Introduction
Ovarian cancer remains the leading cause of
mortality due to gynecological cancer in the world (1). At present, platinum-based chemotherapy
following surgery represents the standard therapy for those cases
of ovarian cancer at advanced and high-risk early stages (2–5). A total
of 70% of patients with ovarian cancer demonstrate a response to
cytoreductive surgery followed by platinum-based chemotherapy
(6); however, the majority of these
patients will experience relapse. Thus, subsequent treatments are
moderate in curative effect and typically short in duration
(7).
Tumor cells that are resistant to a particular
chemotherapeutic drug are also often observed to be not sensitive
to a variety of structurally and functionally unrelated
chemotherapeutic drugs (8). This
phenomenon is known as multidrug resistance (MDR) (9). The mechanisms of MDR remain unclear, but
it may be attributed to increased drug efflux (10–12),
enhanced DNA damage repair (13,14),
resistance to apoptosis (15,16), self-renewing tumor stem cells
(17) and tumor microenvironment
(18–20). Since 1995, when major vault protein
(MVP) was identified to be identical to lung resistance-related
protein (21), numerous studies have
investigated the role of MVP in MDR (22–24).
MVP is the main component of vault, which contains
two additional proteins known as vault poly-(adenosine
diphosphate-ribose) polymerase and telomerase-associated protein 1,
in addition to several small untranslated vault RNAs (25–28). The
amino acid sequence of MVP is highly conserved among eukaryotic
cells (21). MVP is present in normal
tissues, including bronchus, digestive tract and macrophages, and
in malignant cells, including acute myeloid leukemia, ovarian
cancer and colon carcinoma (29). The
high degree of conservation and ubiquitous expression of MVP
suggest that this protein exerts crucial cellular functions
(29). Increasing evidence from
previous studies has demonstrated that high messenger (m)RNA and
protein levels of MVP are associated with resistance to
antineoplastic agents and reduced survival in certain types of
cancer, including ovarian cancer (22,30–32).
Single nucleotide polymorphisms (SNPs) are the most
common type of genetic variation among the human population, and
are often associated with inter-individual diversity in various
malignancies regarding the patient's susceptibility to disease,
drug response, toxicity and survival (33–36).
However, to the best of our knowledge, the role of MVP SNPs in
platinum resistance and prognosis of patients with ovarian cancer
has not been reported thus far. Therefore, in order to assess
whether SNPs in the MVP gene were associated with platinum
resistance and survival in epithelial ovarian cancer (EOC), two
polymorphisms were selected from the genotype data of Chinese Han
population derived from the phase II International (haplotype map)
HapMap Project (date of access to the database, August 14, 2014).
The specific MVP genotypes were subsequently identified in the
patients with EOC recruited for the present study, and the
associations between these genotypes and the response to
platinum-based regimens, progression-free survival (PFS) and
overall survival (OS) exhibited by these patients were analyzed.
Furthermore, the results of the present study confirmed the
feasibility of tetra-primer amplification-refractory mutation
system (ARMS)-polymerase chain reaction (PCR) as a genotyping
tool.
Materials and methods
Patients and clinical data
A total of 116 Chinese Han female patients with EOC
and Karnofsky performance status score ≥70 were recruited between
June 2005 and February 2012, and treated at the Department of
Oncology of The First Affiliated Hospital of Liaoning Medical
University (Jinzhou, China). Written informed consent was obtained
from all the patients included in the study, which was approved by
the Ethics Review Committee of The First Affiliated Hospital of
Liaoning Medical University. Following surgery, the patients were
intravenously treated with taxol (135 mg/m2 d1) or taxetere (75
mg/m2 d1) and cisplatin (30 mg d2-4) or carboplatin (AUC 4–6 d2) at
three or four weeks intervals for at least 3 cycles.
Follow-up examinations
Follow-up examinations were performed every 3 months
or when patients presented with symptoms of progression and
consulted a doctor. The examinations included pelvic examination,
determination of the levels of cancer antigen (CA)125 in serum and
pelvic computed tomography (CT) scanning. In addition, liver
ultrasonography, thoracic or abdominal CT scanning and brain
magnetic resonance imaging were conducted when necessary. Patients
who exhibited persistent or progressive disease during the
treatment or recurred within 6 months of completion of the
platinum-based chemotherapy were defined as platinum-resistant
(37). By contrast, patients who
exhibited disease progression later than 6 months upon completion
of the platinum-based therapy were considered to be
platinum-sensitive. PFS was calculated as the duration, in months,
from the date of histological diagnosis to the first sign of
recurrence detected by physical examination, CA125 evaluation or
radiographic inspection. OS was calculated as the duration, in
months, from the date of histological diagnosis to mortality or
last follow-up.
DNA extraction
Prior to the initiation of chemotherapy, genomic DNA
was obtained from peripheral venous blood using TIANamp Blood DNA
Kit (catalog no., DP318; Tiangen Biotech Co., Ltd., Beijing,
China), according to the manufacturer's protocol. The purity and
concentration of the extracted DNA were assessed by
spectrophotometry using the BioPhotometer Plus (Eppendorf, Hamburg,
Germany). The method yielded DNA of relatively high concentration
(median = 39.3 µg/ml, range = 18.4–60.5 µg/ml) and purity (median
absorbance (A)260/A280 ratio = 1.81, range = 1.46–2.37). The
extracted DNA was stored at −80°C until further use.
Genotyping
The tagging SNPs reference SNP (rs)1057451,
rs4788186 and rs2288043 were selected from the genotype data of
Chinese Han population derived from the International HapMap
Project (HapMap Data Release 24/phase II Nov08, on National Center
for Biotechnology Information B36 assembly, database SNP b126) to
capture the maximum variation based on r2 ≥0.8 and minor allele
frequency ≥0.05. Genotyping of the selected SNPs was performed by
the cost-effective method of tetra-primer ARMS-PCR, as proposed by
Ye et al (38). PCR was
conducted in a total volume of 20 µl, which contained 1 µl template
DNA, 0.5 µl each of the four primers (the concentration of the
working solution was 10 µM; primers were designed by Primer Premier
5.0, Premier Biosoft International, Palo Alto, CA, USA), 10 µl
2XTaq PCR MasterMix (containing 0.1 U/µl Taq polymerase, 500 µM
each deoxynucleotide, 20 mM Tris-HCl pH 8.3, 100 mM KCl, 3 mM MgCl2
and other stabilizers and enhancers; catalog no., KT203; Tiangen
Biotech Co., Ltd.) and 7 µl double-distilled (dd)H2O.
Table I indicates the primer sets
used for the amplification of the three aforementioned
polymorphisms. The reaction was performed on 2720 Thermal Cycler
(Applied Biosystems, Foster City, CA, USA) under the following
conditions: a denaturation step at 95°C for 5 min, followed by 30
cycles of 95°C for 30 sec, 30 sec at the corresponding annealing
temperature (as described in Table I)
and 30 sec at 72°C, and a final extension at 72°C for 10 min. All
PCR products were added to 2% agarose gel (catalog no., 5260;
Takara Biotechnology Co., Ltd., Dalian, China) which was stained
with 1 µl/10 ml DuRed (catalog no., 009–500; Fanbo Biochemicals Co.
Ltd., Beijing, China). DL1,000 DNA marker (catalog no., 3591Q;
Takara Biotechnology Co., Ltd.) was also added into the well as a
reference for the targeted DNA bands. The products and marker were
defined by agarose gel electrophoresis with the PowerPac™ 3000
system (Bio-Rad Laboratories, Inc., Hercules, CA, USA), and
subsequently visualized using the 2500R Gel Imaging System (Tanon
Science and Technology Co., Ltd., Shanghai, China).
 | Table I.PCR primers and conditions. |
Table I.
PCR primers and conditions.
| Polymorphism | PCR primer,
5′-3′ | Ta, °C | Amplicon size,
bp | Sequencing primer,
5′-3′ |
|---|
| rs1057451 |
|
|
|
|
| F inner
primer (G allele) |
ATTGATGAAGATCAGGGaTG | 54 | 288 (G allele) |
TTCCACTTGTCCTCCCTC |
| R inner
primer (T allele) |
CAGGAACCAGGCTTCAaA |
| 420 (T allele) |
|
| F outer
primer |
ATTGAGGGCACTTAACACTAC |
| 671 (outer
primers) |
|
| R outer
primer |
GACTCAGGAATTGCCAACA |
|
|
|
| rs4788186 |
|
|
|
|
| F inner
primer (G allele) |
TAAAGCATAGGAAAGAGtCG | 55 | 449 (G allele) | F outer primer |
| R inner
primer (A allele) |
TGAGCTGTGTCTATGTTCaCT |
| 211 (A allele) |
|
| F outer
primer |
ACCCTACCCTTGCTCACA |
| 620 (outer
primers) |
|
| R outer
primer |
AGCCCATCCTGACCTTAC |
|
|
|
| rs2288043 |
|
|
|
|
| F inner
primer (A allele) |
CATAGATGCCCTCGTTCgCA | 55 | 292 (A allele) | F outer primer |
| R inner
primer (G allele) |
GCCAGGCCATCCCTCTAGtC |
| 80 (G allele) |
|
| F outer
primer |
CTCACTCCCAGCCATTACCTTTC |
| 333 (outer
primers) |
|
| R outer
primer |
GGCACTGACCCTAACCTCACG |
|
|
|
Genotyping validation
To validate the accuracy of the results obtained by
tetra-primer ARMS-PCR analysis, a number of representative samples
of each genotype were selected, and conventional PCR was conducted
in a total volume of 20 µl, which contained 2 µl template DNA, 1 µl
each outer primer, 10 µl 2XTaq PCR MasterMix and 6 µl ddH2O. The
reaction was performed on a 2720 Thermal Cycler (Applied
Biosystems) with a denaturation step at 95°C for 5 min, 30 cycles
of 95°C for 30 sec, the corresponding annealing temperature
(described in Table I) for 30 sec,
and 72°C for 30 sec, followed by a final extension at 72°C for 10
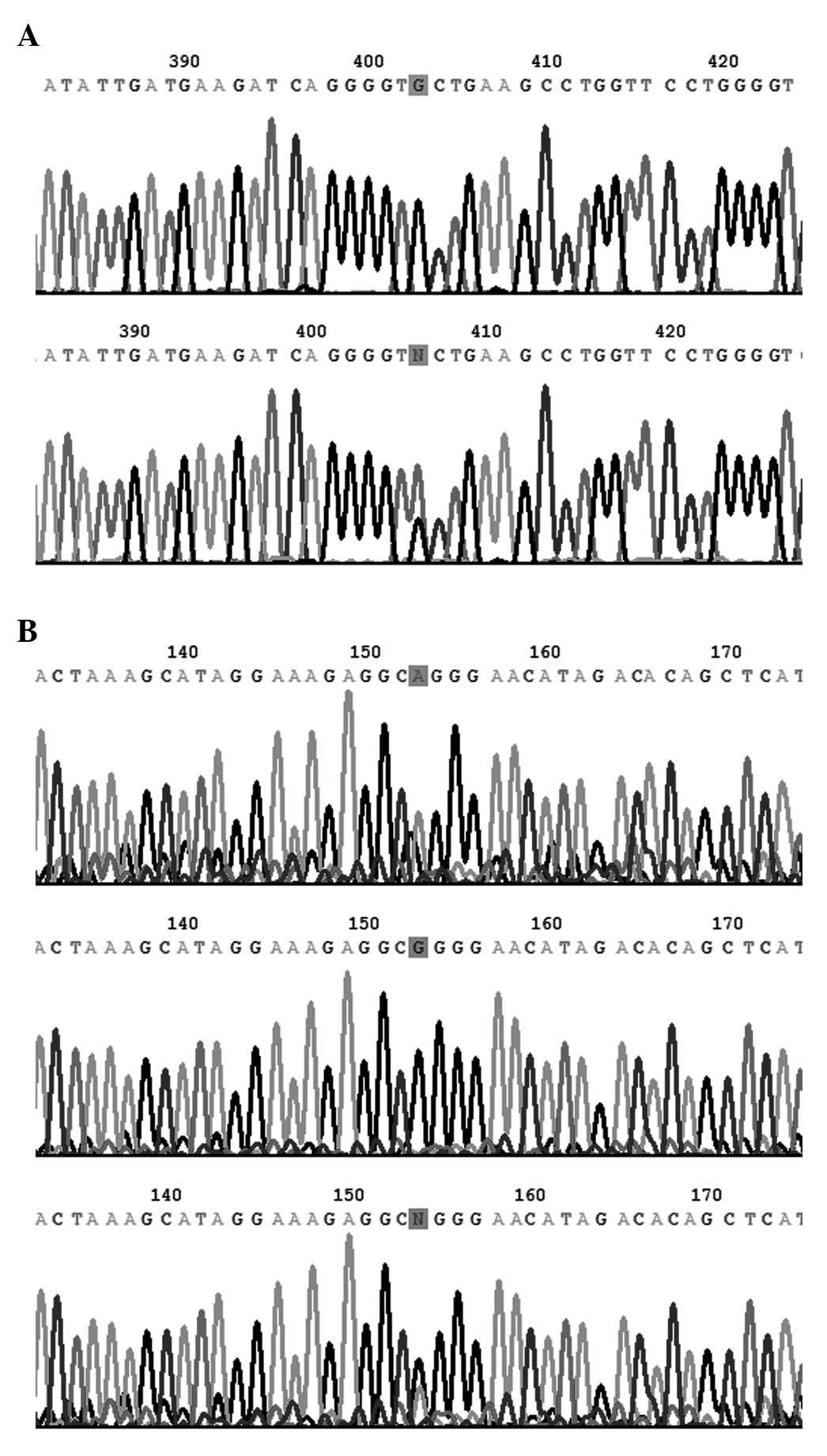
min. The PCR products were then sequenced by the Sanger method
[reagents included the following: BigDye® Direct Cycle Sequencing
kit; BigDye Terminator 5X Sequencing Buffer and Hi-Di Formamide
(all purchased from Invitrogen; Thermo Fisher Scientific, Inc.,
Waltham, MA, USA); the equipment used was a 3730xl DNA Analyzer
(Applied Biosystems)], using their respective forward outer primers
as sequencing primers, with the exception of rs1057451, whose
sequence differs from the others, since it contains two
poly-deoxyribonucleotide structures between the two outer primers.
Therefore, in order to avoid those structures, another reverse
primer was designed for DNA sequencing purposes (Table I). The reverse complement sequence is
presented in Fig. 2A.
Statistics
The association between each polymorphism and the
clinicopathological parameters of the patients was assessed by
Pearson's χ2 test or Fisher's exact test. The allele and genotype
distribution of the investigated SNPs in platinum-resistant and
platinum-responsive cohorts was compared using Pearson's χ2 test or
Fisher's exact test. The combined effect of the polymorphisms on
tumor response was investigated by haplotype analysis using the
SHEsis software platform (date of access, November 3, 2014), which
is available at http://analysis.bio-x.cn/myAnalysis.php (40). Univariate survival analysis was
determined using the Kaplan-Meier method, and survival curves were
compared by log-rank test. Multivariate survival analysis was
performed by the Cox proportional hazards regression model to
adjust for tumor stage, histological type and chemotherapy
response. Statistical analyses were performed using SPSS 13.0
software (SPSS, Inc., Chicago, IL, USA). All statistics were
two-sided, and P<0.05 was considered to indicate a statistically
significant difference.
Results
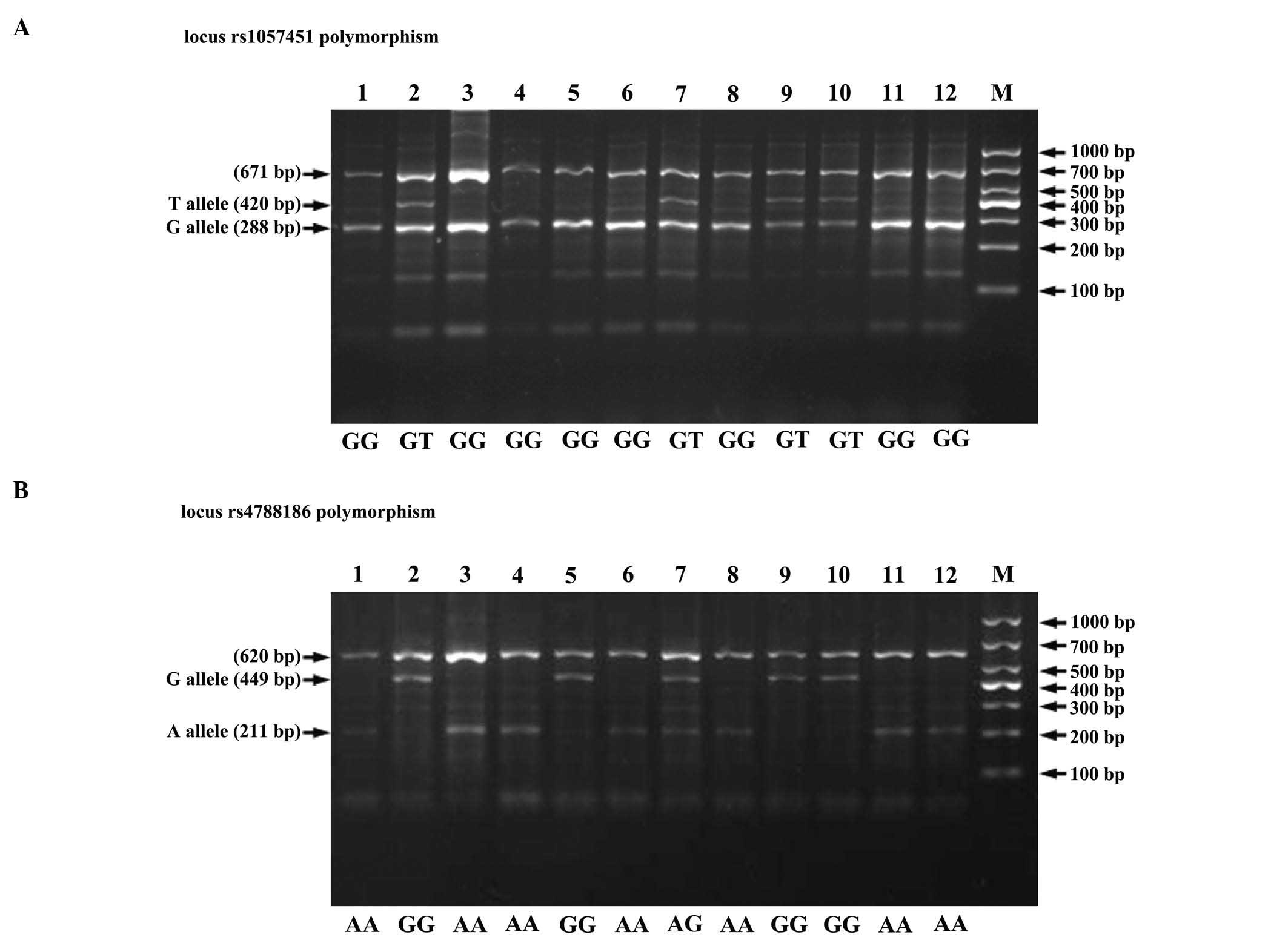
Genotyping by tetra-primer
ARMS-PCR
The tetra-primer ARMS-PCR method was successfully
applied to genotype the selected SNPs (Fig. 1). The results from DNA sequencing were
consistent with those from tetra-primer ARMS-PCR (Fig. 2).
Genotype frequency distribution of the
two polymorphisms and their association with clinicopathological
parameters
The clinicopathological parameters, including the
FIGO stage (41) of the patients are
summarized in Table II. Polymorphism
of locus rs2288043 was not analyzed, since the mutant allele for
this locus was absent in the patients selected for the present
study. By contrast, the other two polymorphisms, rs1057451 and
rs4788186, were evaluated. The genotype frequencies in the studied
patient population were as follows: MVP locus rs1057451, 87.9% GG
and 12.1% GT; and MVP locus rs4788186, 54.3% AA, 38.8% GA and 6.9%
GG. Neither of these two polymorphisms in the MVP gene was
associated with age or tumor stage, grade or histological type
(data not shown). The overall response rate of platinum-based
chemotherapy was 73.3%, with no significant difference in response
rate among all platinum-based regimens (Fisher's exact test,
P=0.696) (data not shown).
 | Table II.Clinicopathological parameters of 116
patients with epithelial ovarian cancer. |
Table II.
Clinicopathological parameters of 116
patients with epithelial ovarian cancer.
|
Characteristics | No. (%) |
|---|
| Age at diagnosis,
years |
|
|
≤50 | 53 (45.7) |
|
>50 | 63 (54.3) |
| FIGO stage |
|
| I | 8 (6.9) |
| II | 17 (14.7) |
|
III | 57 (49.1) |
| IV | 34 (29.3) |
| Tumor grade |
|
| G1 | 3 (2.6) |
| G2 | 29 (25.0) |
| G3 | 38 (32.8) |
|
Unknown | 46 (39.7) |
| Histological
type |
|
|
Serous | 83 (71.6) |
|
Mucinous | 4 (3.4) |
|
Endometrioid | 14 (12.1) |
| Clear
cell | 10 (8.6) |
|
Othera | 5 (4.3) |
| Chemotherapy
regimen |
|
|
TAX+DDP | 54 (46.6) |
|
TAX+CBP | 29 (25.0) |
|
TXT+DDP | 12 (10.3) |
|
TXT+CBP | 11 (9.5) |
| Other
platinum-based regimen | 10 (8.6) |
| Chemotherapy
response |
|
|
Resistance | 31 (26.7) |
|
Sensitivity | 85 (73.3) |
Association between the two
polymorphisms and platinum resistance
There was no significant difference in genotype and
allele distributions of the studied SNPs between platinum-resistant
and platinum-responsive patients (Table
III). Additionally, haplotype analysis did not reveal any
association between haplotypes and platinum resistance (Table IV).
 | Table III.Genotype and allele frequencies of
polymorphisms of the major vault protein gene in platinum-resistant
and platinum-responsive cohorts. |
Table III.
Genotype and allele frequencies of
polymorphisms of the major vault protein gene in platinum-resistant
and platinum-responsive cohorts.
| Polymorphism | Non-responder, no.
(%) | Responder, no.
(%) | P-value |
|---|
| rs1057451 |
|
|
|
| GG | 26 (83.9) | 76 (89.4) | 0.520 |
| GT | 5 (16.1) | 9 (10.6) | 0.520 |
| G | 57 (91.9) | 161 (94.7) | 0.533 |
| T | 5 (8.1) | 9 (5.3) | 0.533 |
| rs4788186 |
|
|
|
| AA | 15 (48.4) | 48 (56.5) | 0.606 |
| AG | 13 (41.9) | 32 (37.6) | 0.606 |
| GG | 3 (9.7) | 5 (5.9) | 0.606 |
|
AG+GG | 16 (51.6) | 37 (43.5) | 0.439 |
|
AA+AG | 28 (90.3) | 80 (94.1) | 0.439 |
| A | 43 (69.4) | 128 (75.3) | 0.363 |
| G | 19 (30.6) | 42 (24.7) | 0.363 |
 | Table IV.Major vault protein gene haplotypes
and patients' response to platinum chemotherapy. |
Table IV.
Major vault protein gene haplotypes
and patients' response to platinum chemotherapy.
|
Haplotypesa | Responder
(frequency) | Non-responder
(frequency) | OR (95% CI) | P-value |
|---|
| GA | 128 (0.753) | 43 (0.694) | 1.347
(0.708–2.561) | 0.363 |
| GG | 33 (0.194) | 14 (0.226) | 0.826
(0.408–1.674) | 0.595 |
| TA | 0
(0.000) | 0
(0.000) | – | – |
| TG | 9
(0.053) | 5
(0.081) | 0.637
(0.205–1.981) | 0.433 |
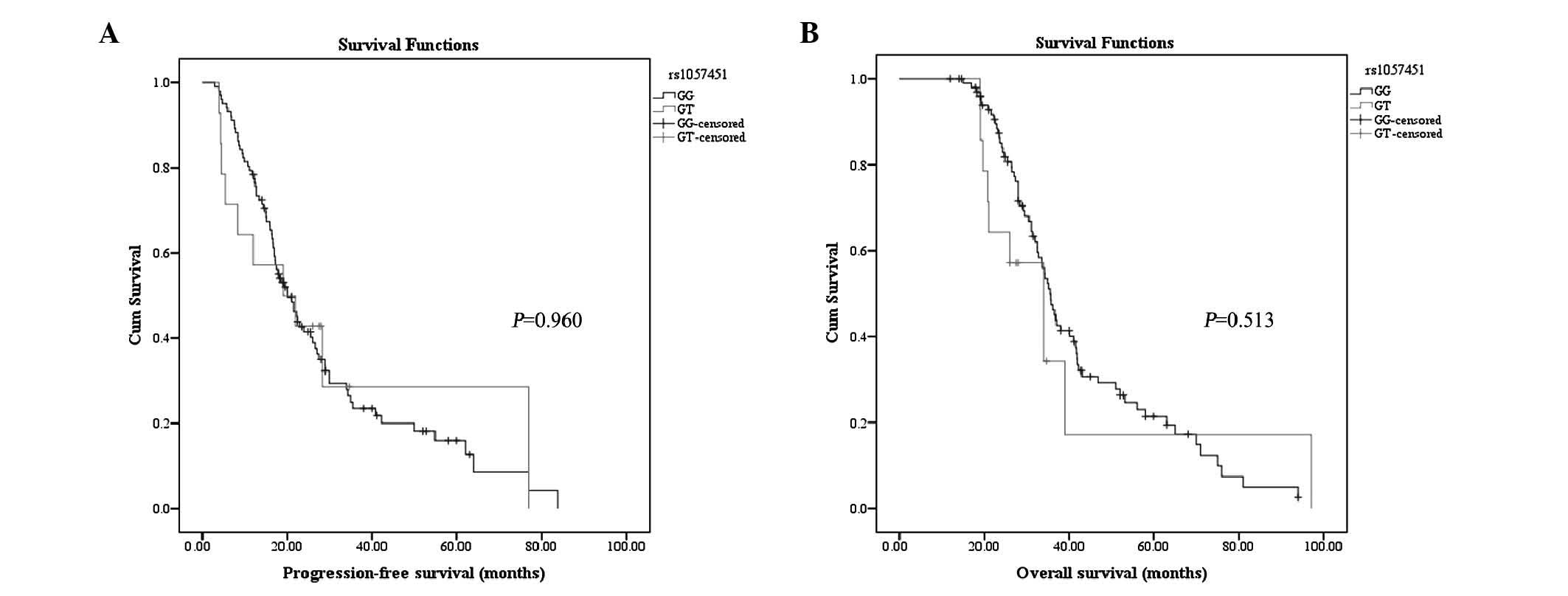
Association between rs1057451
polymorphism and survival
None of the patients was observed to be homozygous
for the minor TT allele. Univariate Kaplan-Meier analysis
demonstrated that PFS did not differ between patients carrying the
GG genotype and those carrying the GT genotype (log-rank test,
P=0.960; Fig. 3A). OS did not differ
either between the two genotypes (P=0.513, Fig. 3B). When adjusting for other potential
confounding variables in a multivariate Cox regression model, the
locus rs1057451 polymorphism had no significant predictive value
for PFS (P=0.102; Table V) or OS
(P=0.231; Table V).
 | Table V.Multivariate survival analysis by Cox
proportional hazards regression model. |
Table V.
Multivariate survival analysis by Cox
proportional hazards regression model.
|
|
| Progression-free
survival | Overall
survival |
|---|
|
|
|
|
|
|---|
| Polymorphism | Genotype | HR | 95% CI | P-value | HR | 95% CI | P-value |
|---|
| rs1057451 | GG | Reference | – | – | Reference | – | – |
|
| GT | 0.508 | 0.226–1.143 | 0.102 | 0.586 | 0.245–1.405 | 0.231 |
| rs4788186 | AA | Reference | – | – | Reference | – | – |
|
| AG | 0.600 | 0.358–1.007 | 0.053 | 0.803 | 0.482–1.337 | 0.399 |
|
| GG | 0.936 | 0.386–2.270 | 0.883 | 1.037 | 0.427–2.516 | 0.937 |
|
| AA | Reference | – | – | Reference | – | – |
|
| AG+GG | 0.650 | 0.402–1.051 | 0.079 | 0.839 | 0.519–1.356 | 0.474 |
|
| GG | Reference | – | – | Reference | – | – |
|
| AA+AG | 0.888 | 0.370–2.129 | 0.790 | 0.882 | 0.371–2.093 | 0.775 |
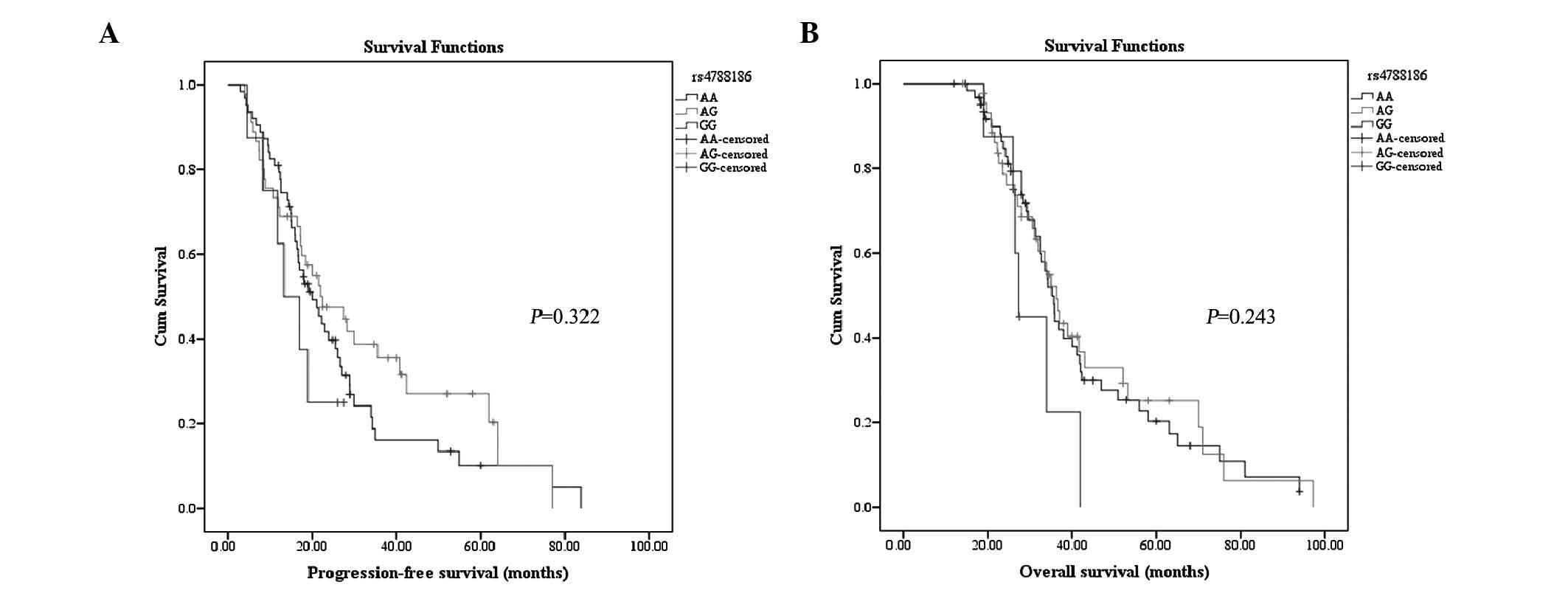
Association between rs4788186
polymorphism and survival
Kaplan-Meier analysis of the rs4788186 polymorphism
did not reveal a significant difference in PFS between genotypes
AA, AG and GG (P=0.322; Fig. 4A).
Similar trends were observed in OS for these genotypes at this
locus (P=0.243; Fig. 4B). When the AG
and GG subgroups were combined, the log-rank test failed to detect
any significant difference in PFS (log-rank test, P=0.453) or OS
(log-rank test, P=0.905). There was no statistically significant
difference in PFS (log-rank test, P=0.278) or OS (log-rank test,
P=0.097) between patients carrying the GG genotype and those
carrying a non-GG genotype (data not shown). Multivariate Cox
regression analysis revealed that rs4788186 variants were neither
associated with PFS nor with OS, once adjusted by International
Federation of Gynecology and Obstetrics stage, histological type
and chemotherapeutic response (Table
V), although a trend toward reduced risk of progression was
observed for patients with the AG genotype, compared with those
exhibiting the AA genotype (hazard ratio, 0.600; 95% confidence
interval, 0.358–1.007; P=0.053; Table
V). The multivariate survival analysis performed for genotypes
AG and GG, compared with the major AA genotype, did not reveal any
statistically significant difference in PFS or OS (Table V). Comparison of PFS and OS between
patients with the GG genotype and those with a non-GG genotype did
not reveal any significant difference (Table V).
Discussion
MVP is considered to be important in the treatment
response and prognosis of various tumors (22,23,42). The
majority of clinical studies published to date have evaluated the
mRNA or protein levels of MVP in order to investigate the
association of MVP with platinum resistance and survival in EOC
(22,43). To the best of our knowledge, there are
no previous studies on the association between genetic variants of
the MVP gene and platinum resistance in patients with EOC. To
investigate the role of MVP polymorphisms in predicting platinum
response and survival, three SNPs from the genotype data derived
from the phase II HapMap Project of Han Chinese population were
selected in the present study. The results indicated that the
mutant allele for locus rs2288043 was not present in any of the
patients enrolled in the study, and neither platinum resistance nor
survival were associated with the other two polymorphisms
(rs1057451 and rs4788186).
These findings may be explained by the following
factors: Firstly, it is important to remember that MDR is certainly
involved in various mechanisms (44).
Thus, the complex mechanisms involved in MDR may represent one of
the obstacles in predicting the treatment response and survival by
merely several genetic polymorphisms. Secondly, although MVP has
been previously implicated in drug resistance, several studies have
reported conflicting results (45,46).
Certain studies have demonstrated that MVP has no influence on
intracellular drug distribution or chemoresistance (47–50).
Similarly, Siva et al (51)
demonstrated that the upregulation of MVP is not sufficient to
confer an MDR phenotype. In addition, recent studies have
correlated MVP with several signaling pathways (52–56) and
immune responses (57–60), which imply that the function of MVP
may be more complex than expected. Thirdly, the inability to find
an association between variants of the MVP gene and platinum
resistance or survival in the present study may be due to the
limited number of patients enrolled in the study, since the number
of patients in the subgroup was below the statistical threshold.
Therefore, to demonstrate the independent effect of the
aforementioned polymorphisms on the chemotherapy response and
prognosis of patients with EOC, a large and homogeneous cohort of
patients, such as advanced stage cases following optimal
cytoreductive surgery, may be required. Finally, the absence of
correlations observed in the present study may be due to the SNPs
selected, which may not tag the SNPs responsible for the
upregulation of the protein levels of MVP.
The tetra-primer ARMS-PCR method applied in the
present study is more cost- and time-effective than other commonly
used genotyping methods such as PCR-restriction fragment length
polymorphism and TaqMan assays (61,62). In
addition, the tetra-primer ARMS-PCR method has also been
demonstrated to possess a high reliability in genotyping (38,61,62), thus
it may be used for detecting polymorphisms. The limitations of the
present study were the absence of subgroups analysis due to the
small number of patients participating in the study, and the
absence of toxicity analysis.
In conclusion, no associations between the two
polymorphisms in the MVP gene analyzed in the present study and
platinum-resistance or survival were observed in the patients with
EOC who were recruited for the study. Furthermore, the present
study has demonstrated that tetra-primer ARMS-PCR is a reliable
method for genotyping.
Acknowledgements
The present study was supported by funds from the
Natural Science Foundation of Liaoning Province (Shenyang, China;
grant no. 2014022011) and the Medical Peak Construction Project of
Liaoning Province (Shenyang, China).
References
|
1
|
Jemal A, Bray F, Center MM, Ferlay J, Ward
E and Forman D: Global cancer statistics. CA Cancer J Clin.
61:69–90. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Omura G, Blessing JA, Ehrlich CE, Miller
A, Yordan E, Creasman WT and Homesley HD: A randomized trial of
cyclophosphamide and doxorubicin with or without cisplatin in
advanced ovarian carcinoma. A Gynecologic Oncology Group Study.
Cancer. 57:1725–1730. 1986. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Omura GA, Bundy BN, Berek JS, Curry S,
Delgado G and Mortel R: Randomized trial of cyclophosphamide plus
cisplatin with or without doxorubicin in ovarian carcinoma: A
Gynecologic Oncology Group Study. J Clin Oncol. 7:457–465.
1989.PubMed/NCBI
|
|
4
|
Aabo K, Adams M, Adnitt P, Alberts DS,
Athanazziou A, Barley V, Bell DR, Bianchi U, Bolis G, Brady MF, et
al: Chemotherapy in advanced ovarian cancer: Four systematic
meta-analyses of individual patient data from 37 randomized trials.
Advanced Ovarian Cancer Trialists' Group. Br J Cancer.
78:1479–1487. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Trimbos JB, Parmar M, Vergote I, Guthrie
D, Bolis G, Colombo N, Vermorken JB, Torri V, Mangioni C, Pecorelli
S, et al: European Organisation for Research and Treatment of
Cancer Collaborators - Adjuvant ChemoTherapy In Ovarian Neoplasm:
International Collaborative Ovarian Neoplasm trial 1 and Adjuvant
ChemoTherapy In Ovarian Neoplasm trial: Two parallel randomized
phase III trials of adjuvant chemotherapy in patients with
early-stage ovarian carcinoma. J Natl Cancer Inst. 95:105–112.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Bookman MA: Developmental chemotherapy and
management of recurrent ovarian cancer. J Clin Oncol. 21(Suppl 10):
149s–167s. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Modesitt SC and Jazaeri AA: Recurrent
epithelial ovarian cancer: Pharmacotherapy and novel therapeutics.
Expert Opin Pharmacother. 8:2293–2305. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Biedler JL and Riehm H: Cellular
resistance to actinomycin D in Chinese hamster cells in vitro:
Cross-resistance, radioautographic, and cytogenetic studies. Cancer
Res. 30:1174–1184. 1970.PubMed/NCBI
|
|
9
|
Fojo A, Hamilton TC, Young RC and Ozols
RF: Multidrug resistance in ovarian cancer. Cancer. 60(Suppl):
2075–2080. 1987. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Scheffer GL, Schroeijers AB, Izquierdo MA,
Wiemer EA and Scheper RJ: Lung resistance-related protein/major
vault protein and vaults in multidrug-resistant cancer. Curr Opin
Oncol. 12:550–556. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Teodori E, Dei S, Martelli C, Scapecchi S
and Gualtieri F: The functions and structure of ABC transporters:
Implications for the design of new inhibitors of Pgp and MRP1 to
control multidrug resistance (MDR). Curr Drug Targets. 7:893–909.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Roy S, Kenny E, Kennedy S, Larkin A,
Ballot J, De Perez Villarreal M, Crown J and O'Driscoll L:
MDR1/P-glycoprotein and MRP-1 mRNA and protein expression in
non-small cell lung cancer. Anticancer Res. 27:1325–1330.
2007.PubMed/NCBI
|
|
13
|
Ahmad A, Robinson AR, Duensing A, van
Drunen E, Beverloo HB, Weisberg DB, Hasty P, Hoeijmakers JH and
Niedernhofer LJ: ERCC1-XPF endonuclease facilitates DNA
double-strand break repair. Mol Cell Biol. 28:5082–5092. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Arora S, Kothandapani A, Tillison K,
Kalman-Maltese V and Patrick SM: Downregulation of XPF-ERCC1
enhances cisplatin efficacy in cancer cells. DNA Repair. 9:745–753.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Kim R, Tanabe K, Uchida Y, Emi M, Inoue H
and Toge T: Current status of the molecular mechanisms of
anticancer drug-induced apoptosis. The contribution of
molecular-level analysis to cancer chemotherapy. Cancer Chemother
Pharmacol. 50:343–352. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Johnstone RW, Cretney E and Smyth MJ:
P-glycoprotein protects leukemia cells against caspase-dependent,
but not caspase-independent, cell death. Blood. 93:1075–1085.
1999.PubMed/NCBI
|
|
17
|
Maugeri-Saccà M, Vigneri P and De Maria R:
Cancer stem cells and chemosensitivity. Clin Cancer Res.
17:4942–4947. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Castells M, Thibault B, Delord JP and
Couderc B: Implication of tumor microenvironment in
chemoresistance: Tumor-associated stromal cells protect tumor cells
from cell death. Int J Mol Sci. 13:9545–9571. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Hanahan D and Weinberg RA: Hallmarks of
cancer: The next generation. Cell. 144:646–674. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Hanahan D and Coussens LM: Accessories to
the crime: Functions of cells recruited to the tumor
microenvironment. Cancer Cell. 21:309–322. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Scheffer GL, Wijngaard PL, Flens MJ,
Izquierdo MA, Slovak ML, Pinedo HM, Meijer CJ, Clevers HC and
Scheper RJ: The drug resistance-related protein LRP is the human
major vault protein. Nat Med. 1:578–582. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Kerr EH, Frederick PJ, Egger ME, Stockard
CR, Sellers J, DellaManna D, Oelschlager DK, Amm HM, Eltoum IE,
Straughn JM, et al: Lung resistance-related protein (LRP)
expression in malignant ascitic cells as a prognostic marker for
advanced ovarian serous carcinoma. Ann Surg Oncol. 20:3059–3065.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Chen ZJ, Le HB, Zhang YK, Qian LY, Sekhar
KR and Li WD: Lung resistance protein and multidrug resistance
protein in non-small cell lung cancer and their clinical
significance. J Int Med Res. 39:1693–1700. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Han M, Lv Q, Tang XJ, Hu YL, Xu DH, Li FZ,
Liang WQ and Gao JQ: Overcoming drug resistance of MCF-7/ADR cells
by altering intracellular distribution of doxorubicin via MVP
knockdown with a novel siRNA polyamidoamine-hyaluronic acid
complex. J Control Release. 163:136–144. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Kedersha NL, Hill DF, Kronquist KE and
Rome LH: Subpopulations of liver coated vesicles resolved by
preparative agarose gel electrophoresis. J Cell Biol. 103:287–297.
1986. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Kickhoefer VA, Searles RP, Kedersha NL,
Garber ME, Johnson DL and Rome LH: Vault ribonucleoprotein
particles from rat and bullfrog contain a related small RNA that is
transcribed by RNA polymerase III. J Biol Chem. 268:7868–7873.
1993.PubMed/NCBI
|
|
27
|
Kickhoefer VA, Siva AC, Kedersha NL, Inman
EM, Ruland C, Streuli M and Rome LH: The 193-kD vault protein,
VPARP, is a novel poly(ADP-ribose) polymerase. J Cell Biol.
146:917–928. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Kickhoefer VA, Stephen AG, Harrington L,
Robinson MO and Rome LH: Vaults and telomerase share a common
subunit, TEP1. J Biol Chem. 274:32712–32717. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Izquierdo MA, Scheffer GL, Flens MJ,
Giaccone G, Broxterman HJ, Meijer CJ, van der Valk P and Scheper
RJ: Broad distribution of the multidrug resistance-related vault
lung resistance protein in normal human tissues and tumors. Am J
Pathol. 148:877–887. 1996.PubMed/NCBI
|
|
30
|
List AF, Spier CS, Grogan TM, Johnson C,
Roe DJ, Greer JP, Wolff SN, Broxterman HJ, Scheffer GL, Scheper RJ
and Dalton WS: Overexpression of the major vault transporter
protein lung-resistance protein predicts treatment outcome in acute
myeloid leukemia. Blood. 87:2464–2469. 1996.PubMed/NCBI
|
|
31
|
Henríquez-Hernández LA, Moreno M, Rey A,
Lloret M and Lara PC: MVP expression in the prediction of clinical
outcome of locally advanced oral squamous cell carcinoma patients
treated with radiotherapy. Radiat Oncol. 7:1472012. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Li J, Li ZN, Yu LC, Bao QL, Wu JR, Shi SB
and Li XQ: Association of expression of MRP1, BCRP, LRP and ERCC1
with outcome of patients with locally advanced non-small cell lung
cancer who received neoadjuvant chemotherapy. Lung Cancer.
69:116–122. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Lu S, Pardini B, Cheng B, Naccarati A,
Huhn S, Vymetalkova V, Vodickova L, Buchler T, Hemminki K, Vodicka
P and Försti A: Single nucleotide polymorphisms within interferon
signaling pathway genes are associated with colorectal cancer
susceptibility and survival. PLoS One. 9:e1110612014. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Bosó V, Herrero MJ, Santaballa A, Palomar
L, Megias JE, de la Cueva H, Rojas L, Marqués MR, Poveda JL,
Montalar J and Aliño SF: SNPs and taxane toxicity in breast cancer
patients. Pharmacogenomics. 15:1845–1858. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Du Y, Su T, Zhao L, Tan X, Chang W, Zhang
H and Cao G: Associations of polymorphisms in DNA repair genes and
MDR1 gene with chemotherapy response and survival of non-small cell
lung cancer. PLoS One. 9:e998432014. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Mendoza J, Martínez J, Hernández C,
Pérez-Montiel D, Castro C, Fabián-Morales E, Santibáñez M,
González-Barrios R, Díaz-Chávez J, Andonegui MA, et al: Association
between ERCC1 and XPA expression and polymorphisms and the response
to cisplatin in testicular germ cell tumours. Br J Cancer.
109:68–75. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Cannistra SA, Matulonis UA, Penson RT,
Hambleton J, Dupont J, Mackey H, Douglas J, Burger RA, Armstrong D,
Wenham R and McGuire W: Phase II study of bevacizumab in patients
with platinum-resistant ovarian cancer or peritoneal serous cancer.
J Clin Oncol. 25:5180–5186. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Ye S, Dhillon S, Ke X, Collins AR and Day
IN: An efficient procedure for genotyping single nucleotide
polymorphisms. Nucleic Acids Res. 29:E88–E89. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Little S: ARMS analysis of point
mutations. Laboratory Methods for the Detection of Mutations and
Polymorphisms in DNA. Taylor GR: CRC Press. 45–51. 1997.
|
|
40
|
Shi YY and He L: SHEsis, a powerful
software platform for analyses of linkage disequilibrium, haplotype
construction and genetic association at polymorphism loci. Cell
Res. 15:97–98. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Mutch DG and Prat J: 2014 FIGO staging for
ovarian, fallopian tube and peritoneal cancer. Gynecol Oncol.
133:401–404. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Filipits M, Pohl G, Stranzl T, Suchomel
RW, Scheper RJ, Jäger U, Geissler K, Lechner K and Pirker R:
Expression of the lung resistance protein predicts poor outcome in
de novo acute myeloid leukemia. Blood. 91:1508–1513.
1998.PubMed/NCBI
|
|
43
|
Szaflarski W, Sujka-Kordowska P, Pula B,
Jaszczyńska-Nowinka K, Andrzejewska M, Zawierucha P, Dziegiel P,
Nowicki M, Ivanov P and Zabel M: Expression profiles of vault
components MVP, TEP1 and vPARP and their correlation to other
multidrug resistance proteins in ovarian cancer. Int J Oncol.
43:513–520. 2013.PubMed/NCBI
|
|
44
|
Baguley BC: Multiple drug resistance
mechanisms in cancer. Mol Biotechnol. 46:308–316. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Goff BA, Paley PJ, Greer BE and Gown AM:
Evaluation of chemoresistance markers in women with epithelial
ovarian carcinoma. Gynecol Oncol. 81:18–24. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Sedláková I, Laco J, Caltová K, Červinka
M, Tošner J, Řezáč A and Špaček J: Clinical significance of the
resistance proteins LRP, Pgp, MRP1, MRP3, and MRP5 in
epithelialovarian cancer. Int J Gynecol Cancer. 25:236–243. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Mossink MH, van Zon A, Fränzel-Luiten E,
Schoester M, Kickhoefer VA, Scheffer GL, Scheper RJ, Sonneveld P
and Wiemer EA: Disruption of the murine major vault protein
(MVP/LRP) gene does not induce hypersensitivity to cytostatics.
Cancer Res. 62:7298–7304. 2002.PubMed/NCBI
|
|
48
|
van Zon A, Mossink MH, Schoester M,
Scheper RJ, Sonneveld P and Wiemer EAC: Efflux kinetics and
intracellular distribution of daunorubicin are not affected by
major vault protein/lung resistance-related protein (vault)
expression. Cancer Res. 64:4887–4892. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Huffman KE and Corey DR: Major vault
protein does not play a role in chemoresistance or drug
localization in a non-small cell lung cancer cell line.
Biochemistry. 44:2253–2261. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Balan S, Radhab SK, Sathyan S, Vijai J,
Banerjee M and Radhakrishnan K: Major vault protein (MVP) gene
polymorphisms and drug resistance in mesial temporal lobe epilepsy
with hippocampal sclerosis. Gene. 526:449–453. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Siva AC, Raval-Fernandes S, Stephen AG,
LaFemina MJ, Scheper RJ, Kickhoefer VA and Rome LH: Up-regulation
of vaults may be necessary but not sufficient for multidrug
resistance. Int J Cancer. 92:195–202. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Yu Z, Fotouhi-Ardakani N, Wu L, Maoui M,
Wang S, Banville D and Shen SH: PTEN associates with the vault
particles in HeLa cells. J Biol Chem. 277:40247–40252. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Steiner E, Holzmann K, Pirker C, Elbling
L, Micksche M, Sutterlüty H and Berger W: The major vault protein
is responsive to and interferes with interferon-gamma-mediated
STAT1 signals. J Cell Sci. 119:459–469. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Losert A, Lötsch D, Lackner A,
Koppensteiner H, Peter-Vörösmarty B, Steiner E, Holzmann K, Grunt
T, Schmid K, Marian B, et al: The major vault protein mediates
resistance to epidermal growth factor receptor inhibition in human
hepatoma cells. Cancer Lett. 319:164–172. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Lötsch D, Steiner E, Holzmann K,
Spiegl-Kreinecker S, Pirker C, Hlavaty J, Petznek H, Hegedus B,
Garay T, Mohr T, et al: Major vault protein supports glioblastoma
survival and migration by upregulating the EGFR/PI3K signalling
axis. Oncotarget. 4:1904–1918. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Tomiyasu H, Watanabe M, Goto-Koshino Y,
Fujino Y, Ohno K, Sugano S and Tsujimoto H: Regulation of
expression of ABCB1 and LRP genes by mitogen-activated protein
kinase/extracellular signal-regulated kinase pathway and its role
in generation of side population cells in canine lymphoma cell
lines. Leuk Lymphoma. 54:1309–1315. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Rivera-Rivera L, Perez-Laspiur J, Colón K
and Meléndez LM: Inhibition of interferon response by cystatin B:
Implication in HIV replication of macrophage reservoirs. J
Neurovirol. 18:20–29. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Liu S, Peng N, Xie J, Hao Q, Zhang M,
Zhang Y, Xia Z, Xu G, Zhao F, Wang Q, et al: Human hepatitis B
virus surface and e antigens inhibit major vault protein signaling
in interferon induction pathways. J Hepatol. 62:1015–1023. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Liu S, Hao Q, Peng N, Yue X, Wang Y, Chen
Y, Wu J and Zhu Y: Major vault protein: A virus-induced host factor
against viral replication through the induction of type-I
interferon. Hepatology. 56:57–66. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Dortet L, Mostowy S, Samba-Louaka A, Gouin
E, Nahori MA, Wiemer EA, Dussurget O and Cossart P: Recruitment of
the major vault protein by InlK: A Listeria monocytogenes strategy
to avoid autophagy. PLoS Pathog. 7:e10021682011. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Etlik O, Koksal V, Arican-Baris ST and
Baris I: Development and validation of a cost-effective in-house
method, tetra-primer ARMS PCR assay, in genotyping of seven
clinically important point mutations. Mol Cell Probes. 25:177–181.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Miranzadeh-Mahabadi H, Miranzadeh-Mahabadi
H, Nikpour P, Emadi-Baygi M and Kelishadi R: Comparison of TaqMan
real-time and tetra-primer ARMS PCR techniques for genotyping of Rs
8066560 variant in children and adolescents with metabolic
syndrome. Adv Clin Exp Med. 24:951–955. 2015. View Article : Google Scholar : PubMed/NCBI
|