Introduction
Cell-cell communication is critical in the
development and function of an organism, in addition to cellular
proliferation, differentiation, and apoptosis. Tumorigenesis is
considered to be a result of abnormal cell-cell communication
(1,2).
The microenvironment of solid tumors is essential for their rapid
growth and metastasis to other sites. As angiogenesis is required
for all stages of tumor growth, proangiogenic factors such as TGF-β
and VEGF are required for tumor formation and development (3–6). Previous
studies have proposed that exosomes are potent communicators
between tumors and their microenvironment, and potentially
contribute to tumorigenesis and subsequent metastasis (7,8).
With a size of 50–100 nm, exosomes are tiny vesicles
from endosomes or multivesicular bodies, and are present within the
extracellular microenvironment and biological fluids, such as the
blood and urine (9,10). A previous study presented evidence
that exosomes have numerous functions and are key in an array of
biological events, including coagulation and intercellular
signaling (11). The contents of
exosomes includes proteins, nuclear acids, and lipids, which
usually vary with the cellular and tissue origins of the exosomes
and are adapted to their functions (12,13).
Indeed, the source of exosomes defines their function.
Antigen-presenting cell-derived exosomes induce an immune response
and tumor-derived exosomes suppress it. Exosomes contain
selectively enriched mRNA and miRNA that regulate gene expression
in target cells (14).
Lung cancer is the most common cause of
cancer-associated mortality globally (15). The majority of lung cancer cases are
carcinomas derived from epithelial cells in the lung, and may be
divided into small cell lung cancer (SCLC) and non-small cell lung
cancer (NSCLC). Smoking has been demonstrated to be a major
contributor to developing lung cancer (16). The prognosis for lung cancer is poor,
with <15% of the inflicted population surviving for 5-years
following diagnosis (17).
Elucidation of the mechanisms underlying the tumorigenesis of lung
cancer may aid the diagnosis, therapy and prevention. Although
certain studies have demonstrated that cancer cell-derived exosomes
are involved in tumor growth, to the best of our knowledge, there
are no previous studies that investigate the roles of exosomes in
the development of lung cancer.
In the present study, 2 lung cancer cell lines were
used to investigate the exosome functions in the processes of
migration and immunoregulation by cancer cells.
Materials and methods
Materials and reagents
TGF-β, IL-10 and MCP-1 were obtained from Peprotech
(Rocky Hill, NJ, USA). Monoclonal mouse anti-human TGF-β (cat. no.
16-9243-85; 1:1,000) and rat anti-human IL-10 antibodies (cat. no.
16-7108-85; 1:1,000) were purchased from eBioscience (San Diego,
CA, USA). The materials for cell culture were obtained from Thermo
Fisher Scientific, Inc., (Carlsbad, CA, USA).
Cell culture and transfection
HEK 293T, NCI-H2228, NCI-H1688 and HMEC-1 cells were
purchased from ATCC (Manassas, VA, USA), and cultured as to the
manufacturer's instructions. Briefly, the cells were cultured in
DMEM supplemented with 10% FBS and 50 U penicillin/50 µg
streptomycin. Prior to exosomes preparation, the serum in the
medium was replaced with 10% Serum Replacement reagent (Thermo
Fisher Scientific, Inc.). For hypoxia exposure, the cells were
cultured in an O2/CO2 incubator at either 1%
O2 at 37°C in a 5% CO2 humidified
condition.
Exosomes preparation
Before the medium was collected for exosomes
preparation, the HEK 293T, NCI-H2228 and NCI-H1688 cells were
cultured in serum-free medium. The exosomes preparation was
performed as previously described (18). Briefly, following 4 days in culture,
100 ml of medium was collected. Exosomes were harvested through
subsequential centrifugation of supernatants (2×10 min, 500 × g;
1×20 min, 2,000 × g; 1×30 min, 10,000 × g), followed by
centrifugation (90 min, 100,000 × g) and wash (PBS, 90 min, 100,000
× g). The pellet was resuspended in 8 ml phosphate buffered saline
(PBS), and pelleted again at 100,000 × g, 60 min. The final pellet
was resuspended in a small volume of PBS and was quantified using a
Pierce BCA Protein Assay kit (Thermo Fisher Scientific, Inc.).
Exosome preparations were stored at 4°C until use.
Migration assay
Migration was assessed using a Transwell migration
assay with HMEC-1 cells. The migration assay was performed as
previous described (19). Briefly,
cells were added to the upper (1×106cells/ml) chamber,
and to the lower chamber DMEM/10% FCS with 10 µg/ml exosomes.
Following 12 h incubation, cells on the lower membrane side were
fixed and stained with crystal violet stained and counted using a
microscope (Eclipse TS100; Nikon Corporation, Tokyo, Japan).
ELISA
To analyze cytokines presence in exosomes by
enzyme-linked immunoassay (ELISA) was performed according to the
kit manufacturer's instructions. The ELISA kits for TGF-β, MCP-1
and IL-10 were purchased from R&D system (Minneapolis, MN,
USA).
Statistical analysis
Statistical analysis was performed using SPSS
software, version 20 (IBM, Armonk, NY, USA). Student t-test or
one-way analysis of variance were used for comparisons to determine
statistical significance, and P<0.05 was considered to indicate
a statistically significant difference.
Results
The exosomes derived from cancer cells
are required for migration
The contents of exosomes include various
biologically active materials, including proteins and RNA. As
cancer cells grows very rapidly, they are often under hypoxic
conditions. As such, the functional differences were assessed in
exosomes derived from cancer cells under hypoxic conditions. The
present study used 2 cells lines representing classic small cell
lung cancer (NCI-H1688) and non-small cell lung cancer (NCI-H2228),
respectively. Exosomes were collected from these 2 cell lines
cultured with regular oxygen levels (21%) or 1% oxygen. Human
non-small lung cancer cells, NCI-H2228, were then used to detect
whether the exosomes influence the migration of HMEC-1. The
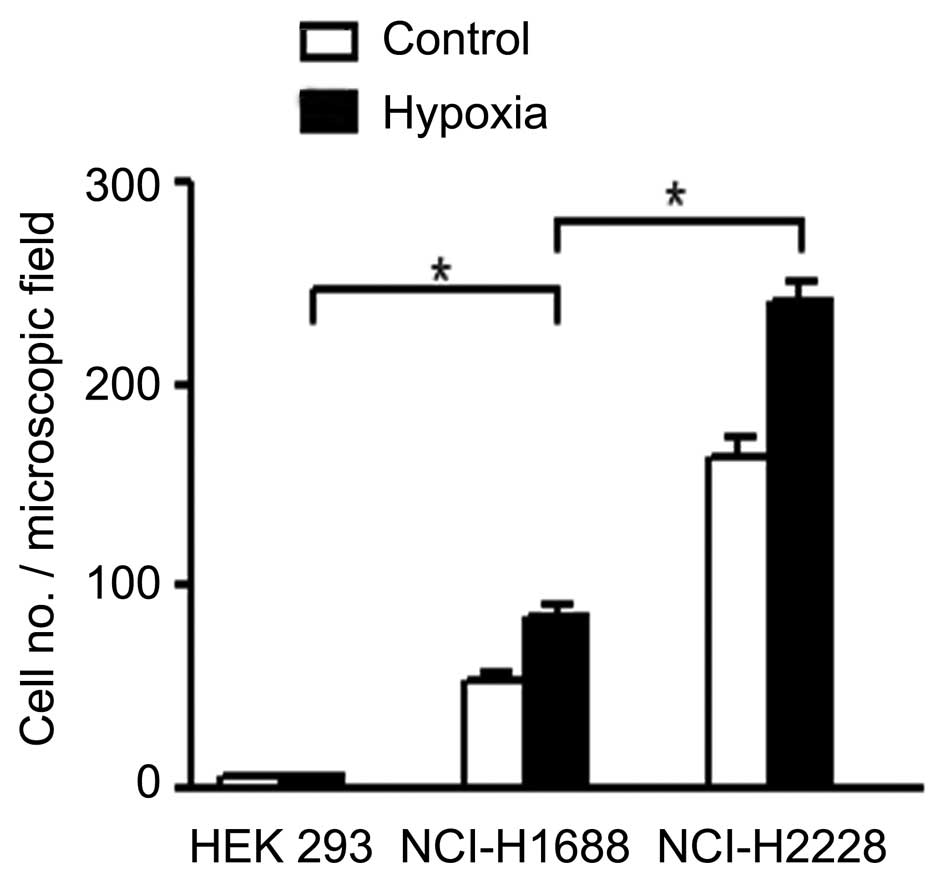
exosomes from 293T cells were used as a control. As presented in
Fig. 1, the migration of HMEC-1 cells
through Transwell filters was considerably increased by exosomes
from lung cancer cells NCI-H1688 (P<0.05) and NCI-H2228
(P<0.05). These results indicate that hypoxia leads to exosomes
promoting the migration of cancer cells. In addition, the exosomes
from non-small cell lung cancer cell NCI-H2228 had more marked
effects on cells migration compared with exosomes from small cell
lung cancer cell NCI-H1688.
The exosomes derived from cancer cells
contain various migration-associated factors
Since the exosomes derived from lung cancer cells
increased the migration of endothelial cells and cancer cells, the
present study then sought to further characterize the contents of
exosomes involved in the regulation of migration. As TGF-β, MCP-1
and IL-10 have been previously been demonstrated to regulate the
cellular migration of A375 human melanoma cells (20), these factors may also affect the
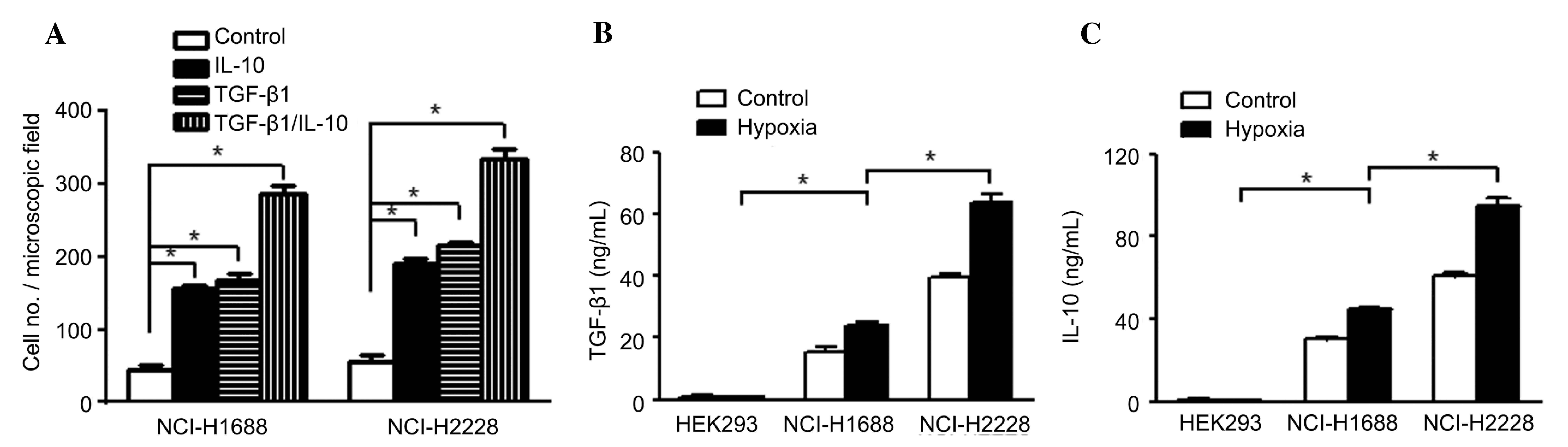
migration of lung cancer cells. The Transwell assay demonstrated
that TGF-β, MCP-1 and IL-10 all promoted the migration of NCI-H1688
and NCI-H2228 cells (Fig. 2A;
P<0.05), supporting the hypothesis that TGF-β, MCP-1 and IL-10
contribute to lung cancer cell migration. Next, it was examined
whether TGF-β, MCP-1 and IL-10 are present in exosomes. Following
concentration of the exosome cell fraction by ultracentrifugation,
repeated freeze-thaw of pellets was performed, and it was
demonstrated that the concentration of TGF-β and IL-10 were
increased in exosomes derived from non-small cell lung cancer cells
NCI-H2288 and small cell lung cancer cells NCI-H1688, and
particularly in hypoxic conditions (Fig.
2B and C; P<0.05). MCP-1, however, was not detected in
exosomes from NCI-H1688 and NCI-H2228, under hypoxic conditions or
not (data not shown). Thus, the data demonstrated that hypoxia
enhanced the TGF-β and IL-10 contents in exosomes released from
lung cancer cells, which promoted the migration of cancer
cells.
The TGF-β and IL-10 in exosomes are
responsible for cellular migration
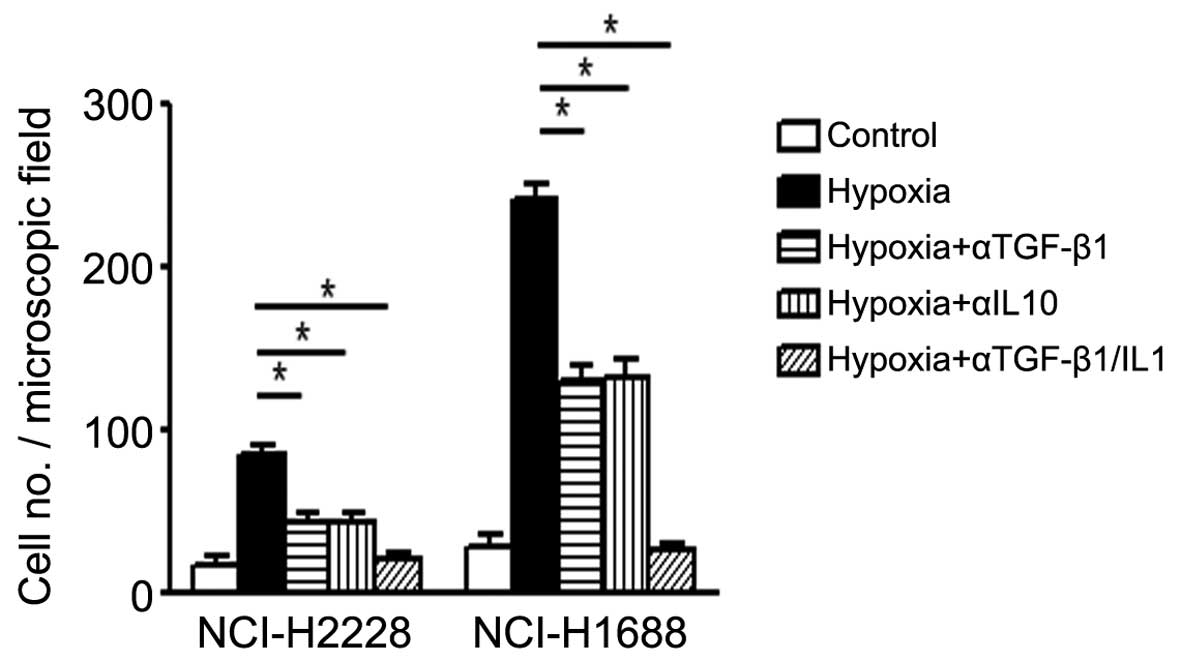
As the exosome released from cells contains
different components, which potentially work with or against each
other, the next experiment specifically blocked the effects of
TGF-β and IL-10 using neutrolizing antibodies individually or
together. The results demonstrated that both TGF-β and IL-10 are
essential for lung cancer cells migration, as blockade of TGF-β or
IL-10 alone only partially (about 40%) reduced the effects of
exosomes, while combined blockade of both TGF-β and IL-10 resulted
in marked reduction (>80%) (Fig.
3; P<0.05), supporting the idea that the release of TGF-β or
IL-10 from lung cancer cells mediate the enhanced cellular
migration, which is essential for tumor metastasis.
Discussion
The contents of exosomes may have important effects
on tumor cell malignancy. Previous studies have revealed that tumor
cells are a dynamic resource for exosomes, such as melanoma
(20), prostate cancer (21) and gliobastoma (22). Exosomes have been detected in human
malignant effusions (12). Exosomes
derived from different types of cancer contain different contents,
therefore they may function distinctively on tumor behavior,
indicating the significance of exosomes in prognosis, diagnosis,
and therapy for cancer (21,23,24).
The contents of exosomes are heterogeneous,
including diverse RNA, protein including cytokines, growth factors,
and lipids (13,21,25). A
number of miRs have been demonstrated to be present in exosomes
from various cells (including cancer cells), which serve essential
roles in the regulation of tumor cell migration (21–27). Other
miRs may be involved in other steps of tumor development: miR-31,
−185, and −34b are involved in melanoma invasion (25).
Proteomic analysis contributes much to the efforts
of investigating exosomes, as exosomes contain bulky proteins,
including growth factors such as VEGF, EGF, and various cytokines
such as MCP-1, IL-4, which serve essential roles in tumor cell
survival, proliferation and migration. TGF-β may induce expression
of MCP-1 and IL-10 expression, which are involved in melanoma tumor
progression (20). The present study
investigated the effects of TGF-β and IL-10 on lung cancer cells,
and attained similar data on migration of NCI-H2228 and NCI-H1688.
TGF-β signaling has widespread crosstalk with multiple signaling
pathways, including SMAD, PI3K (phosphoinositide 3-kinase)/AKT, and
BRAF-MAPK (mitogen activated protein kinase) and therefore
contributes to the expression of MCP-1 and IL-10 (28). However, no MCP-1 expression was
detected in lung cancer cells, both in NCI-H2228 and NCI-H1688
cells.
The present study revealed a novel aspect of
exosomes derived from lung cancer cells, indicating that the
contents of exosomes may promote migration. This finding could be
potentially significant, leading to novel therapeutic methods. Of
course, further detailed information into exosomes contents should
be obtained in order to further evaluate the therapeutic role of
exosomes on anti-tumor effects.
Acknowledgements
The present study was supported by the National
Natural Science Foundation of China (81273814).
References
|
1
|
Trinchieri G: Cancer and inflammation: An
old intuition with rapidly evolving new concepts. Annu Rev Immunol.
30:677–706. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Grivennikov SI, Greten FR and Karin M:
Immunity, inflammation and cancer. Cell. 140:883–899. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Barbera-Guillem E, Nyhus JK, Wolford C,
Friece CR and Sampsel JW: Vascular endothelial growth factor
secretion by tumor-infiltrating macrophages essentially supports
tumor angiogenesis and IgG immune complexes potentiate the process.
Cancer Res. 62:7042–7049. 2002.PubMed/NCBI
|
|
4
|
Jain RK and Carmeliet P: SnapShot: Tumor
angiogenesis. Cell. 149:1408–1408.e1. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Carmeliet P and Jain RK: Molecular
mechanisms and clinical applications of angiogenesis. Nature.
473:298–307. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Facciabene A, Peng X, Hagemann IS, Balint
K, Barchetti A, Wang LP, Gimotty PA, Gilks CB, Lal P, Zhang L, et
al: Tumour hypoxia promotes tolerance and angiogenesis via CCL28
and T (reg) cells. Nature. 475:226–230. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Corrado C, Raimondo S, Chiesi A, Ciccia F,
De Leo G and Alessandro R: Exosomes as intercellular signaling
organelles involved in health and disease: Basic science and
clinical applications. Int J Mol Sci. 14:5338–5366. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Ahmed KA and Xiang J: Mechanisms of
cellular communication through intercellular protein transfer. J
Cell Mol Med. 15:1458–1473. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Denzer K, Kleijmeer MJ, Heijnen HF,
Stoorvogel W and Geuze HJ: Exosome: From internal vesicle of the
multivesicular body to intercellular signaling device. J Cell Sci.
113:3365–3374. 2000.PubMed/NCBI
|
|
10
|
Schorey JS and Bhatnagar S: Exosome
function: From tumor immunology to pathogen biology. Traffic.
9:871–881. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
van Niel G, Porto-Carreiro I, Simoes S and
Raposo G: Exosomes: A common pathway for a specialized function. J
Biochem. 140:13–21. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Bard MP, Hegmans JP, Hemmes A, Luider TM,
Willemsen R, Severijnen LA, van Meerbeeck JP, Burgers SA,
Hoogsteden HC and Lambrecht BN: Proteomic analysis of exosomes
isolated from human malignant pleural effusions. Am J Respir Cell
Mol Biol. 31:114–121. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Street JM, Barran PE, Mackay CL, Weidt S,
Balmforth C, Walsh TS, Chalmers RT, Webb DJ and Dear JW:
Identification and proteomic profiling of exosomes in human
cerebrospinal fluid. J Transl Med. 10:52012. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Kourembanas S: Exosomes: Vehicles of
intercellular signaling, biomarkers, and vectors of cell therapy.
Annu Rev Physiol. 77:13–27. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Lozano R, Naghavi M, Foreman K, Lim S,
Shibuya K, Aboyans V, Abraham J, Adair T, Aggarwal R, Ahn SY, et
al: Global and regional mortality from 235 causes of death for 20
age groups in 1990 and 2010: A systematic analysis for the global
burden of disease study 2010. Lancet. 380:2095–2128. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Yu Y, Liu H, Zheng S, Ding Z, Chen Z, Jin
W, Wang L, Wang Z, Fei Y, Zhang S, et al: Gender susceptibility for
cigarette smoking-attributable lung cancer: A systematic review and
meta-analysis. Lung Cancer. 85:351–360. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Travis WD, Brambilla E and Riely GJ: New
pathologic classification of lung cancer: Relevance for clinical
practice and clinical trials. J Clin Oncol. 31:992–1001. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Epple LM, Griffiths SG, Dechkovskaia AM,
Dusto NL, White J, Ouellette RJ, Anchordoquy TJ, Bemis LT and
Graner MW: Medulloblastoma exosome proteomics yield functional
roles for extracellular vesicles. PLoS One. 7:e420642012.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Xu C, Zhang YH, Thangavel M, Richardson
MM, Liu L, Zhou B, Zheng Y, Ostrom RS and Zhang XA: CD82
endocytosis and cholesterol-dependent reorganization of tetraspanin
webs and lipid rafts. FASEB J. 23:3273–3288. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Diaz-Valdes N, Basagoiti M, Dotor J,
Aranda F, Monreal I, Riezu-Boj JI, Borrás-Cuesta F, Sarobe P and
Feijoó E: Induction of monocyte chemoattractant protein-1 and
interleukin-10 by TGFbeta1 in melanoma enhances tumor infiltration
and immunosuppression. Cancer Res. 71:812–821. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Hessvik NP, Sandvig K and Llorente A:
Exosomal miRNAs as biomarkers for prostate cancer. Front Genet.
4:362013. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Mizoguchi M, Guan Y, Yoshimoto K, Hata N,
Amano T, Nakamizo A and Sasaki T: Clinical implications of
microRNAs in human glioblastoma. Front Oncol. 3:192013. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Gailhouste L, Gomez-Santos L and Ochiya T:
Potential applications of miRNAs as diagnostic and prognostic
markers in liver cancer. Front Biosci (Landmark Ed). 18:199–223.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Kahlert C and Kalluri R: Exosomes in tumor
microenvironment influence cancer progression and metastasis. J Mol
Med (Berl). 91:431–437. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Xiao D, Ohlendorf J, Chen Y, Taylor DD,
Rai SN, Waigel S, Zacharias W, Hao H and McMasters KM: Identifying
mRNA, microRNA and protein profiles of melanoma exosomes. PLoS One.
7:e468742012. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Katakowski M, Buller B, Zheng X, Lu Y,
Rogers T, Osobamiro O, Shu W, Jiang F and Chopp M: Exosomes from
marrow stromal cells expressing miR-146b inhibit glioma growth.
Cancer Lett. 335:201–204. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Chiba M, Kimura M and Asari S: Exosomes
secreted from human colorectal cancer cell lines contain mRNAs,
microRNAs and natural antisense RNAs, that can transfer into the
human hepatoma HepG2 and lung cancer A549 cell lines. Oncol Rep.
28:1551–1558. 2012.PubMed/NCBI
|
|
28
|
Massagué J: TGFβ signalling in context.
Nat Rev Mol Cell Biol. 13:616–630. 2012. View Article : Google Scholar : PubMed/NCBI
|