Introduction
Acute myeloid leukemia (AML) is a highly
heterogeneous hematologic malignancy, which is characterized by the
rapid growth of abnormal white blood cells that accumulate in the
peripheral blood, bone marrow and other tissues (1,2). AML is
the most common form of acute leukemia in adults (3), and is treated initially with
chemotherapy with the aim of inducing a remission (4). Demethylation agents such as azacitidine
and decitabine have been previously used in the treatment of AML
(5). Aberrant CpG island
hypermethylation of tumor suppressor genes has been shown to be
important in inducing transcriptional silencing of genes in
leukemic cells (6). Therefore, the
promoter methylation status of cancer-associated genes may serve as
a predictive and prognostic biomarker in the pathogenesis of
various types of cancer (7).
The O(6)-methylguanine-DNA methyltransferase
(MGMT) gene encodes a unique DNA repair enzyme that removes
mutagenic and cytotoxic alkyl adducts from the O(6) position of guanine (8). Hypermethylation of the MGMT
promoter was identified in certain tumor cell lines such as
non-Hodgkin lymphoma and AML (9).
Targeted therapy was feasible in patients with AML who displayed
low expression levels of MGMT (10). However, the frequency of MGMT
promoter methylation in AML, and its potential prognostic value
remain to be elucidated.
The aim of the present study was to evaluate whether
the promoter methylation status of the MGMT gene would be
influenced by chemotherapeutic agents, and whether the methylation
changes induced by the treatment would be able to predict the
chemotherapeutic outcomes of patients with AML.
Materials and methods
Samples
Bone marrow samples from 30 individuals with AML
were collected by the members of the Department of Hematology of
Yuyao People's Hospital (Ningbo, China) between January 2013 and
June 2014. AML diagnosis was established in accordance with the
revised French-American-British (FAB) classification (11). There were 13 male and 17 female AML
patients, with a mean age of 47.8±15.4 years (range, 19–76 years).
The present study was approved by the Ethics Committee of Yuyao
People's Hospital, and all the donors signed the informed consent
form for participation in the study.
DNA isolation and bisulfite
conversion
Genomic DNA was extracted using a nucleic acid
extraction automatic analyzer (Lab-Aid 820; Zeesan Biotech, Xiamen,
China). DNA concentration was measured with NanoDrop 1000
spectrophotometer (Thermo Fisher Scientific, Inc., Wilmington, DE,
USA). DNA samples were subjected to bisulfite conversion by EZ DNA
Methylation-Gold™ kit (Zymo Research Corporation, Irvine, CA, USA),
according to the manufacturer's protocol.
Methylation-specific polymerase chain
reaction (MSP) and DNA sequencing
The methylation status of the MGMT promoter was
determined by MSP (12). The reaction
contained 1.5 µl sodium bisulfite-converted DNA, 0.5 µl forward
primer, 0.5 µl reverse primer (13),
10 µl ZymoTaq™ PreMix (Zymo Research Corporation) and 7.5 µl
DNAase/RNAase-free water (Zymo Research Corporation), in a final
volume of 20 µl. The details of the methylated and unmethylated
primers used are provided in Table I.
DNA amplification was performed on Veriti® PCR machine (Applied
Biosystems, Thermo Fisher Scientific, Inc.), and the following
amplification conditions were used: 95°C for 10 min, followed by 30
or 35 cycles at 94°C for 30 sec, 55°C for 45 sec and 72°C for 1
min. PCR products were subjected to the Qsep100 DNA Analyzer
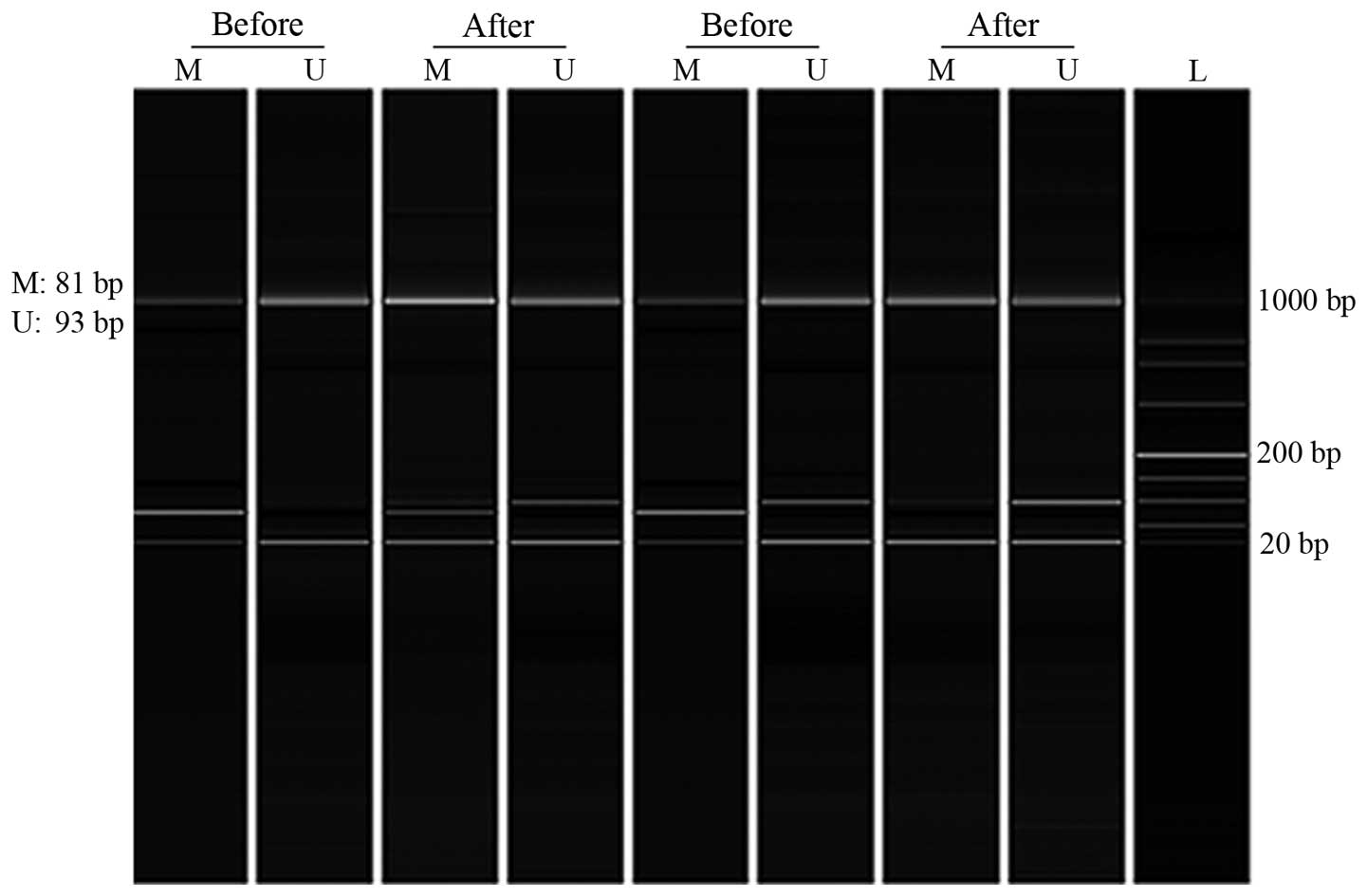
(BiOptic Inc., Taiwan, China). Samples were considered as
methylated or unmethylated when peaks were clearly visible by
Q-Analyzer software (BiOptic Inc.) (Fig.
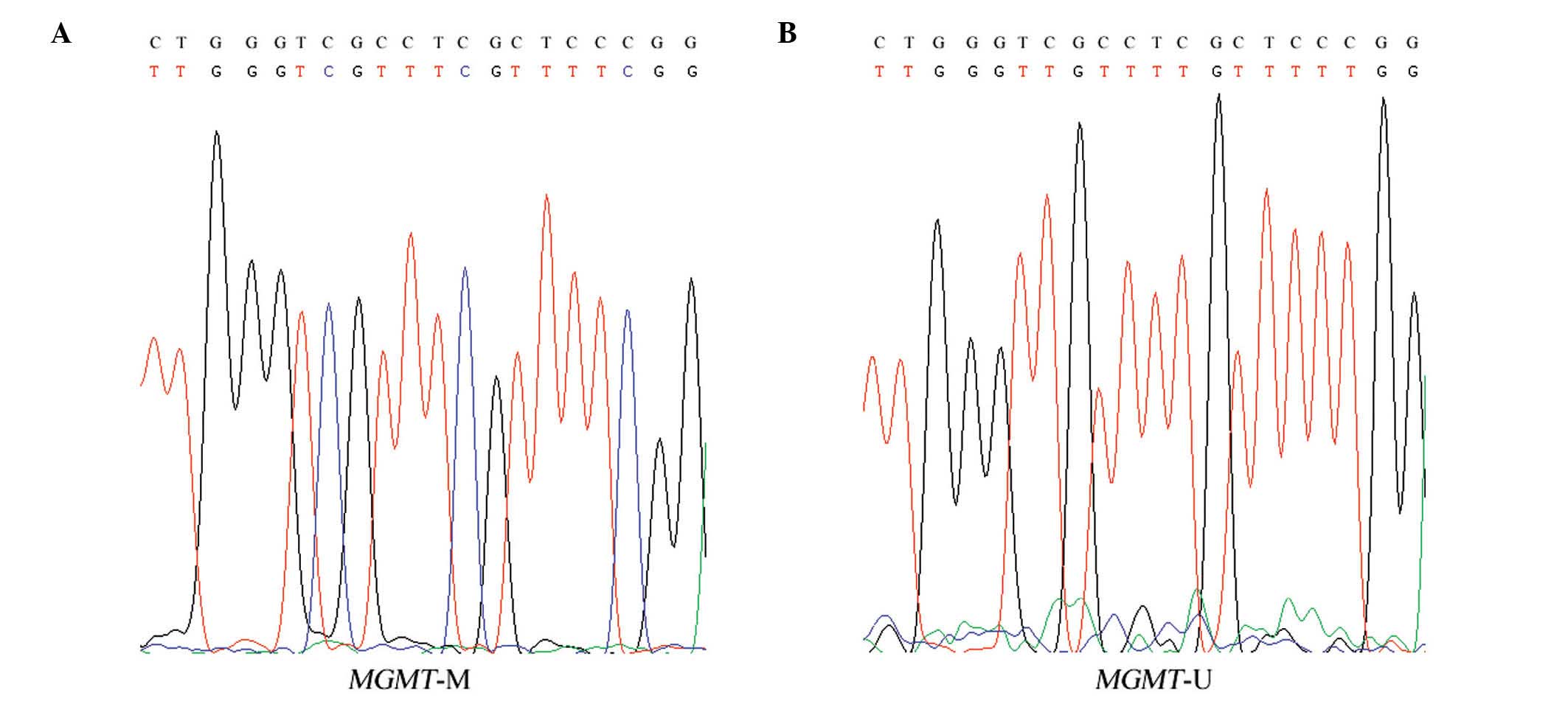
1). A number of DNA samples were randomly sequenced using the
3730 DNA Analyzer (Applied Biosystems; Thermo Fisher Scientific,
Inc.) to confirm a complete bisulfite conversion (Fig. 2).
 | Table I.List of all primers and conditions
used in polymerase chain reaction amplification. |
Table I.
List of all primers and conditions
used in polymerase chain reaction amplification.
| Primer set | Primer sequence,
5′-3′ | Target size, bp | Ta, °C | Cycles, no. |
|---|
| Methylated |
| 81 | 55 | 35 |
| F |
TTTCGACGTTCGTAGGTTTTCGC |
|
|
|
| R |
GCACTCTTCCGAAAACGAAACG |
|
|
|
| Unmethylated |
| 93 | 55 | 30 |
| F |
TTTGTGTTTTGATGTTTGTAGGTTTTTGT |
|
|
|
| R |
AACTCCACACTCTTCCAAAAACAAAACA |
|
|
|
Statistical analysis
Statistical analyses were performed using SPSS
version 18.0 (SPSS, Inc., Chicago, IL, USA). MSP results were
compared between prior and post-chemotherapy. A two-tailed P-value
of <0.05 was considered to indicate a statistically significant
difference.
Results
MSP analysis of bone marrow samples
from AML patients
To determine whether the chemotherapy treatment
would change the methylation status of the MGMT promoter in
patients with AML, the pre- and post-chemotherapy bone marrow
samples of 30 patients with AML were subjected to MSP analysis. The
chemotherapy agents used to treat the patients, including
cytarabine (Ara-C), idarubicin (IDA), arsenic trioxide
(As2O3), all-trans retinoic acid (ATRA),
homoharringtonine (HHT), granulocyte-colony stimulating factor
(G-CSF), aclacinomycin (ACLA) and daunorubicin (DNR), are described
in Table II. The details of the
chemotherapy regimens of all patients are presented in Table III.
 | Table II.Clinical parameters of patients with
acute myeloid leukemia. |
Table II.
Clinical parameters of patients with
acute myeloid leukemia.
| Case | Gender | Age, years | Subtype | Chemotherapy
regimen | Remission | Methylation
level |
|---|
| 1 | Male | 55 | M1 | HHT+Ara-C+ACLA | Yes | M/U to M/U |
| 2 | Male | 49 | M1 | IDA+Ara-C | No | M/U to M |
| 3 | Male | 76 | M2 | Ara-C+ACLA+G-CSF | No | M/U to M/U |
| 4 | Male | 66 | M2 | IDA+Ara-C | Yes | M/U to M/U |
| 5 | Male | 23 | M3 |
ATRA+As2O3 | Yes | M to M/U |
| 6 | Male | 40 | M3 |
As2O3+DNR+ATRA | No | M to M |
| 7 | Male | 59 | M3 | HHT+Ara-C | Yes | M to M |
| 8 | Male | 67 | M4 |
IDA+Ara-C+ACLA+G-CSF+HHT | Yes | M/U to U |
| 9 | Male | 34 | M4 | HHT+Ara-C | Yes | M/U to M/U |
| 10 | Male | 68 | M5 | Ara-C | Yes | M/U to M/U |
| 11 | Male | 59 | M5 | Ara-C+ACLA | Yes | M/U to M/U |
| 12 | Male | 48 | M5 | HHT+Ara-C+ACLA | No | M/U to M/U |
| 13 | Male | 52 | M6 |
HHT+Ara-C+G-CSF | Yes | M to M/U |
| 14 | Female | 59 | M1 |
Ara-C+ACLA+G-CSF | No | M/U to M/U |
| 15 | Female | 66 | M2 |
Ara-C+ACLA+G-CSF | Yes | M/U to M/U |
| 16 | Female | 56 | M2 |
Ara-C+ACLA+G-CSF | Yes | M/U to M/U |
| 17 | Female | 48 | M2 | HHT+Ara-C+ACLA | Yes | M/U to M/U |
| 18 | Female | 50 | M2 |
HHT+Ara-C+G-CSF+IDA | Yes | M/U to M/U |
| 19 | Female | 19 | M2 | HHT+Ara-C+ACLA | Yes | M/U to M/U |
| 20 | Female | 53 | M2 | HHT+Ara-C | Yes | M/U to M/U |
| 21 | Female | 51 | M3 |
ATRA+As2O3+HHT+Ara-C | Yes | M to M/U |
| 22 | Female | 42 | M3 | IDA+Ara-C | No | M to M/U |
| 23 | Female | 30 | M3 | Ara-C | Yes | M/U to M/U |
| 24 | Female | 31 | M3 | Ara-C | Yes | M/U to M/U |
| 25 | Female | 30 | M4 | IDA | Yes | M/U to M/U |
| 26 | Female | 30 | M4 | IDA+Ara-C | No | M to M/U |
| 27 | Female | 19 | M4 | HHT+Ara-C+ACLA | Yes | M to M/U |
| 28 | Female | 42 | M4 | Ara-C | Yes | M/U to M/U |
| 29 | Female | 64 | M6 | HHT+Ara-C | No | M/U to M/U |
| 30 | Female | 50 | M6 | Ara-C | Yes | M/U to M/U |
 | Table III.Details of the chemotherapy regimens
of all the acute myeloid leukemia patients. |
Table III.
Details of the chemotherapy regimens
of all the acute myeloid leukemia patients.
| Case | Chemotherapy
regimen, dose, treatment duration (route of administration,
frequency) |
|---|
| 1 | HHT: 3 mg, D1-D5
(ivgtt, qd); Ara-C: 75 mg, D1-D5 (ih, q12h); ACLA: 20 mg, D1-D5
(ivgtt, qd) |
| 2 | IDA: 15 mg, D1-D3
(ivgtt, qd); Ara-C: 100 mg, D1-D5 (ih, q12h) |
| 3 | Ara-C: 15 mg,
D1-D14 (ih, q12h); ACLA: 10 mg, D1-D4 (ivgtt, qod); G-CSF: 200 µg
(ih, q12h) |
| 4 | IDA: 10 mg, D1-D3
(ivgtt, qd); Ara-C: 75 mg, D1-D7 (ih, q12h) |
| 5 | ATRA: 10 mg, D1-D14
(po, tid); As2O3: 10 mg, D1-D14 (ivgtt,
qod) |
| 6 |
As2O3: 9 mg, D1-D14
(ivgtt, qd); DNR: 60 mg, D1-D3 (ivgtt, qd); ATRA: 10 mg, D1-D14
(po, bid) |
| 7 | HHT: 3.8 mg, D1-D7
(ivgtt, qd); Ara-C: 95 mg, D1-D5 (ih, q12h) |
| 8 | IDA: 10 mg, D1-D2
(ivgtt, qd); Ara-C: 75 mg, D1-D3 (ih, q12h); ACLA: 10 mg, D1-D5
(ivgtt, qod); G-CSF: 300 µg (ih, qd); HHT: 2 mg, D1-D5 (ivgtt,
qd) |
| 9 | HHT: 2 mg, D1-D5
(ivgtt, qd); Ara-C: 100 mg, D1-D5 (ih, q12h) |
| 10 | Ara-C: 22.5 mg,
D1-D3 (ih, q12h) |
| 11 | Ara-C: 100 mg,
D1-D5 (ih, q12h); ACLA: 100 mg, D1-D5 (ivgtt, qod) |
| 12 | HHT: 2 mg, D1-D3
(ivgtt, qd); Ara-C: 100 mg, D1-D3 (ih, q12h); ACLA: 100 mg, D1-D3
(ivgtt, qod) |
| 13 | HHT: 2 mg, D1-D5
(ivgtt, qd); Ara-C: 100 mg, D1-D5 (ih, q12h); G-CSF 400 µg (ih,
qd) |
| 14 | Ara-C: 16 mg,
D1-D14 (ih, q12h); ACLA: 20 mg, D1-D4 (ivgtt, qod); G-CSF: 300 µg
(ih, qd) |
| 15 | Ara-C: 15 mg,
D1-D14 (ih, q12h); ACLA: 20 mg, D1-D7 (ivgtt, qod); G-CSF: 300 µg
(ih, qd) |
| 16 | Ara-C: 16 mg,
D1-D14 (ih, q12h); ACLA: 20 mg, D1-D4 (ivgtt, qod); G-CSF: 300 µg
(ih, qd) |
| 17 | HHT: 2.8 mg, D1-D5
(ivgtt, qd); Ara-C: 70 mg, D1-D7 (ih, q12h); ACLA: 20 mg, D1-D5
(ivgtt, qd) |
| 18 | HHT: 3 mg, D1-D7
(ivgtt, qd); Ara-C: 15 mg, D1-D14 (ih, q12h); G-CSF: 200 mg (ih,
qd); IDA: 10 mg, D1-D4 (ivgtt, qd) |
| 19 | HHT: 3 mg, D1-D5
(ivgtt, qd); Ara-C: 75 mg, D1-D5 (ih, q12h); ACLA: 20 mg, D1-D5
(ivgtt, qd) |
| 20 | HHT: 2 mg, D1-D5
(ivgtt, qd); Ara-C: 100 mg, D1-D5 (ih, q12h) |
| 21 | ATRA: 20 mg, D1-D14
(po, bid); As2O3: 10 mg, D1-D14 (ivgtt, qd);
HHT: 3 mg, D1-D5 (ivgtt, qd); Ara-C: 15 mg, D1-D5 (ivgtt, qd) |
| 22 | IDA: 10 mg, D1-D3
(ivgtt, qd); Ara-C: 146 mg, D1-D5 (ih, q12h) |
| 23 | Ara-C: 12.5 mg,
D1-D3 (ih, q12h) |
| 24 | Ara-C: 100 mg,
D1-D3 (ih, q12h); |
| 25 | IDA: 15 mg, D1-D3
(ivgtt, qd) |
| 26 | IDA: 15 mg, D1-D3
(ivgtt, qd); Ara-C: 400 mg, D1-D7 (ih, q12h) |
Regimen-based subgroup analysis of
methylation changes in patients with AML
A total of 8 patients subjected to 6 different
chemotherapy regimens presented chemotherapy-induced methylation
changes, including 1 patient with induced hypermethylation and 7
patients with induced hypomethylation. Among the 3 patients under
IDA+Ara-C treatment, 1 exhibited induced hypermethylation and poor
prognosis, in contrast to the other 2 patients, who exhibited
induced hypomethylation and poor prognosis. The remaining 5
patients with induced hypomethylation and good prognosis were under
ATRA+As2O3, IDA+Ara-C+ACLA+G-CSF+HHT,
HHT+Ara-C+G-CSF, ATRA+As2O3+HHT+Ara-C and
HHT+Ara-C+ACLA treatment, respectively.
AML subtype-based subgroup analysis of
methylation changes in patients with AML
The present cohort included 3 M1, 8 M2, 7 M3, 6 M4,
3 M5 and 3 M6 AML subtypes. The outcomes of chemotherapy-induced
methylation changes differed among the various AML subtypes.
Chemotherapy-induced methylation changes were more frequent among
M4 subtype patients (50.0%, 3 out of 6 patients), compared with
other subtypes (M1, 33.3%, 1 out of 3 patients; M2, 0.0%, 0 out of
8 patients; M3, 42.9%, 3 out of 7 patients; M5, 0.0%, 0 out of 3
patients; and M6, 33.3%, 1 out of 3 patients). Of the
aforementioned M4 cases, 2 patients who had received
IDA+Ara-C+ACLA+G-CSF+HHT and HHT+Ara-C+ACLA treatment,
respectively, presented chemotherapy-induced hypomethylation and
good prognosis, whereas 1 patient who had been treated with
IDA+Ara-C exhibited chemotherapy-induced hypomethylation and poor
prognosis.
Age-based subgroup analysis of
methylation changes in patients with AML
In the present study, there were 24 AML patients
aged ≤60 years and 6 AML patients aged >60 years. Analysis by
age revealed that chemotherapy-induced methylation changes were
more often observed among AML patients aged ≤60 years (29.2%, 7 out
of 24 patients), compared with those aged >60 years (16.7%, 1
out of 6 patients). Among the patients aged ≤60 years, 1 M1 patient
(IDA+Ara-C, aged 49 years) exhibited chemotherapy-induced
hypermethylation and poor prognosis; 2 M3 patients
(ATRA+As2O3, aged 23 years; and
ATRA+As2O3+HHT+Ara-C, aged 51 years)
presented chemotherapy-induced hypomethylation and remission; 1 M3
patient (aged 42 years) experienced worsening of the condition
following IDA+Ara-C treatment; 1 M4 patient (IDA+Ara-C, aged 30
years) exhibited chemotherapy-induced hypomethylation and
aggravation of the condition; 1 M4 patient (HHT+Ara-C+ACLA, aged 19
years) displayed chemotherapy-induced hypomethylation along with
remission; and 1 M6 patient (HHT+Ara-C+G-CSF, aged 52 years)
presented chemotherapy-induced hypomethylation along with
remission. Only 1 M4 patient aged >60 years
(IDA+Ara-C+ACLA+G-CSF+HHT, aged 67 years) exhibited
chemotherapy-induced hypomethylation and remission.
Gender-based subgroup analysis of
methylation changes in patients with AML
In total, 4 out of 13 male patients and 4 out of 17
female patients who had been subjected to different combined
regimens exhibited chemotherapy-induced methylation changes in the
promoter of the MGMT gene. Among the male patients, 1 M1 patient
(IDA+Ara-C) presented hypermethylation along with poor prognosis,
whereas 1 M3 patient (ATRA+As2O3), 1 M4
patient (IDA+Ara-C+ACLA+G-CSF+HHT) and 1 M6 patient
(HHT+Ara-C+G-CSF) presented chemotherapy-induced hypomethylation
along with remission. The remaining 9 male patients (2 of which had
been treated with HHT+Ara-C+ACLA, 1 with HHT+Ara-C, 1 with
Ara-C+ACLA+G-CSF, 1 with IDA+Ara-C, 1 with
As2O3+DNR+ATRA, 1 with Ara-C and 1 with
Ara-C+ACLA) did not exhibit any methylation changes induced by the
chemotherapeutic agents.
In the female subgroup, 1 M3 patient who had been
treated with ATRA+As2O3+HHT+Ara-C) and 1 M4
patient who had been treated with HHT+Ara-C+ACLA presented
chemotherapy-induced hypomethylation along with good prognosis,
while 2 female patients (1 M3 and 1 M4 who had been treated with
IDA+Ara-C) exhibited chemotherapy-induced hypomethylation and poor
prognosis. The remaining 13 female patients (4 of which had been
treated with Ara-C, 3 with Ara-C+ACLA+G-CSF, 2 with HHT+Ara-C, 2
with Ara-C+ACLA+G-CSF, 1 with HHT+Ara-C+G-CSF+IDA and 1 with IDA)
did not exhibit any induced methylation changes following
chemotherapy.
Discussion
Increasing evidence has demonstrated that
epigenetics, including DNA methylation, is important in the
pathogenesis of cancer (14). The
dysregulation of critical genes involved in cell growth,
differentiation, apoptosis and repair of DNA lesions may increase
the incidence of malignant phenotypes (15). As a DNA repair enzyme, MGMT mainly
participates in maintaining genomic stability if spontaneous
mutagenesis occurs (10). However,
the association between chemotherapy-induced methylation changes in
the MGMT promoter and pathogenesis of AML has not been fully
elucidated thus far. The aim of the present study was to explore
the association between the chemotherapeutic outcomes of patients
with AML and the chemotherapy-induced methylation changes in the
promoter of the MGMT gene that occur in the bone marrow of
AML patients during chemotherapeutic treatment.
The results of the present study demonstrated that
the methylation status of the MGMT promoter changed during
the treatment with different chemotherapeutic regimens. MGMT
was observed to be more often hypomethylated (7 patients out of 8)
than hypermethylated (1 patient out of 8) by the different
chemotherapeutic regimens. Among the AML patients, M4 (3 out of 6
patients) and M3 patients (3 out of 7 patients) exhibited
chemotherapy-induced methylation changes more frequently than
patients with other AML subtypes. Age-based subgroup analysis
revealed that patients aged ≤60 years (7 out of 24 patients)
presented methylation changes more frequently than patients aged
>60 years (1 out of 6 patients). In addition, male patients
appeared to be more susceptible to chemotherapy-induced methylation
changes than female patients.
Different subtypes of AML may exhibit different
responses to multiple chemotherapeutics, which may influence DNA
methylation and remission (16).
According to the FAB classification, M3 is the best curable subtype
of AML, due to its sensitivity toward ATRA treatment, which causes
the differentiation of promyelocytes (17). The present findings revealed 3
hypomethylated patients, of which, 2 (who had been treated with
ATRA+As2O3 and
ATRA+As2O3+HHT+Ara-C, respectively)
experienced emission, while 1 (wo had been subjected to IDA+Ara-C
treatment) experienced aggravation of the condition, suggesting
individual differences during chemotherapy. In addition, M4
patients presented chemotherapy-induced methylation changes more
often than other subtypes, which suggests that the association
between the methylation status of the MGMT promoter and the
chemotherapeutic outcomes of patients with AML may depend on the
AML subtype.
Age may be an important risk factor in the selection
of therapy for AML (18). Decreased
visceral function, drug resistance and poor reactivity among the
elders may lead to poor prognosis and high recurrence rate, which
results in a poor overall 5-year survival rate for elderly AML
patients (3). The analysis by age
conducted in the present study demonstrated that
chemotherapy-induced hypomethylation events were more frequent
among AML patients aged ≤60 years than among those aged >60
years, suggesting that young patients may be more sensitive to
chemotherapeutic drugs.
Gender discrepancy must be considered when referring
to cancer prognosis (19). A previous
study emphasized the significantly higher incidence rates of acute
leukemia in male patients, compared with female patients (20). In the present study,
chemotherapy-induced hypomethylation of MGMT along with good
prognosis occurred more often in males than in females, suggesting
the potential of this gene to serve as a biomarker to predict the
chemotherapeutic outcomes in male patients with AML. Furthermore,
IDA+Ara-C regimen led to worse outcomes among female patients,
compared with males.
In conclusion, the outcomes of chemotherapy-induced
methylation of the MGMT gene in AML patients varied among
the different AML subtypes, gender and age of patients in the
present study. MGMT promoter methylation may serve as a
prognostic value for individualized chemotherapy in the future.
However, further studies in larger sample sets are required to
confirm the present findings.
Acknowledgements
The present study was supported by grants from the
National Natural Science Foundation of China (Beijing, China; grant
nos. 31100919 and 81371469), Zhejiang Provincial Natural Science
Foundation (Hangzhou, China; grant no. LR13H020003), Ningbo City
Medical Science and Technology Projects (Ningbo, China; grant no.
2014A20) and K. C. Wong Magna Fund in Ningbo University, Ningbo,
China.
References
|
1
|
Estey EH: Acute myeloid leukemia: 2013
update on risk-stratification and management. Am J Hematol.
88:318–327. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Davis AS, Viera AJ and Mead MD: Leukemia:
An overview for primary care. Am Fam Physician. 89:731–738.
2014.PubMed/NCBI
|
|
3
|
Thein MS, Ershler WB, Jemal A, Yates JW
and Baer MR: Outcome of older patients with acute myeloid leukemia:
An analysis of SEER data over 3 decades. Cancer. 119:2720–2727.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Stringaris K, Sekine T, Khoder A,
Alsuliman A, Razzaghi B, Sargeant R, Pavlu J, Brisley G, de
Lavallade H, Sarvaria A, et al: Leukemia-induced phenotypic and
functional defects in natural killer cells predict failure to
achieve remission in acute myeloid leukemia. Haematologica.
99:836–847. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Maurillo L, Venditti A, Spagnoli A,
Gaidano G, Ferrero D, Oliva E, Lunghi M, D'Arco AM, Levis A,
Pastore D, et al: Azacitidine for the treatment of patients with
acute myeloid leukemia: Report of 82 patients enrolled in an
Italian Compassionate Program. Cancer. 118:1014–1022. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Boultwood J and Wainscoat JS: Gene
silencing by DNA methylation in haematological malignancies. Br J
Haematol. 138:3–11. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Tuominen R, Jewell R, van den Oord JJ,
Wolter P, Stierner U, Lindholm C, Johansson Hertzman C, Lindén D,
Johansson H, Frostvik Stolt M, et al: MGMT promoter methylation is
associated with temozolomide response and prolonged
progression-free survival in disseminated cutaneous melanoma. Int J
Cancer. 136:2844–2853. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Iyama T and Wilson DM 3rd: DNA repair
mechanisms in dividing and non-dividing cells. DNA Repair (Amst).
12:620–636. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Kraguljac Kurtović N, Krajnović M,
Bogdanović A, Suvajdžić N, Jovanović J, Dimitrijević B, Colović M
and Krtolica K: Concomitant aberrant methylation of p15 and MGMT
genes in acute myeloid leukemia: Association with a particular
immunophenotype of blast cells. Med Oncol. 29:3547–3556. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Brandwein JM, Kassis J, Leber B, Hogge D,
Howson-Jan K, Minden MD, Galarneau A and Pouliot JF: Phase II study
of targeted therapy with temozolomide in acute myeloid leukaemia
and high-risk myelodysplastic syndrome patients pre-screened for
low O (6)-methylguanine DNA methyltransferase expression. Br J
Haematol. 167:664–670. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Fasan A, Alpermann T, Haferlach C,
Grossmann V, Roller A, Kohlmann A, Eder C, Kern W, Haferlach T and
Schnittger S: Frequency and prognostic impact of CEBPA proximal,
distal and core promoter methylation in normal karyotype AML: A
study on 623 cases. PLoS One. 8:e543652013. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Chen C, Wang L, Liao Q, Huang Y, Ye H,
Chen F, Xu L, Ye M and Duan S: Hypermethylation of EDNRB promoter
contributes to the risk of colorectal cancer. Diagn Pathol.
8:1992013. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Vidal DO, Paixão VA, Brait M, Souto EX,
Caballero OL, Lopes LF and Vettore AL: Aberrant methylation in
pediatric myelodysplastic syndrome. Leuk Res. 31:175–181. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Reid T, Oronsky B, Scicinski J, Scribner
CL, Knox SJ, Ning S, Peeh DM, Korn R, Stirn M, Carter CA, et al:
Safety and activity of RRx-001 in patients with advanced cancer: A
first-in-human, open-label, dose-escalation phase 1 study. Lancet
Oncol. 16:1133–1142. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Malonia SK, Sinha S, Lakshminarasimhan P,
Singh K, Jalota-Badhwar A, Rampalli S, Kaul-Ghanekar R and
Chattopadhyay S: Gene regulation by SMAR1: Role in cellular
homeostasis and cancer. Biochim Biophys Acta. 1815:1–12.
2011.PubMed/NCBI
|
|
16
|
Johannes Z, Ina Radtke, Timothy S, Pardee,
Zhen Zhao, Amy R, Rappaport, Weijun Luo, Mila E, McCurrach, Yang
MM, Dolan ME, Kogan SC, et al: Mouse models of human AML accurately
predict chemotherapy response. Genes Dev. 23:877–889. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Poddighe PJ, Wessels H, Merle P, Westers
M, Bhola S, Loonen A, Zweegman S, Ossenkoppele GJ and Wondergem MJ:
Genomic amplification of MYC as double minutes in a patient with
APL-like leukemia. Mol Cytogenet. 7:672014. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Ofran Y and Rowe JM: Acute myeloid
leukemia in adolescents and young adults: Challenging aspects. Acta
Haematol. 132:292–297. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Pui CH: Acute leukemias with the
t(4;11)(q21;q23). Leuk Lymphoma. 7:173–179. 1992. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Shahab F and Raziq F: Clinical
presentations of acute leukemia. J Coll Physicians Surg Pak.
24:472–476. 2014.PubMed/NCBI
|