Introduction
Colorectal cancer (CRC) is the second most commonly
diagnosed cancer in females and the third most commonly diagnosed
cancer in males worldwide (1).
Patients with CRC have a poor prognosis due to cancer recurrence
and metastasis following surgical resection. Numerous patients are
at a high risk of recurrence and may be considered candidates for
targeted therapy or chemotherapy. However, colorectal
carcinogenesis is a complicated process that is associated with
cumulative genomic alterations (2,3).
Therefore, it is necessary to explore the molecular markers that
underlie tumor progression and identify novel targets to improve
therapeutic strategies and extend the survival of CRC patients.
Chemokines are a group of small proinflammatory
cytokines that attract, activate and regulate leukocytes in
inflamed tissues, and recent studies identified the role of
chemokines in the initiation and promotion of carcinogenesis
(3). Chemokines may be classified
into three subfamilies, consisting of C, CC and CXC, on the basis
of the number and arrangement of conserved cysteine residues.
Growth-related oncogene (GRO) is a member of the CXC chemokine
family, which is composed of GRO-α, GRO-β, and GRO-γ, also termed
the CXCL1, CXCL2 and CXCL3 genes, respectively (4,5). The
chemokines have a conserved CXC motif at the NH2
terminus, but vary at the COOH terminus. The three GRO chemokines
vary in binding affinity to CXCR2 or CXCR1 receptors, with GRO-α
having the highest affinity with CXCR2 (6). GRO-α and GRO-β have been identified to
be dysregulated in pre-malignant colonic adenomas (7). However, the impact of GRO-β
overexpression on the clinical outcome in patients with CRC remains
unclear. The present study explored the expression of GRO-β in
human CRC compared with adjacent normal tissue using cDNA
microarray data and tissue microarray (TMA) sections, and assessed
the potential correlation with the critical clinicopathological
features of CRC and patient prognosis.
Materials and methods
cDNA microarray data
CXCL2 mRNA expression data were extracted from six
independent studies and the expression levels in CRC tissues were
compared with matched normal tissues using publicly available gene
expression data in the Oncomine Cancer Microarray database
(http://www.oncomine.org). All data were
log-transformed and median-centered per array, and the standard
deviation was normalized to one per array (8,9).
Patients and samples
Formalin-fixed, paraffin-embedded tumor tissues and
corresponding tumor-adjacent specimens were obtained during surgery
from 198 patients with CRC treated at the Affiliated Hospital of
Nantong University between 2002 and 2008. The clinical data,
including gender, age, differentiation, location, extent of primary
tumor, tumor-node-metastasis (TNM) classification, lymph node
metastasis status, carcinoembryonic antigen (CEA) level and
follow-up, including the 5-year survival rate, were obtained from
the medical records of individual patients. The overall survival
time was calculated as the time between the date of surgery and the
date of mortality or last follow-up. The tumor stage was in
accordance with the Union for International Cancer Control TNM
system, but differentiation was determined following the World
Health Organization standards (10).
Representative 2.0-mm tissue cores from each patient were used to
conduct a tissue microarray (TMA) analysis using the Manual Tissue
Microarrayer (Quick-Ray WI-UT06; Unitma Co., Ltd., Seoul,
Korea).
Ethics statement
The present study was performed according to the
principles expressed in the Declaration of Helsinki (11). Tissue specimens were collected with
full written informed consent of the patients or the patients'
families, in compliance with the institutional guidelines set by
the Human Research Ethics Committee of the Affiliated Hospital of
Nantong University. Ethical approval for the present study was
granted by the Ethics Committee of the Affiliated Hospital of
Nantong University (approval no., 2013-009).
Immunohistochemistry
For the immunohistochemical (IHC) analysis, TMA
sections were deparaffinized in 100% xylene (Beyotime Institute of
Biotechnology, Shanghai, China) and rehydrated in graded ethanol
solutions (80, 95 and 100%; Beyotime Institute of Biotechnology).
Antigen retrieval was performed by boiling the sections under
pressure (90 kPa) in citrate buffer (pH 6.0; Beyotime Institute of
Biotechnology) for 5 min. The non-specific binding site was blocked
by incubating the sections in 5% goat serum and phosphate buffered
saline (PBS; Beyotime Institute of Biotechnology) for 15 min. The
TMA sections were incubated with a primary polyclonal rabbit
anti-human GRO-β antibody (dilution, 1:400; catalog no., 500-P104;
PeproTech, Inc., Rocky Hill, NJ, USA) and subsequently with goat
anti-rabbit horseradish peroxidase-conjugated antibody (dilution,
1:1,000; catalog no., sc-2004; Santa Cruz Biotechnology, Inc.,
Dallas, TX, USA). GRO-β immunostaining underwent two independent
evaluations under blind experimental conditions. The percentage of
GRO-β-positive cells was recorded as 0–100%, and the staining
intensity was graded as follows: 0, no staining; 1, mild intensity;
2, moderate intensity; and 3, strong intensity. The final GRO-β
staining score was a product of the intensity grading and
percentage of positive cells.
The threshold for the statistically significant
GRO-β expression scores in terms of overall survival (OS) time was
set using the X-tile software program (http://www.tissuearray.org/rimmlab; Rimm Lab, Yale
University, New Haven, CT, USA), as previously described (12). The degree of staining was quantified
using a two-level grading system and the final GRO-β staining score
was defined as follows: 0–150, low expression; 150–300, high
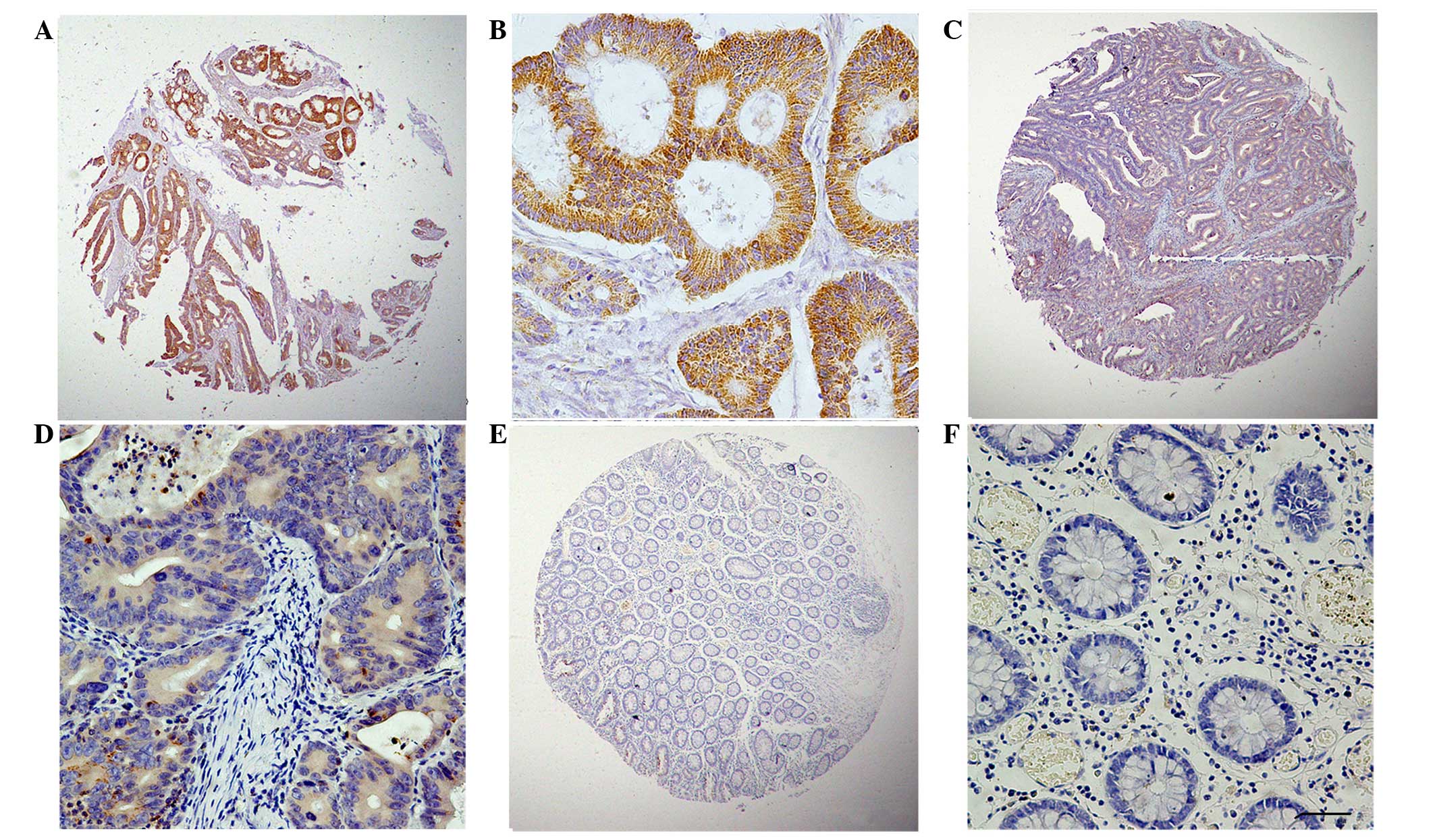
expression. The typical expression patterns of GRO-β are
illustrated in Fig. 1.
Statistical analysis
SPSS version 20.0 software (IBM Corporation, Armonk,
NY, USA) was used to conduct the statistical analyses. The
comparison between the CXCL2 mRNA levels in the microarray was
performed using the Mann-Whitney U or Student's t-tests. A
χ2 test was used to analyze the association between
GRO-β expression and clinicopathological parameters, based on the
IHC analysis. For the TMA slides, the gender, age, differentiation,
location, extent of primary tumor, TNM classification, lymph node
metastasis status and CEA level were assessed. The survival rate
was calculated using the Kaplan-Meier method and Cox proportional
hazards regression model, and the statistical differences were
examined using the log-rank test. P<0.05 was considered to
indicate a statistically significant difference. All statistical
tests were two-sided.
Results
CXCL2 mRNA is overexpressed in
CRC
Using the Oncomine microarray database, the
expression levels of CXCL2 mRNA in CRC tissues were increased
compared with normal tissues from six independent studies (13–18). There
was a significant difference between tumor tissue and normal tissue
according to the mean expression value (P<0.0001 in all six
datasets; Fig. 2).
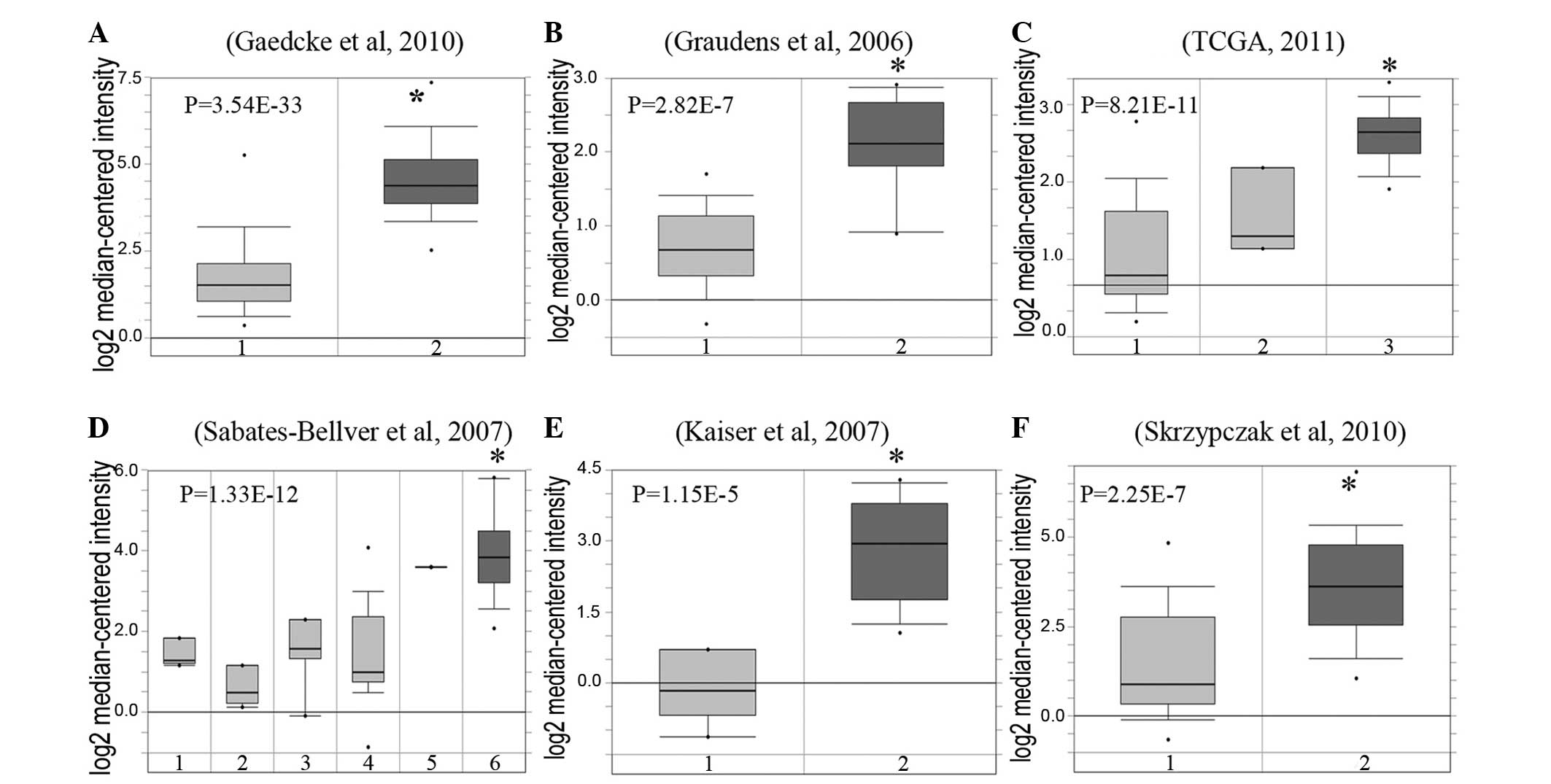 | Figure 2.GRO-β mRNA is overexpressed in
colorectal cancer. GRO-β expression in colorectal cancer tissues
and normal tissues. Data sets in a single panel were from the same
study. GEP data are log transformed and normalized. (A) Gaedcke
et al (16): 1, 65 rectum
samples; 2, 65 rectal adenocarcinoma samples. (B) Graudens et
al (13): 1, 12 colon samples; 2,
18 colorectal carcinoma samples. (C) TCGA (18): 1, 19 colon samples; 2, 3 rectum
samples; 3 22 colon mucinous adenocarcinoma samples. (D)
Sabates-Bellver et al (15):
1, 4 ascending colon samples; 2, 5 descending colon samples; 3, 7
rectum samples; 4, 15 sigmoid colon samples; 5, 1 transverse colon
sample; 6, 25 colon adenoma samples. (E) Kaiser et al
(14): 1, 5 colon samples; 2, 17
cecum adenocarcinoma samples. (F) Skrzypczak et al (17): 1, 24 colorectal tissues samples; 2, 45
colorectal adenocarcinoma samples. GRO-β, growth-related
oncogene-β; GEP, gene expression profiling. *P<0.05. |
Expression of GRO-β in CRC and
peritumoral tissues, determined by IHC analysis
In order to investigate the expression of GRO-β
protein in colorectal carcinoma and the corresponding adjacent
tissues, an IHC analysis on the primary colorectal tumors and
normal colorectal mucosa was performed. As shown in Fig. 1, GRO-β staining was detected at
various levels, primarily in the cytoplasm of CRC cells. The
staining index of cytoplasmic expression of GRO-β in all 86 normal
colorectal mucosa tissues was <150. High GRO-β expression was
detected in 31.31% (62/198) of CRC samples. Therefore, in
accordance with the data of the CXCL2 mRNA analysis, the GRO-β
protein was confirmed to be highly expressed in CRC tissues.
Association between GRO-β expression
and clinicopathological features
The association between GRO-β cytoplasmic expression
and clinicopathological features of 198 cases of CRC was studied
(Table I). The results revealed that
high GRO-β cytoplasmic expression in the primary CRC was
significantly associated with the tumor location (P=0.022), extent
of primary tumor (P=0.005) and lymph node metastasis (P=0.017).
However, no significant association between GRO-β levels and other
clinicopathological characteristics, including patient gender,
patient age, tumor differentiation, TNM stage and CEA level, were
observed (Table I).
 | Table I.Correlation of growth-related
oncogene-β expression in tumor tissues of colorectal cancer
patients by clinicopathological characteristic. |
Table I.
Correlation of growth-related
oncogene-β expression in tumor tissues of colorectal cancer
patients by clinicopathological characteristic.
| Characteristic | N | Low expression, n
(%) | High expression, n
(%) | Pearson
χ2 | P-value |
|---|
| Total | 198 | 136 (68.69) | 62 (31.31) |
|
|
| Gender |
|
|
|
|
|
| Male | 126 | 86
(68.25) | 40 (31.75) | 0.030 | 0.862 |
|
Female | 72 | 50
(69.44) | 22 (30.56) |
|
|
| Age, years |
|
|
|
|
|
|
<60 | 65 | 45
(69.23) | 20 (30.77) | 0.013 | 0.908 |
| ≥60 | 133 | 91
(68.42) | 42 (31.58) |
|
|
| Location |
|
|
|
|
|
|
Colon | 145 | 93
(64.14) | 52 (35.86) | 5.212 | 0.022* |
|
Rectum | 53 | 43
(81.13) | 10 (18.87) |
|
|
| Differentiation |
|
|
|
|
|
| Well -
middle | 166 | 113 (68.07) | 53 (31.93) | 0.180 | 0.671 |
| Poor | 32 | 23
(71.88) | 9
(28.13) |
|
|
| TNM stage |
|
|
|
|
|
| I | 46 | 38
(82.61) | 8
(17.39) | 5.462 | 0.065 |
| II | 78 | 51
(65.38) | 27 (34.62) |
|
|
|
III+IV | 74 | 47
(63.51) | 27 (36.49) |
|
|
| T |
|
|
|
|
|
|
Tis+1+2 | 51 | 43
(84.31) | 8
(15.69) | 7.799 | 0.005* |
|
3+4 | 147 | 93
(63.27) | 54 (36.73) |
|
|
| N |
|
|
|
|
|
| 0 | 124 | 89
(71.77) | 35 (28.23) | 8.176 | 0.017* |
|
1a+1b | 57 | 40
(70.18) | 17 (29.82) |
|
|
|
2a+2b | 17 |
7 (41.18) | 10 (58.82) |
|
|
| CEA |
|
|
|
|
|
| No | 119 | 81
(68.07) | 38 (31.93) | 1.713 | 0.191 |
|
Yes | 24 | 13
(54.17) | 11 (45.83) |
|
|
| Unknown | 55 | 42
(76.36) | 13 (23.64) |
|
|
Prognostic value of high GRO-β
expression in CRC
In total, 68 patients succumbed during the
postoperative follow-up period. The prognostic value of various
factors was investigated using Kaplan-Meier analysis, and
differentiation, TNM classification, extent of primary tumor, lymph
node metastasis, CEA level and high GRO-β expression were
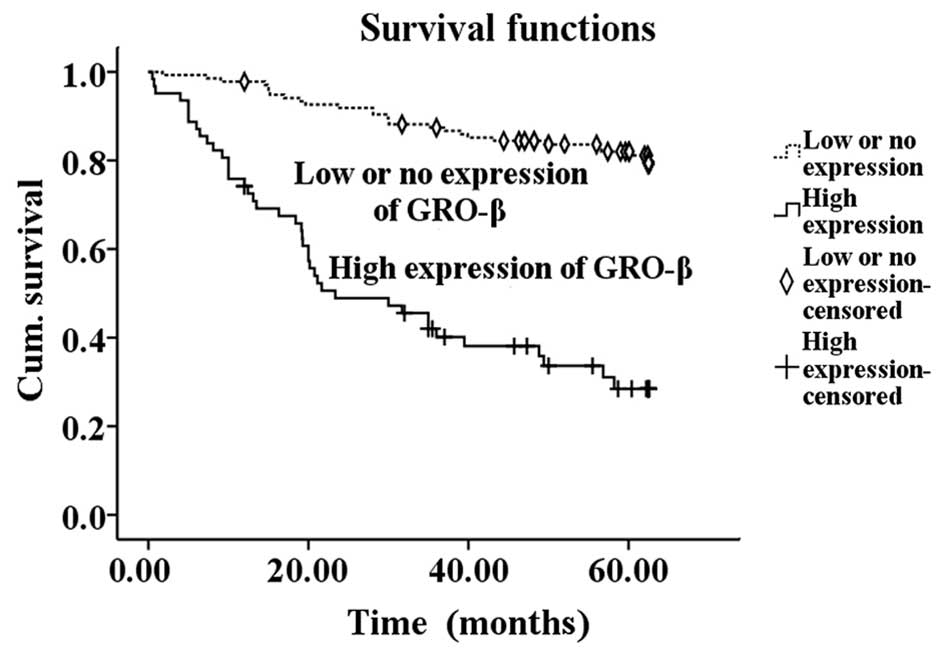
correlated with overall survival (P<0.05) (Table II). Kaplan-Meier survival curves
demonstrated that patients with high GRO-β expression possessed a
significantly shorter survival time compared with the low GRO-β
expression group (P<0.001) (Fig.
3). In the additional multivariate Cox regression analyses,
subsequent to adjustment for gender, age, location,
differentiation, tumor stage and lymph node metastasis, high GRO-β
expression (P<0.001), TNM classification (P=0.002) and CEA level
(P=0.027) remained as independent predictive factors of a poor
outcome in CRC (Table III).
Overall, the results indicate that high GRO-β expression may act as
a prognostic marker for CRC.
 | Table II.Kaplan-Meier univariate analysis of
the overall survival time of colorectal cancer patients following
surgery. |
Table II.
Kaplan-Meier univariate analysis of
the overall survival time of colorectal cancer patients following
surgery.
|
| Univariate
analysis |
|---|
|
|
|
|---|
| Variable | Survival in months
± standard error | 95% confidence
interval | P-value |
|---|
| GRO-β
expression |
|
|
|
|
Low | 56.24±1.29 | 53.71–58.77 |
<0.001a |
|
High | 33.02±2.99 | 27.16–38.88 |
|
| Gender |
|
|
|
|
Female | 51.68±2.35 | 47.07–56.29 |
0.153 |
|
Male | 47.67±1.92 | 43.91–51.42 |
|
| Age, years |
|
|
|
|
<60 | 48.97±2.64 | 43.80–54.14 |
0.929 |
|
≥60 | 49.21±1.84 | 45.61–52.81 |
|
| Location |
|
|
|
|
Rectum | 50.11±1.74 | 46.70–53.52 |
0.169 |
|
Colon | 46.59±2.89 | 40.92–52.27 |
|
|
Differentiation |
|
|
|
| Well -
middle | 50.53±1.57 | 47.45–53.61 |
0.044a |
|
Poor | 41.70±4.18 | 33.51–49.89 |
|
| TNM stage |
|
|
|
| I | 59.73±1.55 | 56.69–62.76 |
<0.001a |
| II | 49.46±2.30 | 44.95–53.97 |
|
| III
+IV | 42.08±2.75 | 36.68–47.48 |
|
| T |
|
|
|
|
Tis+1+2 | 59.36±1.52 | 56.39–62.33 |
<0.001a |
|
3+4 | 45.52±1.86 | 41.88–49.17 |
|
| N |
|
|
|
| 0 | 53.34±1.62 | 50.18–56.51 |
<0.001a |
|
1a+1b | 45.46±3.14 | 39.29–51.62 |
|
|
2a+2b | 30.85±4.91 | 21.22–40.47 |
|
| CEA |
|
|
|
| No | 51.59±1.73 | 48.20–54.98 |
0.001a |
|
Yes | 38.13±5.12 | 28.10–48.15 |
|
 | Table III.Results of the Cox multivariate
regression analysis of the overall survival of colorectal cancer
patients. |
Table III.
Results of the Cox multivariate
regression analysis of the overall survival of colorectal cancer
patients.
| Factor | β | SE(β) | Wald | P-value | eβ
hazard ratio | 95.0% CI for
eβ hazard ratio |
|---|
| GRO-β | 1.778 | 0.306 | 33.702 | 0.000 | 5.920 |
3.248–10.791 |
| TNM | 0.653 | 0.222 |
8.613 | 0.003 | 1.921 | 1.242–2.970 |
| CEA | 0.758 | 0.325 |
5.461 | 0.019 | 2.135 | 1.130–4.032 |
Discussion
Previously, the potential oncogenic role of GRO-β in
the promotion of several human cancers has been examined, including
in esophageal squamous cell carcinoma and melanoma (19,20).
Little is understood regarding the role of GRO-β in CRC. To the
best of our knowledge, only two studies have investigated the
potential role of GRO-β in colorectal tumors at present. These
studies indicated that the expression of CXCL2 mRNA was
significantly increased in CRC compared with normal colon tissue
using quantitative reverse transcription-polymerase chain reaction
(6) and that CXCL2 mRNA was also
enhanced in premalignant adenomas in a previous small cohort study
(7), which suggests that the
dysregulation of GRO-β may be an early event in the tumorigenesis
of CRC. Additional statistical analyses revealed that CXCL2 mRNA
was overexpressed in malignant colorectal tissues compared with
normal adjacent tissues using microarray data from six independent
datasets (13–18).
The present study explored the expression of the
GRO-β protein in human CRC compared with adjacent tissues, and
assessed the potential correlation with the critical
clinicopathological features of CRC and the patient outcome.
Several notable observations were made from the results.
First, the GRO-β protein was demonstrated to be
highly expressed in a series of 198 CRCs comprising all stages of
disease using an IHC approach. In addition, overexpression was
found to be associated with neoplastic epithelial cells rather than
inflammatory cells or non-epithelial stroma. Notably, the division
of the data into two categories according to localization indicated
that increased GRO-β expression in the colon was significantly more
frequent compared with the rectum (P=0.022). Previously, cancers of
the rectum and colon have been implicated as distinct tumors, as
they have a dissimilar prevalence and variations in the clinical
presentation, prognosis and possibly genetic and environmental
epidemiology (21,22). Therefore, GRO-β expression may affect
carcinogenesis of the colorectal tissues in a site-specific manner.
The findings of the present study provide additional evidence that
colon and rectal cancers possessed varied etiologies. In addition,
GRO-β expression was significantly associated with the extent of
the primary tumor and lymph node metastasis. Increased GRO-β
expression is associated with a more advanced stage of disease and
the propensity to develop lymph node metastasis. These results
suggest that the overexpression of cytoplasmic GRO-β in CRC may
facilitate cancer cell invasion and metastasis. Previous studies
have also reported that the increased expression of GRO-β was
involved in the development and invasion of several types of
carcinomas, including esophageal squamous cell carcinoma (19) and melanoma (20). The data in the present study clearly
revealed that a high cytoplasmic expression of GRO-β was associated
with significantly poorer survival time. Multivariate analyses
revealed that GRO-β expression was regarded as an independent
prognostic factor for CRC patients. In addition to increased GRO-β
expression, TNM classification and high CEA levels are also
considered to be independent factors for a poor prognosis in CRC.
Overall, GRO-β expression may be an important prognostic factor for
aggressive human CRC. GRO-β has potential value as a therapeutic
target in patients with CRC.
In conclusion, the role of GRO-β in the
pathophysiology of CRC carcinogenesis and progression is unclear.
GRO-β is a classical neutrophil chemoattractant and was the first
chemokine to be identified as a product of neutrophils, and to be
demonstrated to mediate neutrophil recruitment, the release of
granule enzymes and the expression of adhesion molecules, which
multiply inflammatory effects (7).
Chronic inflammation may increase the risk of cancer development
(23,24). GRO-β is excessively expressed during
inflammation and is chemotactic for neutrophils in combination with
the CXCR2 receptor. GRO-β is crucial in the initiation and
progression of colitis-associated colon cancer, and the GRO-β-CXCR2
axis may be useful in decreasing the risk of ulcerative
colitis-associated colon cancer (6,7).
Sufficient evidence demonstrates that chemokines are also
significant in cancer, in addition to having a role in the
development and inflammatory responses (25,26). GRO
increases matrix metalloproteinase production in oral squamous cell
carcinoma by binding to CXCR2, which may participate in cancer
progression (26). Wang et al
(27) also indicated that GRO-β/CXCR2
forms an autocrine loop by activating the ERK1/2 pathway, and
contributes significantly to proliferation in primary esophageal
squamous cell carcinoma.
The findings of the present study suggest that GRO-β
was overexpressed in the cytoplasm of CRC cells rather than
inflammatory cells. However, the association between GRO-β protein
expression and the clinical characteristics of CRC was confined to
clinical observations using a tumor tissue microarray. The
association between GRO-β in colorectal carcinogenesis and the
autocrine or paracrine mechanisms and the signal pathways involved
remains to be elucidated. Additional in vitro and in
vivo studies are required in order to investigate the
biological functions of GRO-β in CRC.
Acknowledgements
This study was supported by grants from the
Translational Medicine Program of Affiliated Hospital of Nantong
University (grant no. TDFzh2014014), the Six Talent Peaks Program
of Jiangsu Province (grant no. 2012-ws-064) and the 226 Talent
Training Project of Nantong City (The Fourth Batch).
References
|
1
|
Siegel R, Naishadham D and Jemal A: Cancer
statistics, 2013. CA Cancer J Clin. 63:11–30. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Rao CV and Yamada HY: Genomic instability
and colon carcinogenesis: From the perspective of genes. Front
Oncol. 3:1302013. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Khan S, Cameron S, Blaschke M, Moriconi F,
Naz N, Amanzada A, Ramadori G and Malik IA: Differential gene
expression of chemokines in KRAS and BRAF mutated colorectal cell
lines: Role of cytokines. World J Gastroenterol. 20:2979–2994.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Modi WS and Yoshimura T: Isolation of
novel GRO genes and a phylogenetic analysis of the CXC chemokine
subfamily in mammals. Mol Biol Evol. 16:180–193. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Wang D and Richmond A: Nuclear
factor-kappa B activation by the CXC chemokine melanoma
growth-stimulatory activity/growth-regulated protein involves the
MEKK1/p38 mitogen-activated protein kinase pathway. J Biol Chem.
276:3650–3659. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Doll D, Keller L, Maak M, Boulesteix AL,
Siewert JR, Holzmann B and Janssen KP: Differential expression of
the chemokines GRO-2, GRO-3, and interleukin-8 in colon cancer and
their impact on metastatic disease and survival. Int J Colorectal
Dis. 25:573–581. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
McLean MH, Murray GI, Stewart KN, Norrie
G, Mayer C, Hold GL, Thomson J, Fyfe N, Hope M, Mowat NA, et al:
The inflammatory microenvironment in colorectal neoplasia. PLoS
One. 6:e153662011. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Rhodes DR, Yu J, Shanker K, Deshpande N,
Varambally R, Ghosh D, Barrette T, Pandey A and Chinnaiyan AM:
Large-scale meta-analysis of cancer microarray data identifies
common transcriptional profiles of neoplastic transformation and
progression. Proc Natl Acad Sci USA. 101:9309–9314. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Rhodes DR, Yu J, Shanker K, Deshpande N,
Varambally R, Ghosh D, Barrette T, Pandey A and Chinnaiyan AM:
ONCOMINE. A cancer microarray database and integrated data-mining
platform. Neoplasia. 6:1–6. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Ueno H, Mochizuki H, Akagi Y, Kusumi T,
Yamada K, Ikegami M, Kawachi H, Kameoka S, Ohkura Y, Masaki T, et
al: Optimal colorectal cancer staging criteria in TNM
classification. J Clin Oncol. 30:1519–1526. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Williams JR: The Declaration of Helsinki
and public health. Bull World Health Organ. 86:650–652. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Sun R, Wang X, Zhu H, Mei H, Wang W, Zhang
S and Huang J: Prognostic value of LAMP3 and TP53 overexpression in
benign and malignant gastrointestinal tissues. Oncotarget.
5:12398–12409. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Graudens E, Boulanger V, Mollard C,
Mariage-Samson R, Barlet X, Grémy G, Couillault C, Lajémi M,
Piatier-Tonneau D, Zaborski P, et al: Deciphering cellular states
of innate tumor drug responses. Genome Biol. 7:R192006. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Kaiser S, Park YK, Franklin JL, Halberg
RB, Yu M, Jessen WJ, Freudenberg J, Chen X, Haigis K, Jegga AG, et
al: Transcriptional recapitulation and subversion of embryonic
colon development by mouse colon tumor models and human colon
cancer. Genome Biol. 8:R1312007. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Sabates-Bellver J, Van der Flier LG, de
Palo M, Cattaneo E, Maake C, Rehrauer H, Laczko E, Kurowski MA,
Bujnicki JM, Menigatti M, et al: Transcriptome profile of human
colorectal adenomas. Mol Cancer Res. 5:1263–1275. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Gaedcke J, Grade M, Jung K, Camps J, Jo P,
Emons G, Gehoff A, Sax U, Schirmer M, Becker H, et al: Mutated KRAS
results in overexpression of DUSP4, a MAP-kinase phosphatase, and
SMYD3, a histone methyltransferase, in rectal carcinomas. Genes
Chromosomes Cancer. 49:1024–1034. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Skrzypczak M, Goryca K, Rubel T, Paziewska
A, Mikula M, Jarosz D, Pachlewski J, Oledzki J and Ostrowski J:
Modeling oncogenic signaling in colon tumors by multidirectional
analyses of microarray data directed for maximization of analytical
reliability. PLoS One. 5:e130912010. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Cancer Genome Atlas Network: Comprehensive
molecular characterization of human colon and rectal cancer.
Nature. 487:330–337. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Dong QM, Zhang JQ, Li Q, Bracher JC,
Hendricks DT and Zhao XH: Clinical significance of serum expression
of GROβ in esophageal squamous cell carcinoma. World J
Gastroenterol. 17:2658–2662. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Owen JD, Strieter R, Burdick M,
Haghnegahdar H, Nanney L, Shattuck-Brandt R and Richmond A:
Enhanced tumor-forming capacity for immortalized melanocytes
expressing melanoma growth stimulatory activity/growth-regulated
cytokine beta and gamma proteins. Int J Cancer. 73:94–103. 1997.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Buhmeida A, Bendardaf R, Hilska M, Laine
J, Collan Y, Laato M, Syrjänen K and Pyrhönen S: PLA2 (group IIA
phospholipase A2) as a prognostic determinant in stage II
colorectal carcinoma. Ann Oncol. 20:1230–1235. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Saif MW and Chu E: Biology of colorectal
cancer. Cancer J. 16:196–201. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Inoue Y, Iwata T, Okugawa Y, Kawamoto A,
Hiro J, Toiyama Y, Tanaka K, Uchida K, Mohri Y, Miki C and Kusunoki
M: Prognostic significance of a systemic inflammatory response in
patients undergoing multimodality therapy for advanced colorectal
cancer. Oncology. 84:100–107. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Ishizuka M, Nagata H, Takagi K, Iwasaki Y
and Kubota K: Inflammation-based prognostic system predicts
survival after surgery for stage IV colorectal cancer. Am J Surg.
205:22–28. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Dorhoi A, Iannaccone M, Farinacci M, Faé
KC, Schreiber J, Moura-Alves P, Nouailles G, Mollenkopf HJ,
Oberbeck-Müller D, Jörg S, et al: MicroRNA-223 controls
susceptibility to tuberculosis by regulating lung neutrophil
recruitment. J Clin Invest. 123:4836–4848. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Shang K, Bai YP, Wang C, Wang Z, Gu HY, Du
X, Zhou XY, Zheng CL, Chi YY, Mukaida N and Li YY: Crucial
involvement of tumor-associated neutrophils in the regulation of
chronic colitis-associated carcinogenesis in mice. PLoS One.
7:e518482012. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Wang B, Khachigian LM, Esau L, Birrer MJ,
Zhao X, Parker MI and Hendricks DT: A key role for early growth
response-1 and nuclear factor-kappaB in mediating and maintaining
GRO/CXCR2 proliferative signaling in esophageal cancer. Mol Cancer
Res. 7:755–764. 2009. View Article : Google Scholar : PubMed/NCBI
|