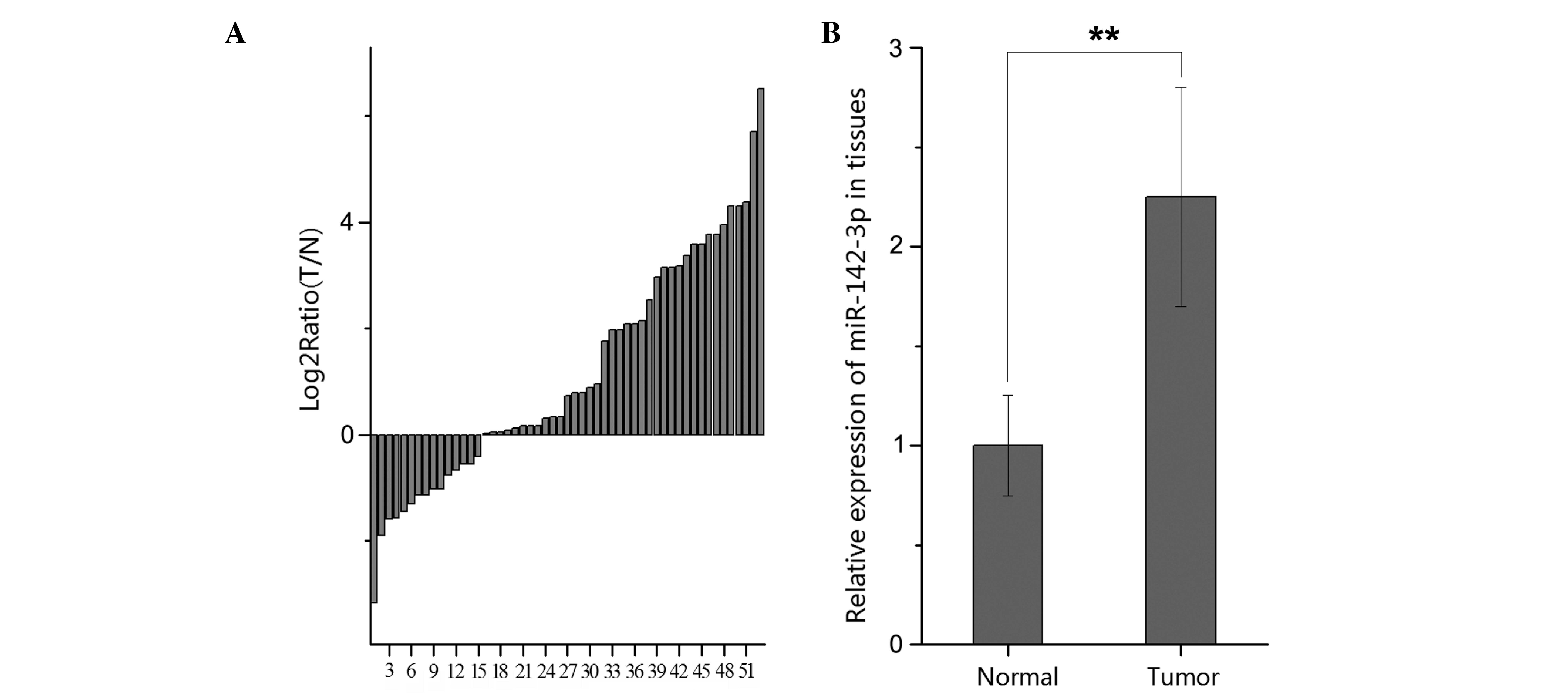
miR-142-3p is upregulated in RCC
tissues compared with adjacent normal tissues
A total of 53 paired RCC and adjacent normal tissues
were analyzed using qPCR in order to quantify the expression
pattern of miR-142-3p in each tissue type. The results demonstrated
that the expression of miR-142-3p [Log2 (T/N)] was generally
increased (38/53) in the tumors in comparison with the paired
normal samples (Fig. 1A). The present
study demonstrated that the relative expression of miR-142-3p in
the RCC tissues was significantly overexpressed when compared with
the adjacent normal tissues (P<0.01), as presented in Fig. 1B. Such results indicated that
miR-142-3p may act as an oncogene during RCC development. However,
the function of miR-142-3p required further investigation.
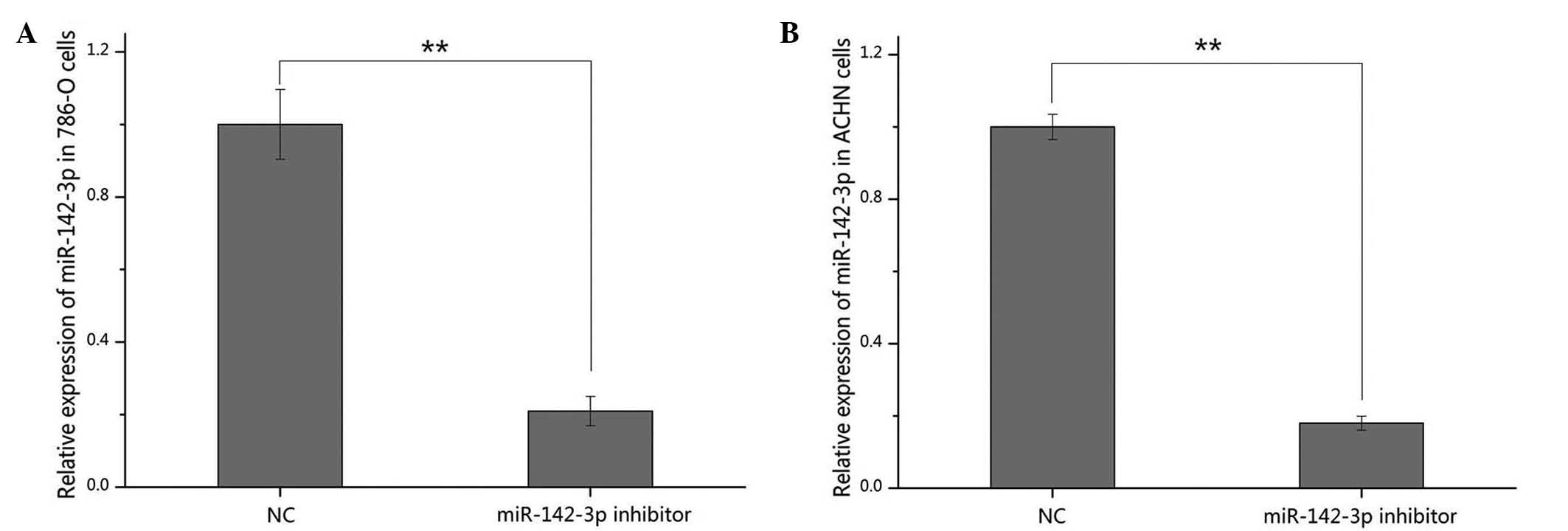
Validation of cell transfection
efficiency
The transfection efficiency of miR-142-3p inhibitor
was quantified by qPCR, whilst comparisons were made with a
negative control. The results indicated that miR-142-3p was
downregulated by 79.04 and 82.02% compared with the negative
control, following transfection in the 786-O (P<0.01; Fig. 2A) and ACHN cells (P<0.01; Fig. 2B), respectively.
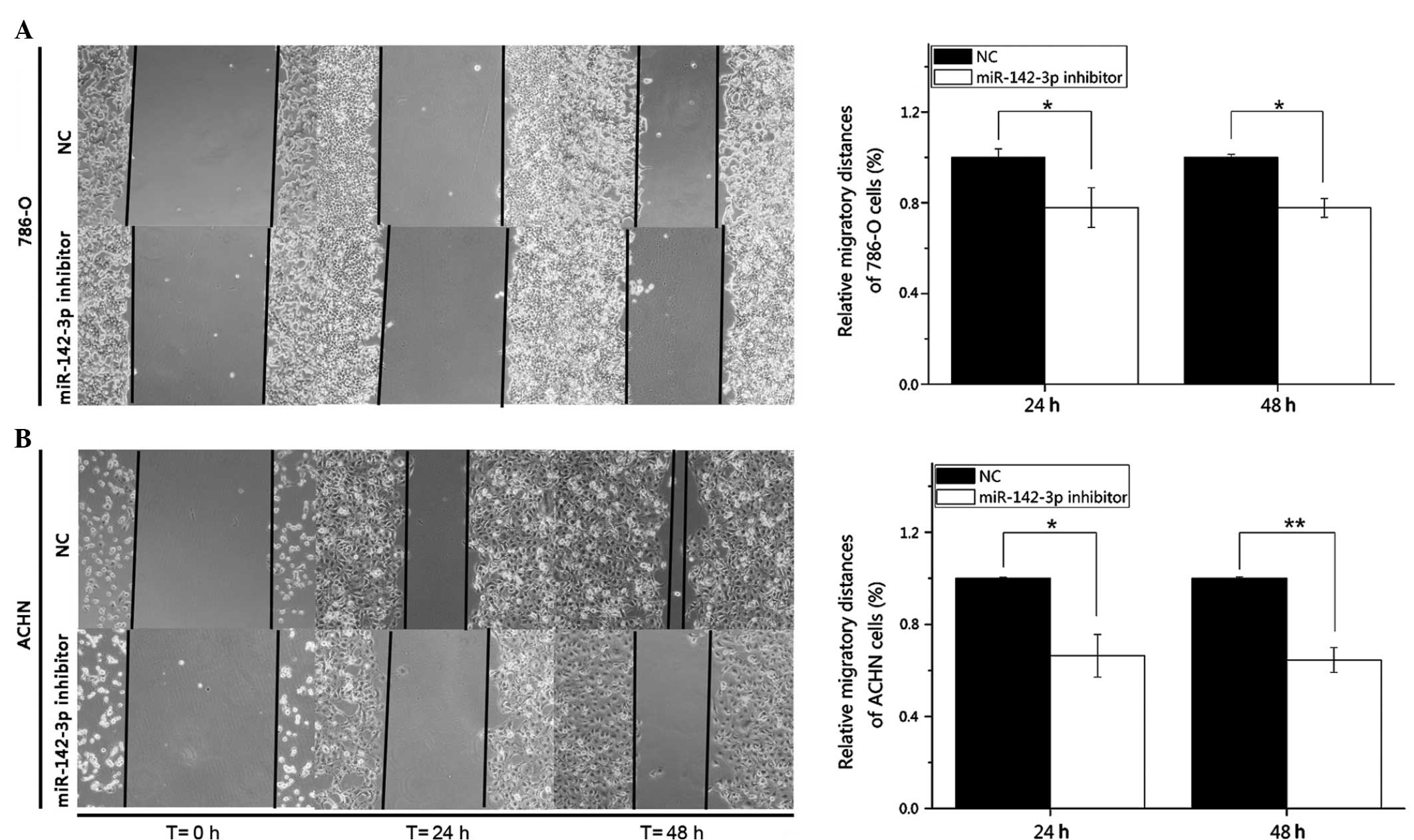
miR-142-3p inhibitor suppresses 786-O
and ACHN cell migration
Wound healing assays were performed to observe the
function of miR-142-3p in cell migration. Images of each wound were
captured at 0, 24, and 48 h post-transfection using a digital
camera system (Fig. 3). The wounds of
cells transfected with miR-142-3p inhibitor were wider than those
of cells transfected with the negative control. Statistical
analysis demonstrated that the migratory distances of the
miR-142-3p inhibitor group were significantly decreased by 22.11%
(P<0.05) and 22.26% (P<0.05) for the 786-O cells, and by
33.66% (P<0.05) and 35.47% (P<0.01) for the ACHN cells at 24
and 48 h post-transfection, in comparison to the negative control
group. Such results suggested that the downregulation of miR-142-3p
inhibited the migratory ability of the RCC cells.
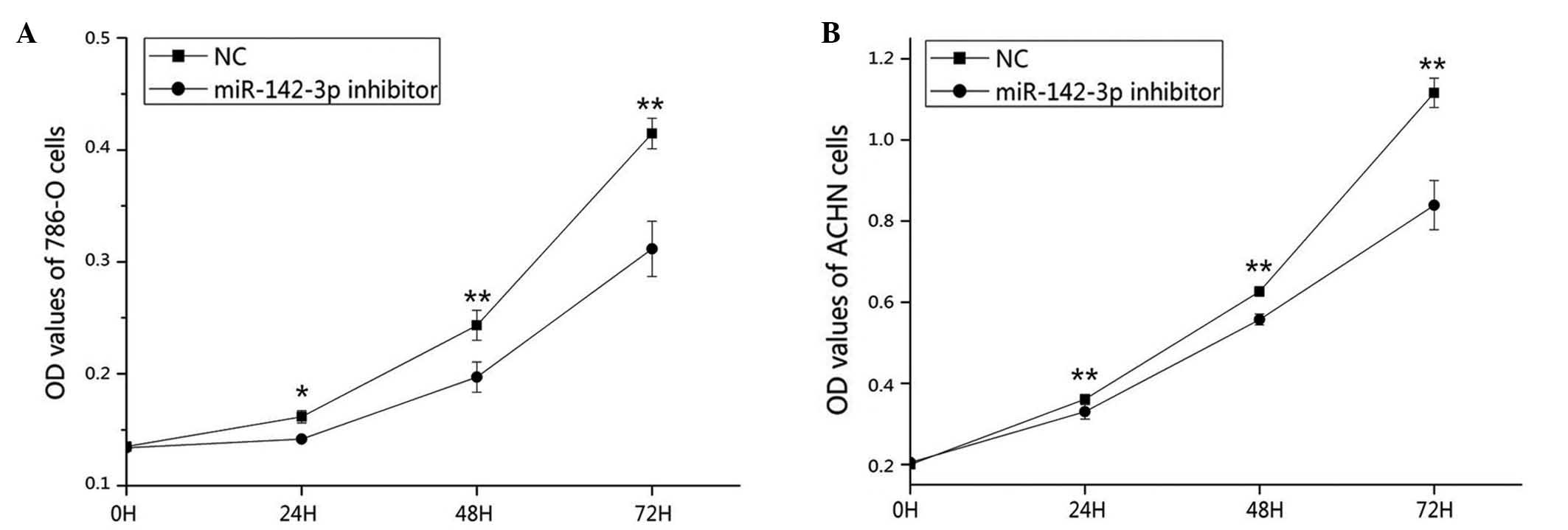
miR-142-3p inhibitor inhibits 786-O
and ACHN cell proliferation
MTT assays were performed to determine if the
downregulation of miR-142-3p had an impact on the proliferation of
the RCC cells. The results demonstrated that the proliferation of
the 786-O cells decreased by 10.15% (P<0.05), 19.04% (P<0.01)
and 24.84% (P<0.01), and that the proliferation of the ACHN
cells decreased by 8.59% (P<0.01), 11.02% (P<0.01) and 24.82%
(P<0.01), at 24, 48 and 72 h post-transfection of the miR-142-3p
inhibitor, as compared with the negative control. The results
indicated that the inhibition of miR-142-3p expression
significantly reduced the proliferation of the RCC cells (Fig. 4).
miR-142-3p inhibitor promotes 786-O
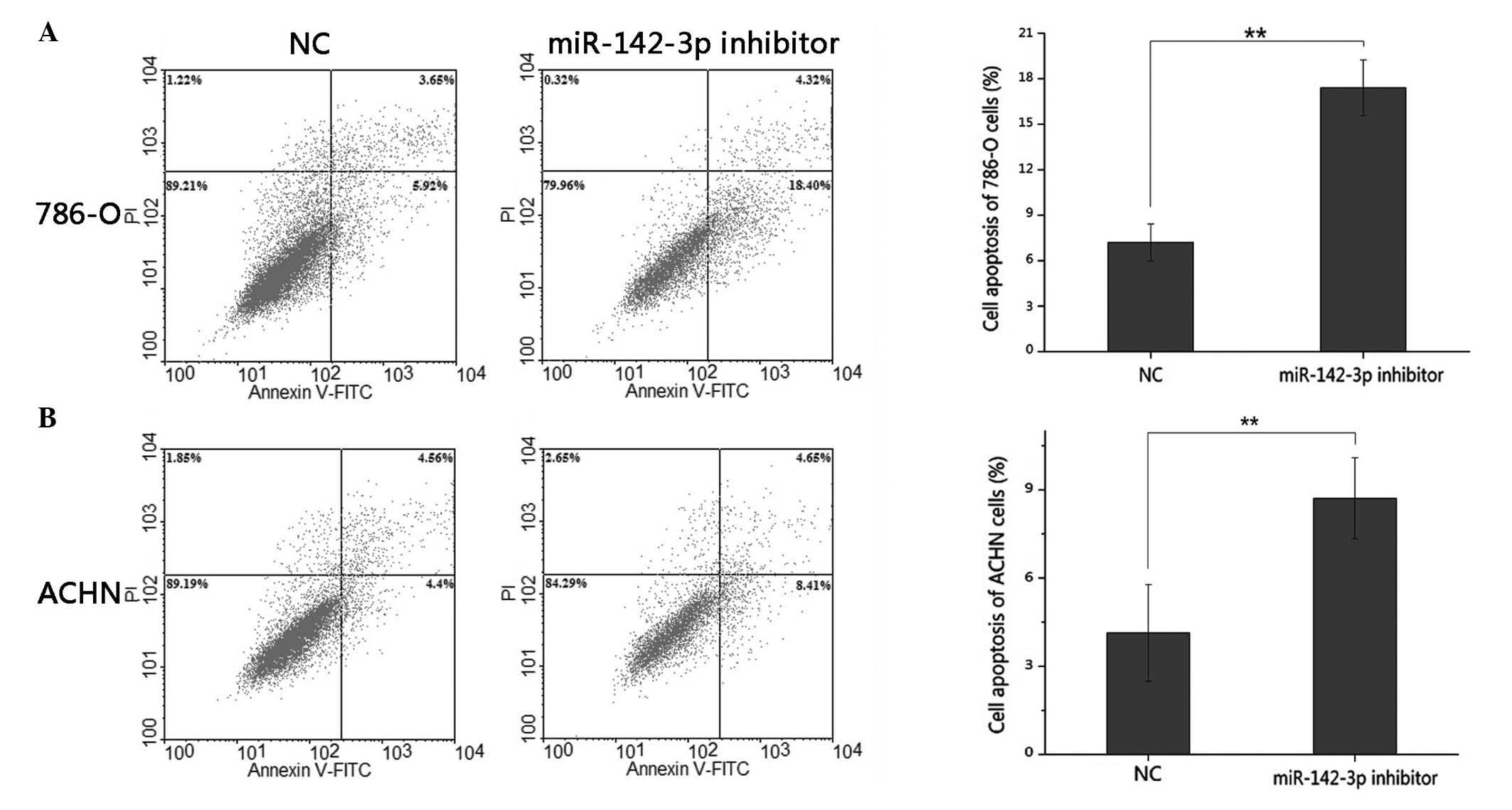
and ACHN cell apoptosis
The effects of the miR-142-3p inhibitor on apoptosis
were determined by flow cytometric analysis. The results
demonstrated that the average early apoptosis rate of the 786-O
cells, transfected with miR-142-3p inhibitor or negative control,
was 17.40 vs. 7.20% (P<0.01), whilst the average apoptotic rate
of the ACHN cells was 8.71 vs. 4.14% (P<0.01). This data
indicated that the downregulation of miR-142-3p promoted RCC cell
apoptosis (Fig. 5).
Discussion
The initiation and development of cancer involves
the activation of a number of oncogenes and the dysfunction of
numerous tumor suppressor genes. miRNAs have been identified to
serve important roles in various types of cancer, regulating ~50%
of human genes (5). Recent genomic
studies of ccRCC have advanced our understanding of the molecular
background and pathways, and the possible genetic alterations,
implicated in ccRCC tumorigenesis (35). The most notable gene pathways involved
in RCC tumorigenesis include the Von Hippel-Lindau (VHL)/hypoxia
inducible factor (HIF), vascular endothelial growth factor (VEGF)
and mechanistic target of rapamycin (mTOR) pathways. Therapies
targeting these genes have been introduced into clinical practice.
miRNAs target numerous genes, including HIF, mTOR, VEGF and VHL,
through translational inhibition or the induction of mRNA
degradation, which suggests that miRNAs function as important
factors in RCC initiation and development (36,37).
Previous studies have reported that miR-142-3p is
dysregulated in various types of cancer (22–33), and
may therefore function as a biomarker. The expression of miR-142-3p
in RCC has not yet been validated by qPCR. However, recent
microarray analyses comparing RCC and adjacent normal or benign
kidney tissues demonstrated that miR-142-3p was significantly
overexpressed in tumors when compared with paired control groups
(18–21), suggesting that miR-142-3p may function
as an oncogene in RCC development. Therefore, in the present study,
qPCR was performed to quantify the relative expression of
miR-142-3p in 53 paired RCC and adjacent normal tissues.
Furthermore, the impacts of miR-142-3p on cellular migration,
proliferation and apoptosis were analyzed through the use of wound
healing, MTT and flow cytometry assays. The results demonstrated
that miR-142-3p was significantly upregulated in RCC tissues in
comparison to adjacent normal tissues. Downregulation of
miR-142-3p, induced by the chemically synthesized miR-142-3p
inhibitor, significantly suppressed cell migration and
proliferation, and induced cell apoptosis in the 786-O and ACHN
cells, suggesting that miR-142-3p may function as an oncogene
during RCC tumorigenesis. However, the miR-142-3p-mediated
molecular pathways that affect cell migration, proliferation and
apoptosis remain unclear.
The various molecular mechanisms of miR-142-3p have
been reported to affect tumor initiation, development, metastasis
and invasion differentially, depending on the type of cancer that
is present. This is believed to be due to the target genes involved
being dependent on the type of cancer. In addition, different
stages of cancer development exhibit the tissue- and
temporal-specific manner of the function of miRNAs (8). For example, in breast cancer,
overexpressed miR-142-3p regulates the properties of breast cancer
stem cells (BCSCs), at least in part by activating the WNT
signaling pathway and miR-150 expression. Previous in vivo
experiments demonstrated that the enforced expression of miR-142 in
normal mouse mammary stem cells resulted in the regeneration of
hyperproliferative mammary glands, whilst the knockdown of
endogenous miR-142 effectively suppressed organoid formation by
BCSCs, and also slowed tumor growth initiated by human BCSCs
(22). Downregulation of miR-142-3p
in thyroid neoplastic tissues contributes to thyroid follicular
tumorigenesis through the targeting of ASH1L and MLL1, and
subsequently promotes its tumor suppressive function (23). In non-small cell lung cancer,
overexpression of miR-142-3p represses transforming growth factor
(TGF)-induced growth inhibition through the repression of TGFβ
receptor 1 (26). However, a previous
study investigating miR-142-3p in human acute lymphoblastic
leukemia demonstrated that miR-142-3p inhibits cell proliferation
by targeting the MLL-AF4 oncogene (27). miR-142-3p-mediated loss of protein
tyrosine phosphatase non-receptor type 23 (PTPN23) expression may
also be a key event in the pathogenesis of testicular germ cell
tumors (30). By targeting the
oncogenes cluster of differentiation 133, ATP-binding cassette,
subfamily G, member 3 and leucine-rich repeat-containing G
protein-coupled receptor 5, miR-142-3p functions as a tumor
suppressor in colon cancer cells (38). Other than its roles in tumorigenesis,
miR-142 is also important for megakaryopoiesis, with the genetic
ablation of miR-142 responsible for impaired megakaryocyte
maturation, the inhibition of polyploidization, abnormal
proplatelet formation and thrombocytopenia, through the
orchestration of an actin cytoskeleton network (39).
In clinical application, the reduced expression of
miR-142-3p in hepatocellular carcinoma and its overexpression in
esophageal squamous cell carcinoma was significantly associated
with reduced survival, suggesting that miR-142-3p may function as a
prognostic predictor (24,31). Serum miR-142-3p was identified to be
associated with a high recurrence risk in patients with early-stage
lung adenocarcinoma, and was a putative serum marker for risk
assessment (40). The expression
levels of miR-142-3p and miR-29a in peripheral blood mononuclear
cells may be used as novel diagnostic markers with ~90% sensitivity
and ~100% specificity for the diagnosis of acute myeloid leukemia
(41). During human bronchial
squamous carcinogenesis, miR-142-3p, which is typically upregulated
during lung development, was first downregulated at the earliest
stages of carcinogenesis, and was also subsequently upregulated
during later stages, suggesting that the expression of miR-142-3p
may be monitored to assess cancer development (42).
In conclusion, the present study demonstrated the
expression and function of miR-142-3p in RCC tumorigenesis. The
oncogenic function of miR-142-3p was indicated by its upregulated
expression in RCC tissues, its inhibition of cellular migration and
proliferation, and the reduction in cell apoptosis induced by an
miR-142-3p inhibitor. Further investigation is required to analyze
the miR-142-3p-mediated molecular pathway and its role in RCC
development, with its potential to aid early disease detection and
prognosis prediction, whilst serving as a therapeutic target, thus
proving its clinical significance.
Acknowledgements
This study was supported by the National Natural
Science Foundation of China (no. 81101922), the Science and
Technology Development Fund Project of Shenzhen (nos.
JCYJ20130402114702124 and JCYJ20130402113131201) and the Fund of
Guangdong Key Medical Subject.
References
|
1
|
Schaefer A, Stephan C, Busch J, Yousef GM
and Jung K: Diagnostic, prognostic and therapeutic implications of
microRNAs in urologic tumors. Nat Rev Urol. 7:286–297. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Garzon R, Calin GA and Croce CM: MicroRNAs
in cancer. Annu Rev Med. 60:167–179. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Huntzinger E and Izaurralde E: Gene
silencing by microRNAs: Contributions of translational repression
and mRNA decay. Nat Rev Genet. 12:99–110. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Bartel DP: MicroRNAs: T arget recognition
and regulatory functions. Cell. 136:215–233. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Krol J, Loedige I and Filipowicz W: The
widespread regulation of microRNA biogenesis, function and decay.
Nat Rev Genet. 11:597–610. 2010.PubMed/NCBI
|
|
6
|
Carthew RW and Sontheimer EJ: Origins and
mechanisms of miRNAs and siRNAs. Cell. 136:642–655. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Shenouda SK and Alahari SK: MicroRNA
function in cancer: Oncogene or a tumor suppressor? Cancer
Metastasis Rev. 28:369–378. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Guil S and Esteller M: DNA methylomes
histone codes and miRNAs: Tying it all together. Int J Biochem Cell
Biol. 41:87–95. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Ridge CA, Pua BB and Madoff DC:
Epidemiology and staging of renal cell carcinoma. Semin Intervent
Radiol. 31:3–8. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Ferlay J, Shin HR, Bray F, Forman D,
Mathers C and Parkin DM: Estimates of worldwide burden of cancer in
2008: GLOBOCAN 2008. Int J Cancer. 127:2893–2917. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Tavani A: LaV ecchia C: Epidemiology of
renal-cell carcinoma. J Nephrol. 10:93–106. 1997.PubMed/NCBI
|
|
12
|
National Cancer Institute: Surveillance,
Epidemiology, and End Results Program. SEER Stat Fact Sheets:
Kidney and Renal Pelvis Cancer. http://seer.cancer.gov/statfacts/html/kidrp.htmlAccessed.
March 30–2013
|
|
13
|
Patel C, Ahmed A and Ellsworth P: Renal
cell carcinoma, A reappraisal. Urol Nurs. 32:182–190.
2012.PubMed/NCBI
|
|
14
|
Lam JS, Leppert JT, Figlin RA and
Belldegrun AS: Role of molecular markers in the diagnosis and
therapy of renal cell carcinoma. Urology 66 (Suppl). 1–9. 2005.
View Article : Google Scholar
|
|
15
|
Rini BI, Campbell SC and Escudier B: Renal
cell carcinoma. Lancet. 373:1119–1132. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Dias F, Teixeira AL, Santos JI, Gomes M,
Nogueira A, Assis J and Medeiros R: Renal cell carcinoma
development and miRNAs, A possible link to the EGFR pathway.
Pharmacogenomics. 14:1793–1803. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Al-Ali BM, Ress AL, Gerger A and Pichler
M: MicroRNAs in renal cell carcinoma, Implications for
pathogenesis, diagnosis, prognosis and therapy. Anticancer Res.
32:3727–3732. 2012.PubMed/NCBI
|
|
18
|
Liu H, Brannon AR, Reddy AR, Alexe G,
Seiler MW, Arreola A, Oza JH, Yao M, Juan D, Liou LS, et al:
Identifying mRNA targets of microRNA dysregulated in cancer: With
application to clear cell renal cell carcinoma. BMC Syst Biol.
4:512010. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Juan D, Alexe G, Antes T, Liu H,
Madabhushi A, Delisi C, Ganesan S, Bhanot G and Liou LS:
Identification of a microRNA panel for clear-cell kidney cancer.
Urology. 75:835–841. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Yi Z, Fu Y, Zhao S, Zhang X and Ma C:
Differential expression of miRNA patterns in renal cell carcinoma
and nontumorous tissues. J Cancer Res Clin Oncol. 136:855–862.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Wu X, Weng L, Li X, Guo C, Pal SK, Jin JM,
Li Y, Nelson RA, Mu B, Onami SH, et al: Identification of a
4-microRNA signature for clear cell renal cell carcinoma metastasis
and prognosis. PLoS One. 7:e356612012. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Isobe T, Hisamori S, Hogan DJ, Zabala M,
Hendrickson DG, Dalerba P, Cai S, Scheeren F, Kuo AH, Sikandar SS,
et al: miR-142 regulates the tumorigenicity of human breast cancer
stem cells through the canonical WNT signaling pathway. eLife.
3:019772014. View Article : Google Scholar
|
|
23
|
Colamaio M, Puca F, Ragozzino E, Gemei M,
Decaussin-Petrucci M, Aiello C, Bastos AU, Federico A, Chiappetta
G, Del Vecchio L, et al: miR-142-3p down-regulation contributes to
thyroid follicular tumorigenesis by targeting ASH1L and MLL1. J
Clin Endocrinol Metab. 100:E59–E69. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Chai S, Tong M, Ng KY, Kwan PS, Chan YP,
Fung TM, Lee TK, Wong N, Xie D, Yuan YF, et al: Regulatory role of
miR-142-3p on the functional hepatic cancer stem cell marker CD133.
Oncotarget. 5:5725–5735. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Han J, Yu J and Ling Z: Screening of
specific miRNA in early gastric cancer. Zhonghua Wei Chang Wai Ke
Za Zhi. 17:175–179. 2014.(In Chinese). PubMed/NCBI
|
|
26
|
Lei Z, Xu G, Wang L, Yang H, Liu X, Zhao J
and Zhang HT: MiR-142-3p represses TGF-β-induced growth inhibition
through repression of TGFβR1 in non-small cell lung cancer. FASEB
J. 28:2696–2704. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Dou L, Li J, Zheng D, Li Y, Gao X, Xu C,
Gao L, Wang L and Yu L: MicroRNA-142-3p inhibits cell proliferation
in human acute lymphoblastic leukemia by targeting the MLL-AF4
oncogene. Mol Biol Rep. 40:6811–6819. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Lv M, Zhang X, Jia H, Li D, Zhang B, Zhang
H, Hong M, Jiang T, Jiang Q, Lu J, et al: An oncogenic role of
miR-142-3p in human T-cell acute lymphoblastic leukemia (T-ALL) by
targeting glucocorticoid receptor-α and cAMP/PKA pathways.
Leukemia. 26:769–777. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Kanaan Z, Roberts H, Eichenberger MR,
Billeter A, Ocheretner G, Pan J, Rai SN, Jorden J, Williford A and
Galandiuk S: A plasma microRNA panel for detection of colorectal
adenomas, A step toward more precise screening for colorectal
cancer. Ann Surg. 258:400–408. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Tanaka K, Kondo K, Kitajima K, Muraoka M,
Nozawa A and Hara T: Tumor-suppressive function of protein-tyrosine
phosphatase non-receptor type 23 in testicular germ cell tumors is
lost upon overexpression of miR142-3p microRNA. J Biol Chem.
288:23990–23999. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Lin RJ, Xiao DW, Liao LD, Chen T, Xie ZF,
Huang WZ, Wang WS, Jiang TF, Wu BL, Li EM and Xu LY: miR-142-3p as
a potential prognostic biomarker for esophageal squamous cell
carcinoma. J Surg Oncol. 105:175–182. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Hui AB, Lenarduzzi M, Krushel T, Waldron
L, Pintilie M, Shi W, Perez-Ordonez B, Jurisica I, O'Sullivan B,
Waldron J, et al: Comprehensive microRNA profiling for head and
neck squamous cell carcinomas. Clin Cancer Res. 16:1129–1139. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Namløs HM, Meza-Zepeda LA, Barøy T,
Østensen IH, Kresse SH, Kuijjer ML, Serra M, Bürger H,
Cleton-Jansen AM and Myklebost O: Modulation of the osteosarcoma
expression phenotype by microRNAs. PLoS One. 7:e480862012.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Edge SB, Byrd DR, Compton CC, et al: AJCC
Cancer Staging Manual (7th). New York, NY, USA: Springer Verlag.
547–560. 2009.
|
|
35
|
Rydzanicz M, Wrzesiński T, Bluyssen HA and
Wesoły J: Genomics and epigenomics of clear cell renal cell
carcinoma. Recent developments and potential applications. Cancer
Lett. 341:111–126. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Maher ER: Genomics and epigenomics of
renal cell carcinoma. Semin Cancer Biol. 23:10–17. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Chow TF, Mankaruos M, Scorilas A, Youssef
Y, Girgis A, Mossad S, Metias S, Rofael Y, Honey RJ, Stewart R, et
al: The miR-17-92 cluster is over expressed in and has an oncogenic
effect on renal cell carcinoma. J Urol. 183:743–751. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Shen WW, Zeng Z, Zhu WX and Fu GH:
miR-142-3p functions as a tumor suppressor by targeting CD133,
ABCG2, and Lgr5 in colon cancer cells. J Mol Med (Berl).
91:989–1000. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Chapnik E, Rivkin N, Mildner A, Beck G,
Pasvolsky R, Metzl-Raz E, Birger Y, Amir G, Tirosh I, Porat Z, et
al: miR-142 orchestrates a network of actin cytoskeleton regulators
during megakaryopoiesis. eLife. 3:e019642014. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Kaduthanam S, Gade S, Meister M, Brase JC,
Johannes M, Dienemann H, Warth A, Schnabel PA, Herth FJ, Sültmann
H, et al: Serum miR-142-3p is associated with early relapse in
operable lung adenocarcinoma patients. Lung Cancer. 80:223–227.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Wang F, Wang XS, Yang GH, Zhai PF, Xiao Z,
Xia LY, Chen LR, Wang Y, Wang XZ, Bi LX, et al: miR-29a and
miR-142-3p downregulation and diagnostic implication in human acute
myeloid leukemia. Mol Biol Rep. 39:2713–2722. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Mascaux C, Laes JF, Anthoine G, Haller A,
Ninane V, Burny A and Sculier JP: Evolution of microRNA expression
during human bronchial squamous carcinogenesis. Eur Respir J.
33:352–359. 2009. View Article : Google Scholar : PubMed/NCBI
|