Introduction
Phosphoinositide 3-kinases (PI3Ks) are an
evolutionarily conserved family of lipid kinases that promote
various cellular functions, including cell growth, metabolism and
survival (1,2). The lipid second messengers that are
generated in this reaction interact with specialized lipid-binding
domains that are present in a wide variety of signaling molecules.
PI3Ks may be classified into one of three classes, each of which
possesses different structures and characteristics (3). The PI3K pathway may be activated by
upstream receptor tyrosine kinases, leading to the generation of
phosphatidylinositol-3,4,5-trisphosphate (PIP3) via the
phosphorylation of phosphatidylinositol-4,5-bisphosphate. The
phosphatase and tensin homolog (PTEN) may dephosphorylate
PIP3, which terminates PI3K signaling. The accumulation
of PIP3 activates a signaling cascade, commencing with
the phosphorylation (activation) of the protein serine-threonine
kinase AKT (also known as protein kinase B) at threonine 308 by
phosphoinositide-dependent kinase 1. Activation of AKT serves a
crucial role in essential cellular functions, including cell
proliferation and survival, via the phosphorylation of a variety of
substrates (Fig. 1) (4).
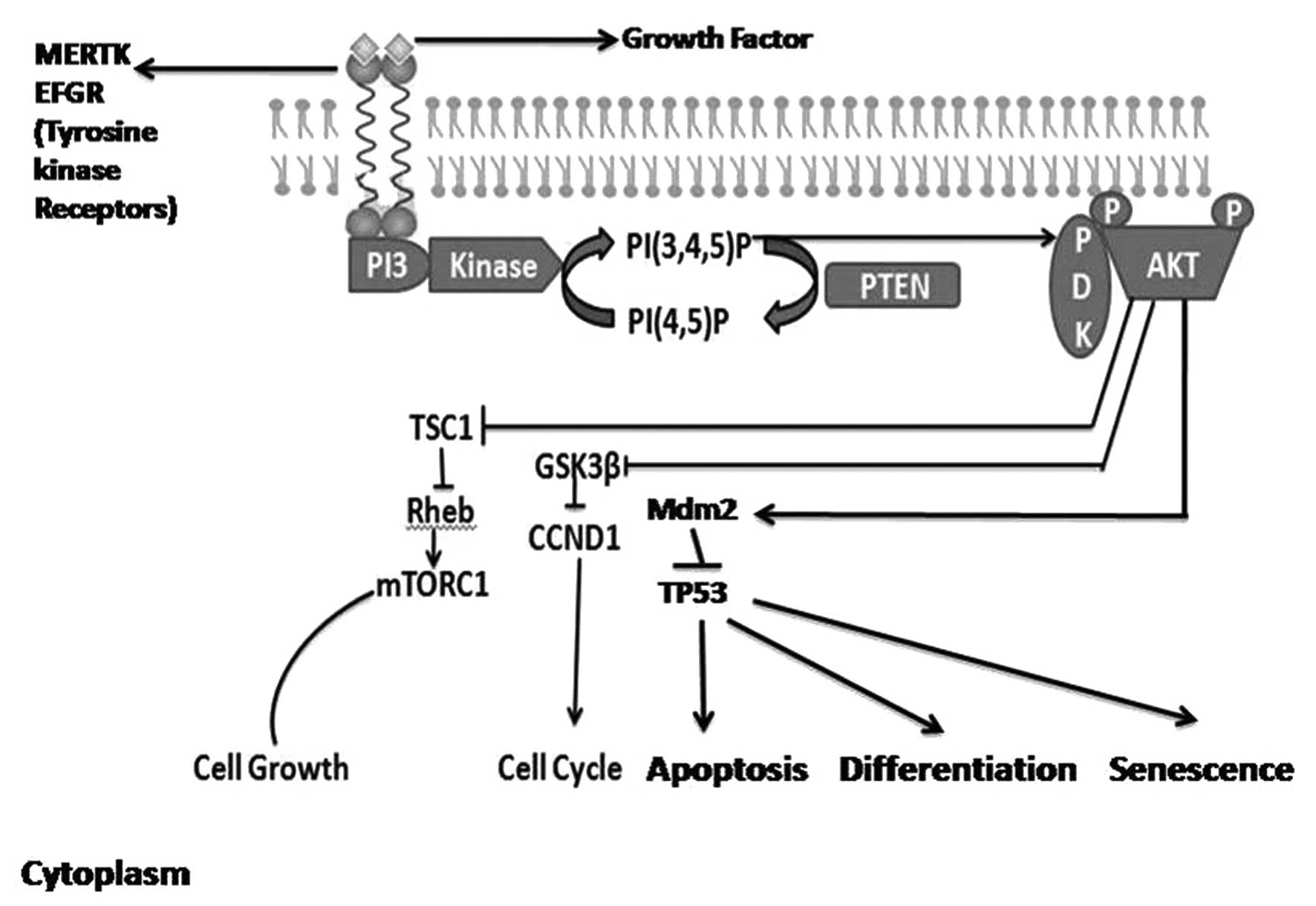 | Figure 1.PI3K/AKT signaling pathway. Binding of
the ligand to membrane receptor tyrosine kinases activates PI3K,
which phosphorylates PIP2 to produce PIP3.
PIP3 recruits PDK1 to the plasma membrane. PDK1
phosphorylates and activates AKT, which regulates various cellular
processes. The lipid phosphate activity of cytoplasmic PTEN
dephosphorylates PIP3, thereby decreasing
PIP3 levels and increasing levels of PIP2,
resulting in a concomitant decrease in AKT activity. PI3K,
phosphoinositide 3-kinase; AKT, protein kinase B; PIP2
[PI(4,5)P], phosphatidylinositol 4,5-bisphosphate;
PIP3 [PI(3,4,5)P],
phosphatidylinositol (3,4,5)-trisphosphate; PDK1,
phosphoinositide-dependent kinase 1; PTEN, phosphatase and tensin
homolog. |
Although the PI3K/AKT pathway has been extensively
investigated in detail in distinct in vitro and in
vivo systems (5), its role in
molecular targeted therapy for cancer required further study.
Molecular targeted therapies (e.g. inhibitors of target molecules
with critical roles in tumor growth and progression) have been
investigated in various cancer models, particularly hematological
malignancies, such as leukemia, lymphoma and myeloma, due to the
ease in obtaining samples for examination (6). The PI3K/AKT pathway has been reported to
be activated in numerous types of malignancy (7), and inhibitors associated with this
pathway have been shown to induce apoptosis in targeted tumor cells
(8).
Aberrant activation of the PI3K pathway may promote
carcinogenesis and tumor angiogenesis (9,10). For
example, a previous study reported that ~30% of breast cancer cases
demonstrated activating missense mutations of
phosphatidylinositol-4,5-bisphosphate 3-kinase, catalytic subunit α
(PIK3CA), the gene encoding the catalytic p110α subunit of class I
PI3K (2); this mutated gene provides
cells with a growth advantage and promotes tumorigenesis (11). In addition, dysregulated PI3K pathway
signaling has been implicated in conferring resistance to
conventional therapies, including biologics, hormonal therapy,
tyrosine kinase inhibitors, radiation and cytotoxic drugs in breast
cancer, glioblastoma and non-small cell lung cancer (12).
Wet laboratory research has revealed enormous data
in the field of cancer research, and expression levels of certain
proteins can be found at the Human Protein Atlas (www.proteinatlas.org). However, these proteins are not
classified according to a specific disease or disorder. The aim of
the present study was to utilize data deposited in the Human
Protein Atlas to investigate the protein expression level of 25
proteins that are known to be implicated in the PI3K pathway in
various cancer tissues. The proteins investigated were as follows:
AKTIP, ARP1, BAD, GSK3A, GSK3B, MERTK-1, PIK3CA, PRR5, PSTPIP2,
PTEN, FOX1, RHEB, RPS6KB1, TSC1, TP53, BCL2, CCND1, WFIKKN2,
CREBBP, capase-9, PTK2, EGFR, FAS, CDKN1A and XIAP. The analysis
reveals a pronounced expression of specific proteins in distinct
cancer tissues, which may be potential targets for cancer treatment
and provide insights into the molecular basis of cancer.
Materials and methods
Data were collected from the Human Protein Atlas
database (www.proteinatlas.org) via manual
searches of the desired gene names. The expression levels of 25
specific proteins that are known to be involved in the PI3K pathway
were investigated in 20 different cancer tissues types: Carcinoid,
glioma, liver cancer, lymphoma, melanoma, ovarian cancer,
pancreatic cancer, skin cancer, testis, urothelial, lung cancer,
breast cancer, cervical cancer, colorectal cancer, head and neck,
renal, thyroid, prostate, endometrial and stomach cancer.
The expression of the 25 proteins in the different
cancer tissues were reported as high, medium or low (excluding no
expression, which was considered as a separate category) relative
to normal tissues as shown in the database. Thereafter, the
percentage of high, medium and low expression in each tissue type
was calculated by dividing the number of patients exhibiting high
expression, for example, over the total number of patients in the
sample for each tissue type. The number of patients per sample
ranged from 8–18. Furthermore, high and medium percentages were
combined as the biological impact of high and medium expression was
believed to be similar. Graphs were created using Microsoft Excel
12.0 (Microsoft Corporation, Redmond, WA, USA) to represent the
percentage of each level of protein expression as it was expressed
in these patients.
Results and Discussion
In this study, the expression levels of 25 proteins
in tissues from 20 cancer types were analyzed utilizing the Human
Protein Atlas (www.proteinatlas.org). The following proteins
examined: AKTIP, ARP1, BAD, GSK3A, GSK3B, MERTK-1, PIK3CA, PRR5,
PSTPIP2, FOX1, RHEB, TSC1, TP53, BCL2, CCND1, WFIKKN2, CREBBP,
RPS6KB1, caspase-9, EGFR, PIK2, FAS, CDKN1A, XIAP and PTEN. The
physiological activity and full name of these proteins, as well as
their role in cancer initiation and control, is summarized in
Table I (13–43).
 | Table I.Summary of the physiological function
and associated signaling pathways of the 25 proteins studied. |
Table I.
Summary of the physiological function
and associated signaling pathways of the 25 proteins studied.
| Protein | Description/full
name | Main function | References |
|---|
| TSC1 | Tuberous sclerosis
1 | Tumor suppressor;
stimulates specific GTPases | (13) |
| EGFR | Epidermal growth
factor receptor | Activates PI3K,
which activates Akt (promotes cell survival and proliferation) | (14) |
| MERTK | MER proto oncogene
tyrosine kinase | Tyrosine kinase
receptor with oncogenic properties that is often overexpressed or
activated in various types of malignancy; member of the
MER/AXL/TYRO3 receptor kinase family; regulates melanoma cell
migration and survival | (15,16) |
| RHEB | Ras homolog
enriched in brain | Regulates growth
and cell cycle progression via the insulin/TOR/S6K signaling
pathway | (17) |
| RPS6KB1 | Ribosomal protein
S6 β1 | mTOR downstream
protein; promotes protein synthesis, cell growth, and cell
proliferation | (18) |
| CCND1 | Cyclin D1 | Functions as a
regulatory subunit of cyclin-dependent kinase (CDK)4 or CDK6, whose
activity is required for cell cycle G1/S transition | (19) |
| TP53 | Tumor protein
p53 | Tumor suppressor;
induces cell cycle arrest, apoptosis, senescence, DNA repair or
changes in metabolism | (20) |
| PTEN | Phosphatase and
tensin homolog | Tumor suppressor;
mostly mutated in a large number of cancers | (21,22) |
| PIK3CA |
Phosphatidylinositol-4,5-bisphosphate
3-kinase, catalytic subunit α | Belongs to the PI3K
family; has roles in various signaling pathways; involved in
proliferation, oncogenic transformation, cell survival and
migration, and intracellular protein trafficking | (23,24) |
| GSK3A | Glycogen synthase
kinase 3α | Control of several
regulatory proteins including glycogen synthase and transcription
factors | (25) |
| BAD | BCL2-associated
agonist of cell death | Regulator of
programmed cell death; proapoptotic activity of this protein is
regulated through its phosphorylation; AKT and mitogen-activated
protein kinase are involved in the regulation of this protein | (26,27) |
| AKTIP | AKT interacting
protein | Mediates
insulin-induced translocation of glucose transporter-4 to the cell
surface | (28) |
| GSK3B | Glycogen synthase
kinase 3β | Involved in energy
metabolism, neuronal cell development and body pattern
formation | (29) |
| ARP1 | Actin-related
protein 1 | Transcriptional
regulator involved in basal and hormone-regulated activity of
prolactin; regulates the cell-specific trafficking of a receptor
protein involved in apoptosis | (30,31) |
| PRR5 | Proline rich 5
(renal) | Regulates
platelet-derived growth factor receptor | (32) |
| FOX1 | Fox-1 homolog. Also
known as: ataxin 2-binding protein 1 (A2BP1); or
hexaribonucleotide-binding | Possesses RNA
binding motif; associated with spinocerebellar ataxia due to its
binding to ataxin-2; regulator of tissue-specific alternative
splicing in mammals protein 1 (HRNBP1) | (33) |
| BCL2 | B-cell lymphoma
2 | Oncogene; blocks
apoptosis | (34,35) |
| WFIKKN2 | WAP,
follistatin/kazal, immunoglobulin, kunitz | Protease-inhibitor
that contains distinct protease inhibitor domains and netrin domain
containing 2 | (36) |
| CREBBP | CREB binding
protein | Regulates embryonic
development, growth control, and homeostasis by coupling chromatin
remodeling to transcription factor recognition | (37) |
| Caspase-9 | Apoptosis-related
cysteine peptidase | Plays a central
role in the execution-phase of cell apoptosis | (38) |
| PTK2 | Protein tyrosine
kinase 2. Also known as: focal adhesion kinase (FAK) | Focal
adhesion-associated protein involved in cellular adhesion and
spreading processes; when it is blocked, cancer cells become less
metastatic due to decreased mobility | (38,39) |
| FAS | Cell surface death
receptor. Also known as: apoptosis antigen 1 (APO-1 or APT) | Plays a central
role in the physiological regulation of programmed cell death | (40) |
| CDKN1A | Cyclin-dependent
kinase inhibitor 1A. Also known as: P21; CIP1; SDI1; WAF1; CAP20;
CDKN1; MDA-6; or p21CIP1 | Inhibits the
activity of cyclin-CDK2 or -CDK4 complexes, and thus functions as a
regulator of cell cycle progression at G1 phase | (41) |
| PSTPIP2 |
Proline-serine-threonine
phosphatase-interacting protein 2 | Substrate for a
number of important signaling proteins | (42) |
| XIAP | X-linked inhibitor
of apoptosis. Also known as: IAP3 and BIRC | Inhibits
apoptosis | (43) |
The results revealed that 9 of the 25 proteins
tested exhibited high expression levels in various cancer tissues.
These proteins were PIK3CA, RPS6KB1, MERTK, RHEB, EGFR, TSC1,
CCND1, TP53 and PTEN. The other 16 proteins exhibited low or no
expression in tumor tissues (data not shown).
The expression level for each protein tested was
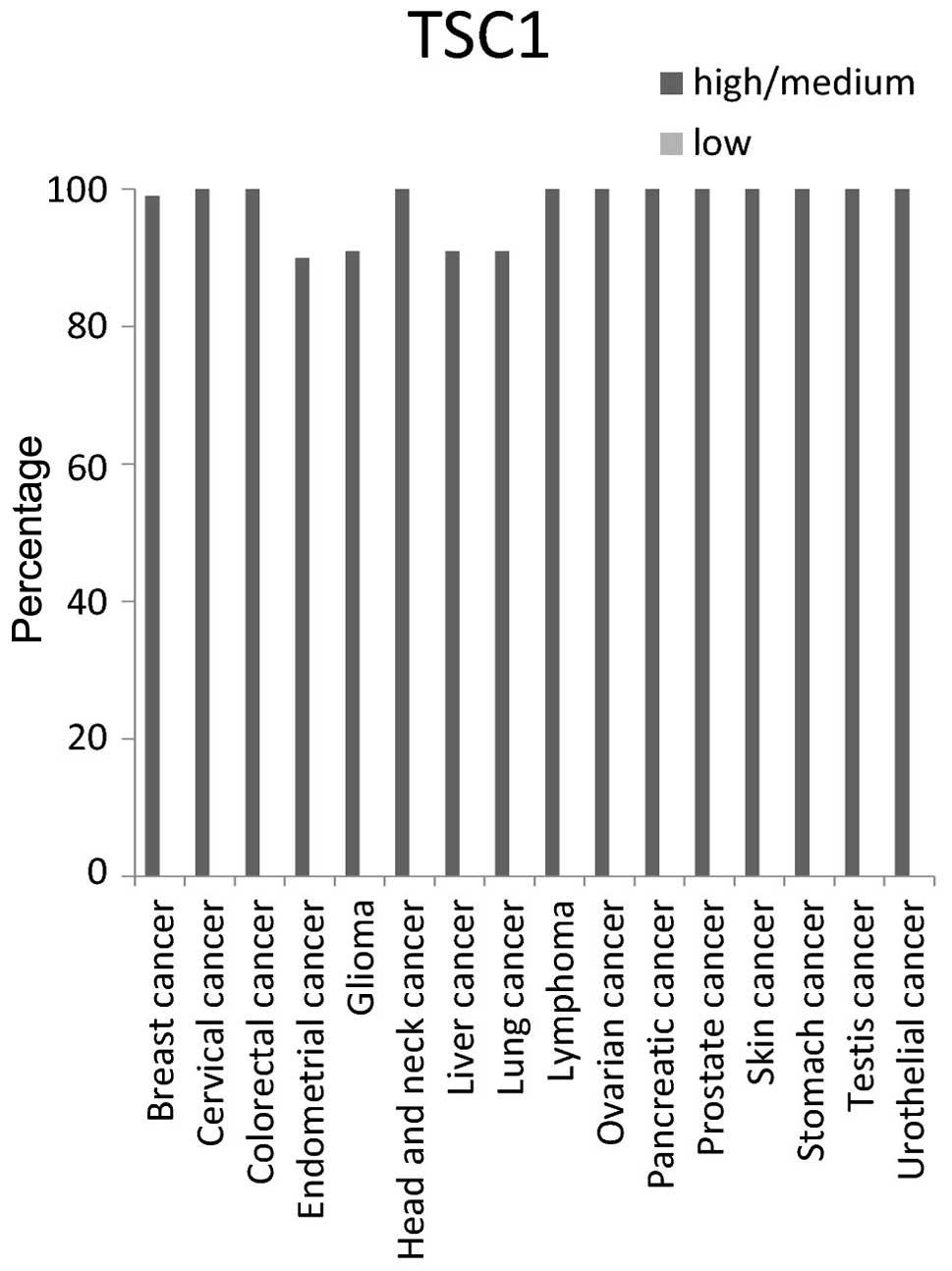
categorized as either high/medium or low. The protein TSC1
exhibited high/medium expression in all types of cancer tissue
tested. TSC1 exhibited ~100% high/medium expression in breast,
cervical, colorectal, head and neck, lymphoma, ovarian, pancreatic,
prostate, skin, stomach, testis and urothelial cancer tissues. It
expression was ~90% high/medium in endometrial, glioma, liver and
lung cancers (Fig. 2).
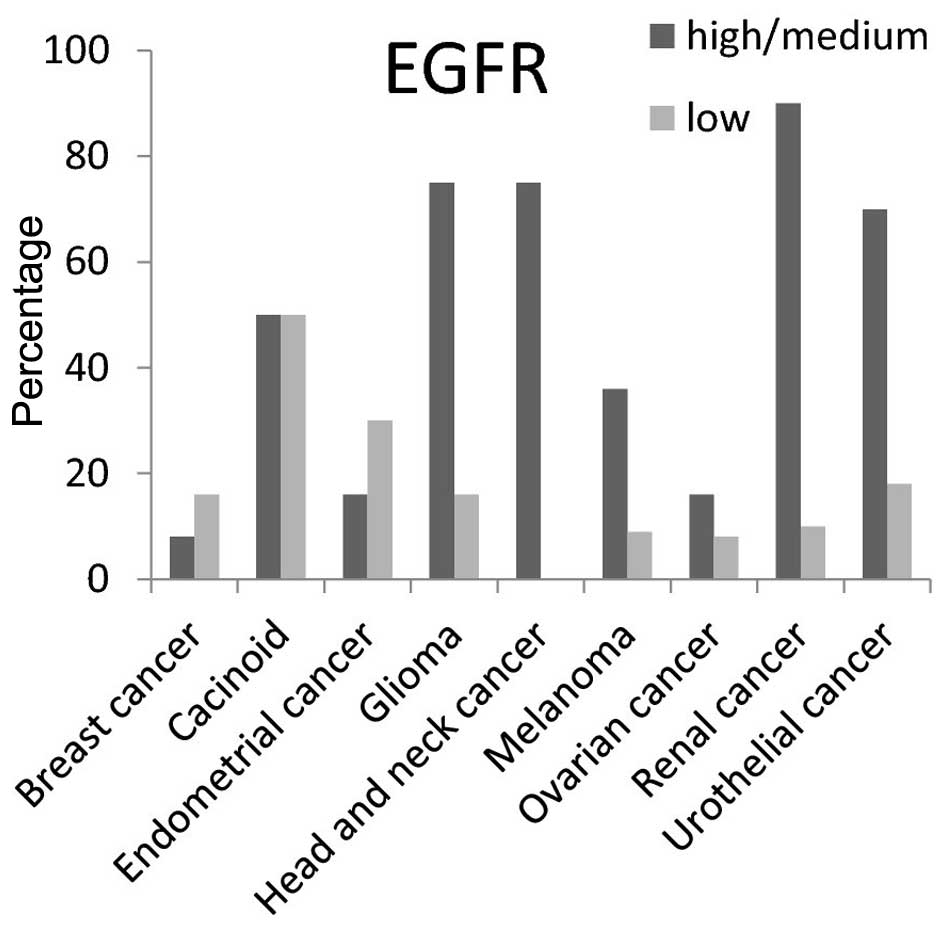
EGFR protein had high/medium expression level in
>50% of carcinoid, head and neck, glioma, renal and urothelial
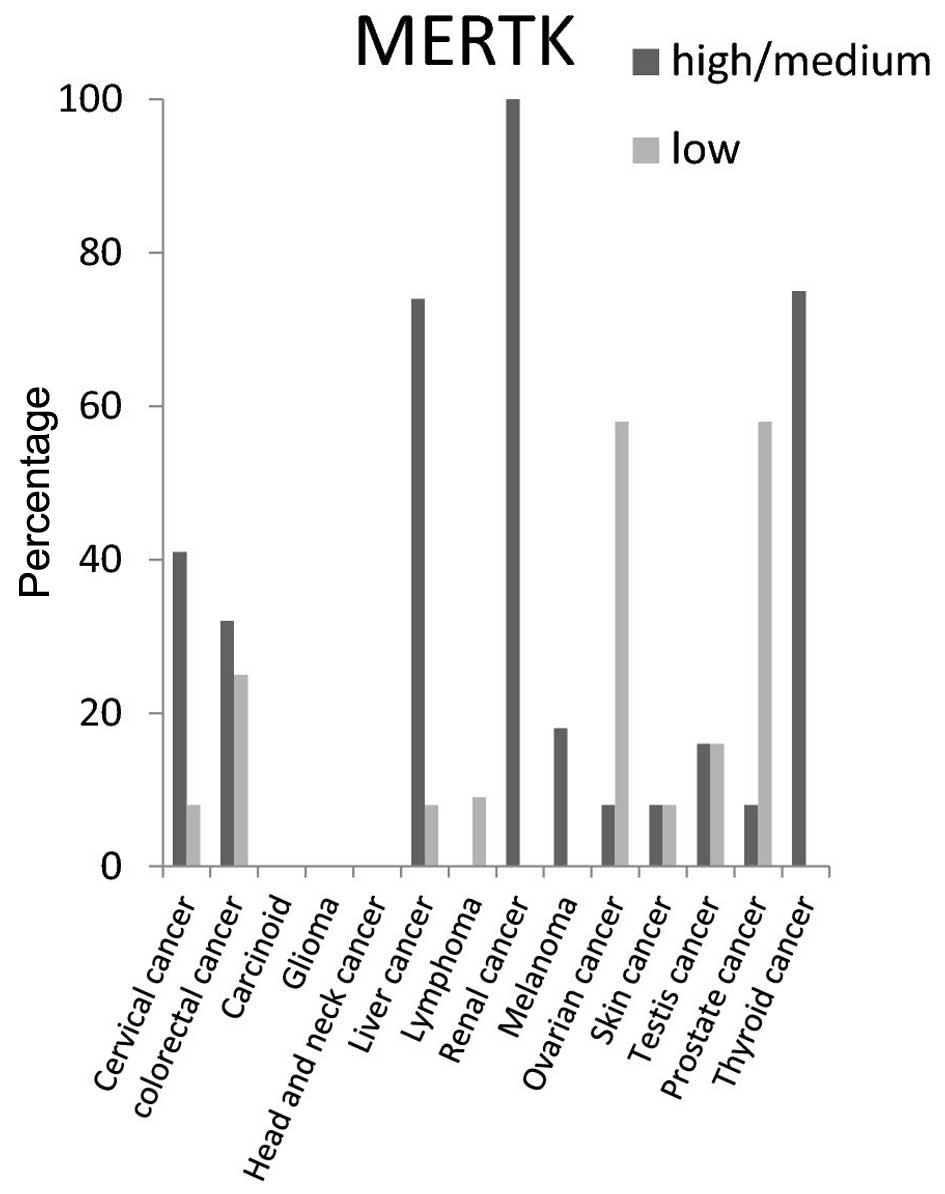
cancer tissues (Fig. 3). For MERTK
protein the high/medium expression rate was >50% in liver and
thyroid cancer tissues, and 100% in renal cancer tissues. It was
not detected in carcinoid, glioma, or head and neck cancer tissues
(Fig. 4).
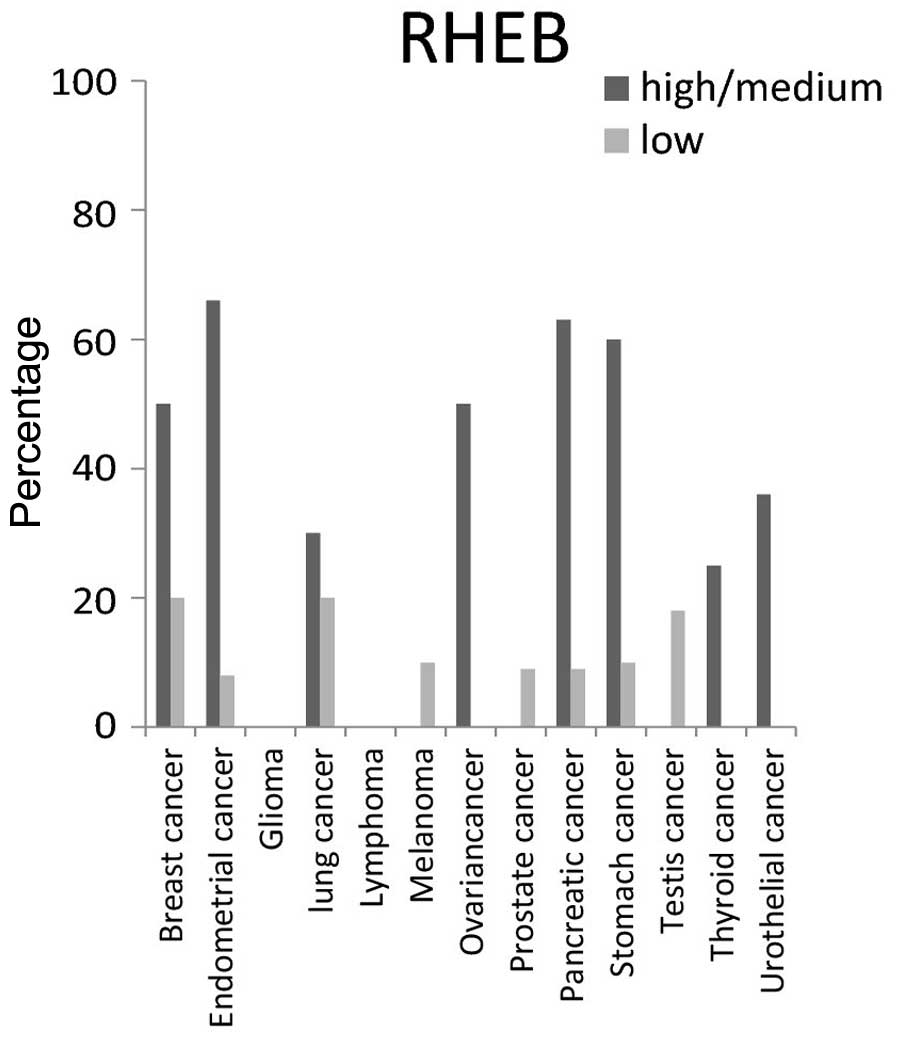
For RHEB protein, the highest expression level was
present in >50% of breast, endometrial, ovarian, pancreatic and
stomach cancer tissues, but was not detected in glioma and lymphoma
cancer tissues (Fig. 5).
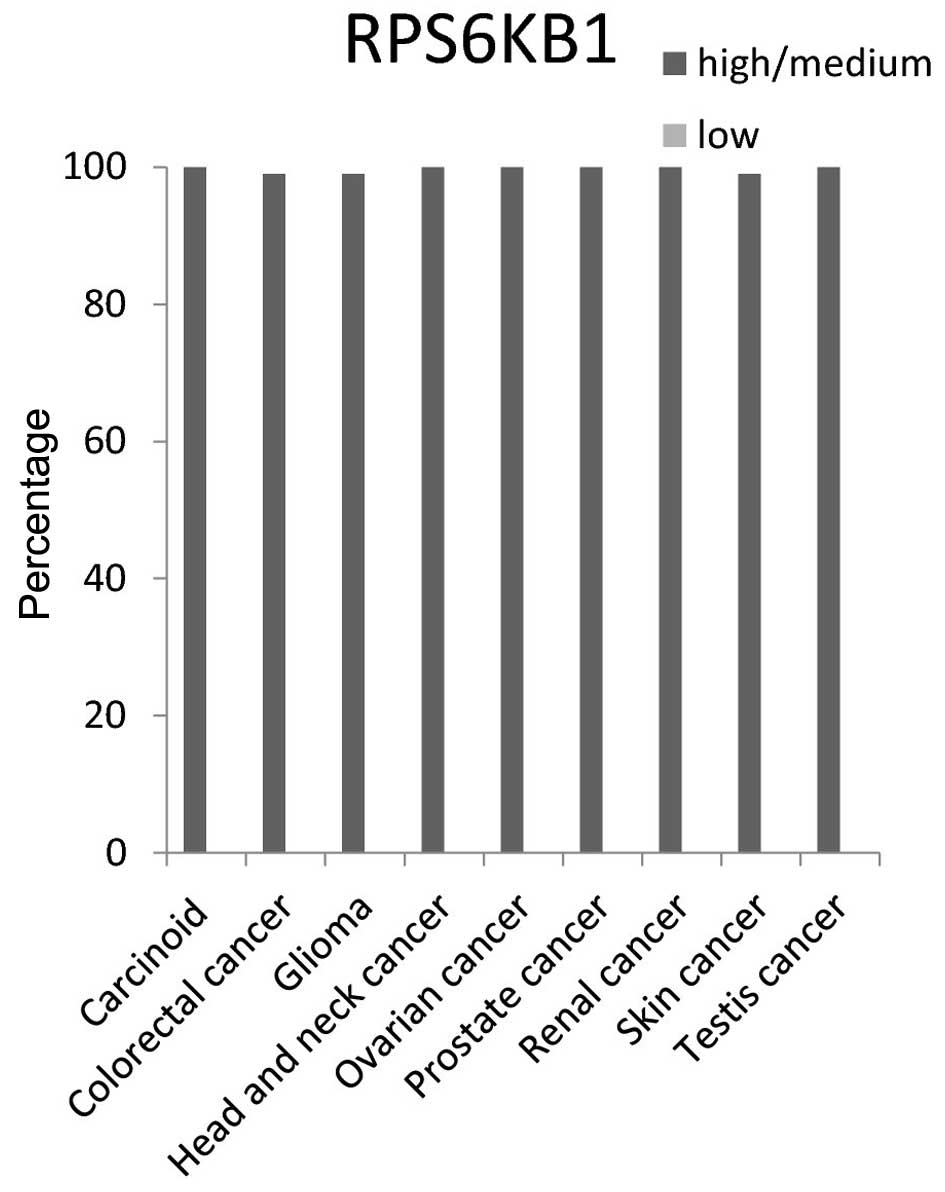
The RPS6KB1 protein expression level had ~100%
high/medium in 9 cancer tissue tested: Carcinoid, colorectal,
glioma, head and neck, ovarian, prostate, renal, skin and testis
cancer tissues (Fig. 6).
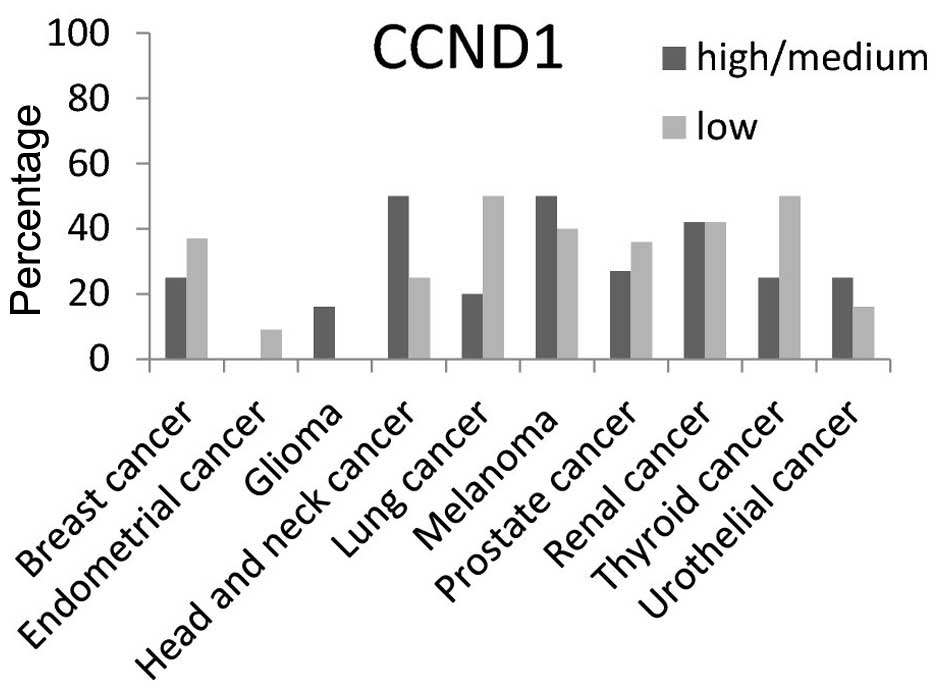
CCND1 protein high/medium expression level was
present in ~50% of head and neck cancer and melanoma tissues
(Fig. 7). The high/medium expression
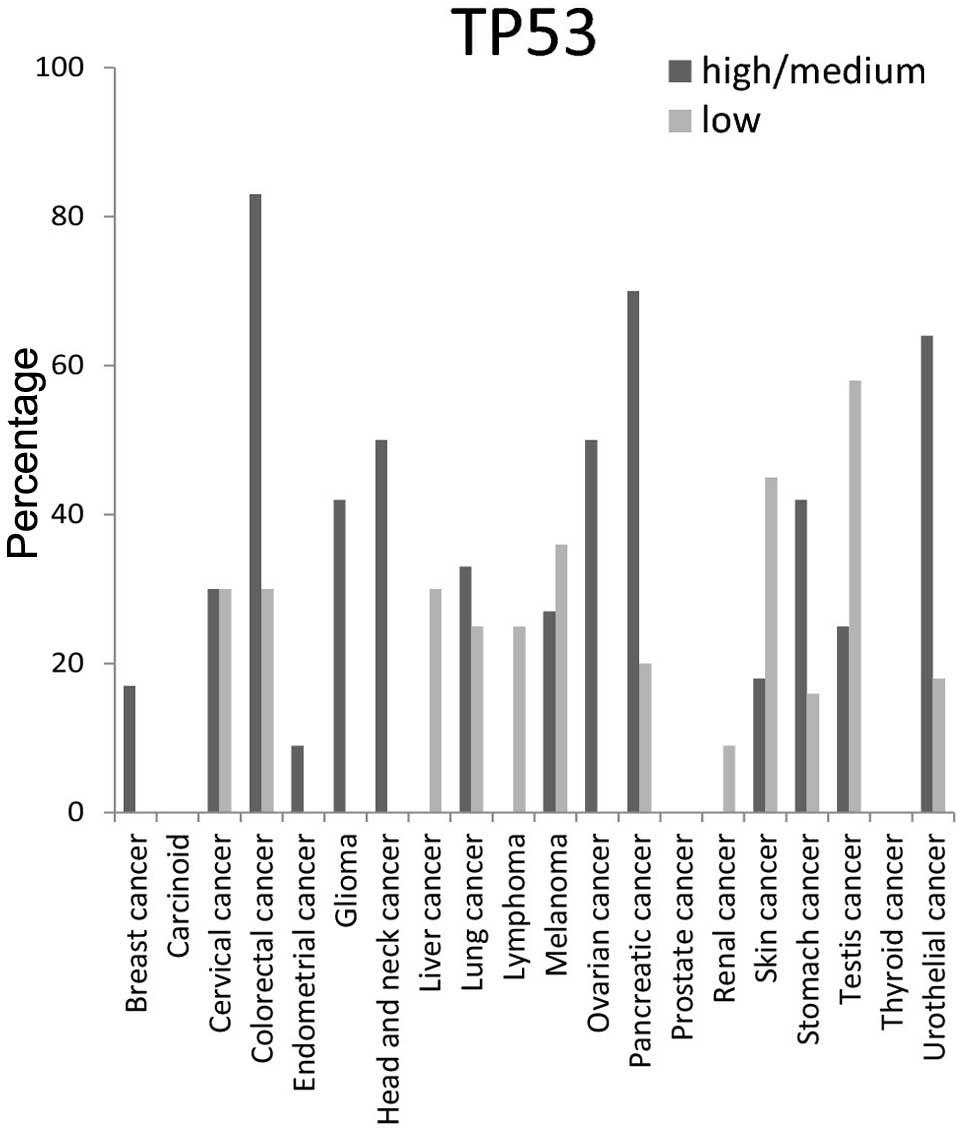
percentage of TP53 protein was ≥50% in colorectal, head and neck,
ovarian, pancreatic and urothelial tissues, but was not detected at
all in carcinoid, prostate and thyroid cancer tissues (Fig. 8).
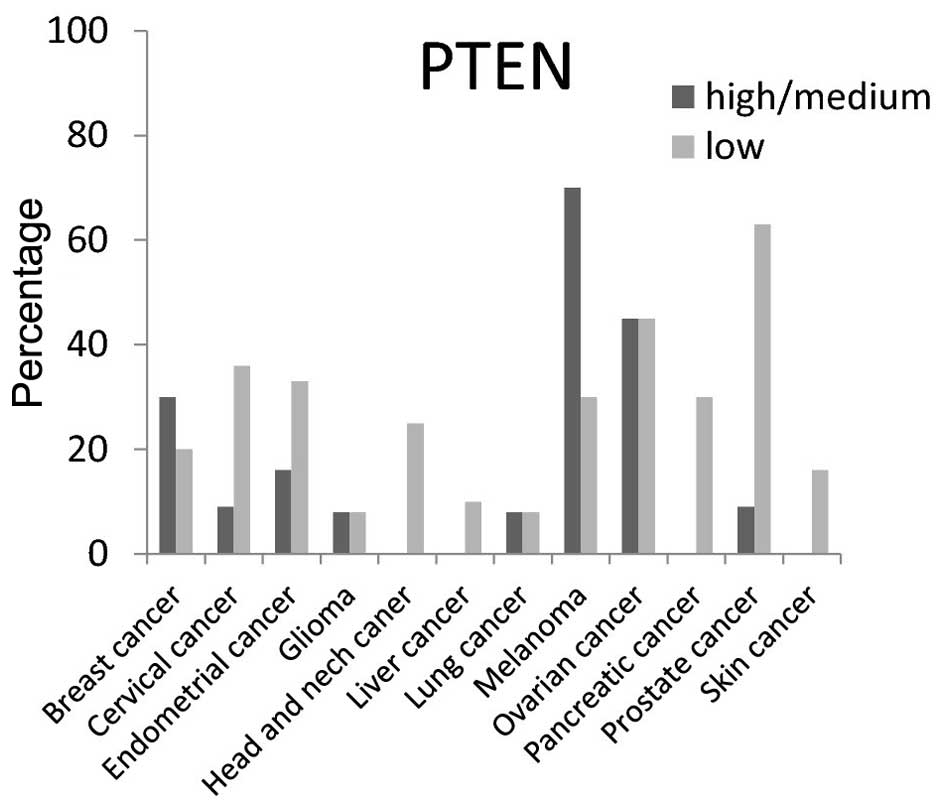
The expression level of PTEN protein (a tumor
suppressor gene) was low in various cancer tissues as was expected.
A high/medium PTEN expression level was present in <50% of
breast, cervical, endometrial, glioma, head and neck, liver,
pancreatic and skin cancer tissues; however, high/medium expression
was present at a rate of ~75% in melanoma (Fig. 9).
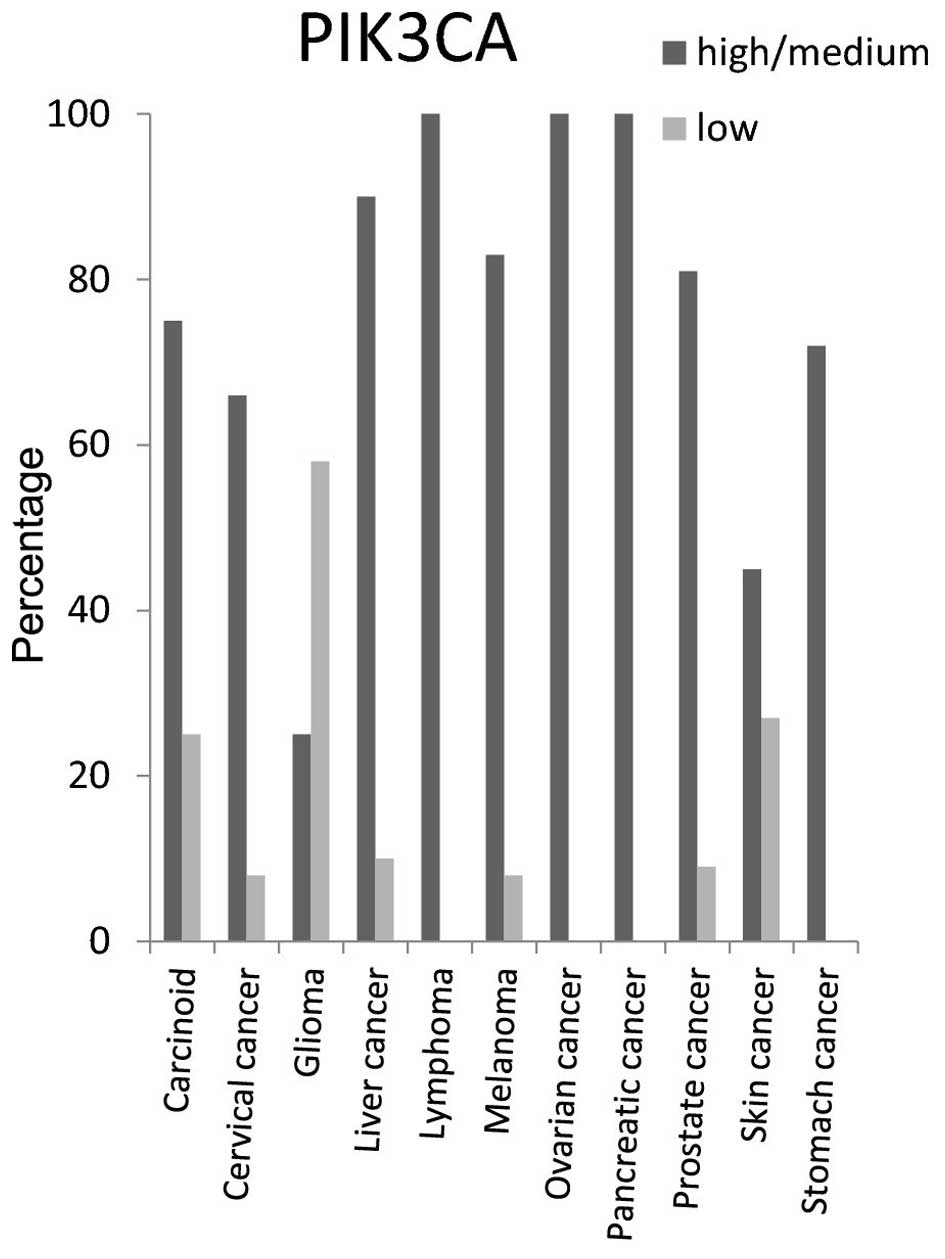
PIK3CA protein expression level was high/medium in
around 100% of lymphoma, ovarian and pancreatic cancer tissues, 90%
of liver cancer tissues, 85% of melanoma and prostate cancer
tissues, 70% of carcinoid and stomach cancer tissues and 65% of
cervical cancer tissues (Fig.
10).
Taking this data together, the current analysis
reveals a pronounced expression of specific proteins in distinct
cancer tissues. These proteins may be potential candidates to serve
as targets for cancer treatments and provide insights into the
molecular basis of cancer. PI3Ks initiate signaling through a
network of downstream effector pathways. Due to the direct
implication of the pathway in numerous cancer types, this pathway
has become the target for novel cancer therapies. This bird's-eye
view study highlights 9 proteins that are involved in the PI3K
pathway and which may be potential targets for cancer treatment.
These proteins are highly expressed in several cancer tissues as
indicated. Designing new drugs that modulate the activity of these
proteins may decrease cancer growth, migration and metastasis.
Acknowledgements
This study was supported by Alqasemi Research Fund
and the Association of Arab Universities Research Fund.
References
|
1
|
Cantley LC: The phosphoinositide 3-kinase
pathway. Science. 296:1655–1657. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Vogelstein B and Kinzler KW: Cancer genes
and the pathways they control. Nat Med. 10:789–799. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Akinleye A, Avvaru P, Furqan M, Song Y and
Liu D: Phosphatidylinositol 3-kinase (PI3K) inhibitors as cancer
therapeutics. J Hematol Oncol. 6:882013. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Ocana A, Vera-Badillo F, Al-Mubarak M,
Templeton AJ, Corrales-Sanchez V, Diez-Gonzalez L, Cuenca-Lopez MD,
Seruga B, Pandiella A and Amir E: Activation of the PI3K/mTOR/AKT
pathway and survival in solid tumors: Systematic review and
meta-analysis. PLoS One. 9:e952192014. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Ott PA and Adams S: Small-molecule protein
kinase inhibitors and their effects on the immune system:
Implications for cancer treatment. Immunotherapy. 3:213–227. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Kummar S, Kinders R, Gutierrez ME,
Rubinstein L, Parchment RE, Phillips LR, Ji J, Monks A, Low JA,
Chen A, et al: Phase 0 clinical trial of the poly (ADP-ribose)
polymerase inhibitor ABT-888 in patients with advanced
malignancies. J Clin Oncol. 27:2705–2711. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Vara Fresno JA, Casado E, de Castro J,
Cejas P, Belda-Iniesta C and González-Barón M: PI3K/Akt signalling
pathway and cancer. Cancer Treat Rev. 30:193–204. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Wilson JM, Kunnimalaiyaan S,
Kunnimalaiyaan M and Gamblin TC: Inhibition of the AKT pathway in
cholangiocarcinoma by MK2206 reduces cellular viability via
induction of apoptosis. Cancer Cell Int. 15:132015. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Osaki M, Oshimura M and Ito H: PI3K-Akt
pathway: Its functions and alterations in human cancer. Apoptosis.
9:667–676. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Patel S: Exploring novel therapeutic
targets in GIST: Focus on the PI3K/Akt/mTOR pathway. Curr Oncol
Rep. 15:386–395. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Liu P, Cheng H, Roberts TM and Zhao JJ:
Targeting the phosphoinositide 3-kinase pathway in cancer. Nat Rev
Drug Discov. 8:627–644. 2009. View
Article : Google Scholar : PubMed/NCBI
|
|
12
|
Samuels Y, Wang Z, Bardelli A, Silliman N,
Ptak J, Szabo S, Yan H, Gazdar A, Powell SM, Riggins GJ, et al:
High frequency of mutations of the PIK3CA gene in human cancers.
Science. 304:5542004. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Demetriades C, Doumpas N and Teleman AA:
Regulation of TORC1 in response to amino acid starvation via
lysosomal recruitment of TSC2. Cell. 156:786–799. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Yewale C, Baradia D, Vhora I, Patil S and
Misra A: Epidermal growth factor receptor targeting in cancer: A
review of trends and strategies. Biomaterials. 34:8690–8707. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Schlegel J, Sambade MJ, Sather S, Moschos
SJ, Tan AC, Winges A, DeRyckere D, Carson CC, Trembath DG, Tentler
JJ, et al: MERTK receptor tyrosine kinase is a therapeutic target
in melanoma. J Clin Invest. 123:2257–2267. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Cummings CT, DeRyckere D, Earp HS and
Graham DK: Molecular pathways: MERTK signaling in cancer. Clin
Cancer Res. 19:5275–5280. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Karassek S, Berghaus C, Schwarten M,
Goemans CG, Ohse N, Kock G, Jockers K, Neumann S, Gottfried S,
Herrmann C, et al: Ras homolog enriched in brain (Rheb) enhances
apoptotic signaling. J Biol Chem. 285:33979–33991. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Du W, Gerald D, Perruzzi CA,
Rodriguez-Waitkus P, Enayati L, Krishnan B, Edmonds J, Hochman ML,
Lev DC and Phung TL: Vascular tumors have increased p70 S6-kinase
activation and are inhibited by topical rapamycin. Lab Invest.
93:1115–1127. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Juskevicius D, Ruiz C, Dirnhofer S and
Tzankov A: Clinical, morphologic, phenotypic and genetic evidence
of cyclin D1-positive diffuse large B-cell lymphomas with cyclin D1
gene rearrangements. Am J Surg Pathol. 38:719–727. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Hejnold M, Dyduch G, Białas M, Demczuk S,
Ryś J, Chłosta P, Szopiński T and Okoń K: Selected
immunohistochemical features of conventional renal cell carcinomas
coexpressing P53 and MDM2. Pol J Pathol. 65:113–119. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Wang H, Chen P, Liu XX, Zhao W, Shi L, Gu
XW, Zhu CR, Zhu HH and Zong L: Prognostic impact of
gastrointestinal bleeding and expression of PTEN and Ki-67 on
primary gastrointestinal stromal tumors. World J Surg Oncol.
12:892014. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Luo J, Manning BD and Cantley LC:
Targeting the PI3K-Akt pathway in human cancer: Rationale and
promise. Cancer Cell. 4:257–262. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Qiu W, Tong GX, Turk AT, Close LG, Caruana
SM and Su GH: Oncogenic PIK3CA mutation and dysregulation in human
salivary duct carcinoma. Biomed Res Int. 2014:8104872014.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Cho DC: Targeting the PI3K/Akt/mTOR
pathway in malignancy: Rationale and clinical outlook. BioDrugs.
28:373–381. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Li X and Jope RS: Is glycogen synthase
kinase-3 a central modulator in mood regulation?
Neuropsychopharmacology. 35:2143–2154. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Huang N, Zhu J, Liu D, Li YL, Chen BJ, He
YQ, Liu K, Mo XM and Li WM: Overexpression of Bcl-2-associated
death inhibits A549 cell growth in vitro and in vivo.
Cancer Biother Radiopharm. 27:164–168. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Cooray S: The pivotal role of phosphatidyl
inositol 3-kinase-Akt signal transduction in virus survival. J Gen
Virol. 85:1065–1076. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Yang F, Deng R, Qian XJ, Chang SH, Wu XQ,
Qin J, Feng GK, Ding K and Zhu XF: Feedback loops blockade
potentiates apoptosis induction and antitumor activity of a novel
AKT inhibitor DC120 in human liver cancer. Cell Death Dis.
5:e11142014. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Taelman VF, Dobrowolski R, Plouhinec JL,
Fuentealba LC, Vorwald PP, Gumper I, Sabatini DD and De Robertis
EM: Wnt signaling requires sequestration of gycogen synthase kinase
3 inside multivesicular endosomes. Cell. 143:1136–1148. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Wang J, Xin YF, Xu WJ, Liu ZM, Qiu XB, Qu
XK, Xu L, Li X and Yang YQ: Prevalence and spectrum of PITX2c
mutations associated with congenital heart disease. DNA Cell Biol.
32:708–716. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Símová S, Klíma M, Cermak L, Sourková V
and Andera L: Arf and Rho GAP adapter protein ARAP1 participates in
the mobilization of TRAIL-R1/DR4 to the plasma membrane. Apoptosis.
13:423–436. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Gan X, Wang J, Wang C, Sommer E, Kozasa T,
Srinivasula S, Alessi D, Offermanns S, Simon MI and Wu D: PRR5L
degradation promotes mTORC2-mediated PKC-delta phosphorylation and
cell migration downstream of G α 12. Nat Cell Biol. 14:686–696.
2012. View
Article : Google Scholar : PubMed/NCBI
|
|
33
|
Kuroyanagi H: Fox-1 family of RNA-binding
proteins. Cell Mol Life Sci. 66:3895–3907. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Greenberg EF, Lavik AR and Distelhorst CW:
Bcl-2 regulation of the inositol 1,4,5-trisphosphate receptor and
calcium signaling in normal and malignant lymphocytes: Potential
new target for cancer treatment. Biochim Biophys Acta.
1843:2205–2210. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Bose P, Rahmani M and Grant S: Coordinate
PI3K pathway and Bcl-2 family disruption in AML. Oncotarget.
3:1499–1500. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Szlama G, Trexler M and Patthy L: Latent
myostatin has significant activity and this activity is controlled
more efficiently by WFIKKN1 than by WFIKKN2. FEBS J. 280:3822–3839.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Kim MS, Yoo NJ and Lee SH: Expressional
and mutational analysis of CREBBP gene in gastric and colorectal
cancers with microsatellite instability. Pathol Oncol Res.
20:221–222. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Unno H, Futamura K, Morita H, Kojima R,
Arae K, Nakae S, Ida H, Saito H, Matsumoto K and Matsuda A: Silica
and double-stranded RNA synergistically induce bronchial epithelial
apoptosis and airway inflammation. Am J Respir Cell Mol Biol.
51:344–353. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Lackie JM, Coote JG and Lloyd CW: The
Dictionary of Cell & Molecular Biology (5th). Academic Press.
San Diego, CA: 2013. View Article : Google Scholar
|
|
40
|
Villa-Morales M, Cobos MA, González-Gugel
E, Álvarez-Iglesias V, Martínez B, Piris MA, Carracedo A, Benítez J
and Fernández-Piqueras J: FAS system deregulation in T-cell
lymphoblastic lymphoma. Cell Death Dis. 5:e11102014. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Gupta R, Dong Y, Solomon PD, Wettersten
HI, Cheng CJ, Min JN, Henson J, Dogra SK, Hwang SH, Hammock BD, et
al: Synergistic tumor suppression by combined inhibition of
telomerase and CDKN1A. Proc Natl Acad Sci USA. 111:E3062–E3071.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Thapa N and Anderson RA: PIP2 signaling,
an integrator of cell polarity and vesicle trafficking in
directionally migrating cells. Cell Adh Migr. 6:409–412. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Miyamoto M, Takano M, Iwaya K, Shinomiya
N, Kato M, Aoyama T, Sasaki N, Goto T, Suzuki A, Hitrata J and
Furuya K: X-chromosome-linked inhibitor of apoptosis as a key
factor for chemoresistance in clear cell carcinoma of the ovary. Br
J Cancer. 110:2881–2886. 2014. View Article : Google Scholar : PubMed/NCBI
|