Introduction
Hepatocellular carcinoma (HCC) is one of the most
frequently occurring malignancies around the world, accounting for
~700,000 mortalities per year (1).
Surgical resection remains the main curative treatment for HCC as
well as the first treatment choice for patients with smaller tumor
size, preserved liver function, and no evidence of vascular
invasion or extrahepatic spread (2).
However, postoperative tumor recurrence, as a result of intra- or
extrahepatic metastasis, occurs in ~50% of patients within 2 years
of surgery (3). Recurrence
significantly worsens the prognosis of HCC patients (4). Understanding what causes recurrence may
help physicians predict or even prevent it.
Evidence indicates that in numerous types of cancer,
including HCC, low-abundance cancer stem cells (CSCs) are
responsible for tumor recurrence and metastasis (5). CSCs detach from the primary mass and
enter the lymph and peripheral blood. A chemoattractant gradient
directs CSCs to a particular point, where they attach to the
endothelium and penetrate the microvessel wall. Finally, CSCs enter
an environmental niche that protects them from damage and allows
them to give rise to a recurrent or metastatic tumor (6,7). This
makes CSCs a potential therapeutic target in efforts to reduce
tumor recurrence and metastasis, and improve outcomes for HCC
patients (8).
CSCs associated with HCC have been identified using
a variety of cell surface markers, including CD133, CD90,
epithelial cell adhesion molecule (EpCAM), CD13, and CD44 (9–12). CSC
populations expressing different markers seem to possess similar
CSC properties, but the lack of a standard set of markers makes the
isolation of liver CSCs a challenge. One approach to obtaining CSCs
is to isolate them from preparations of side population (SP) cells.
SP cells are lower-abundance cells that coexist with, and may
demonstrate significant functional differences from, main
population (MP) cells, and may be considered as a substitute for
CSCs in scientific studies. SP cells have been isolated from
numerous types of tumor, including glioma (13), mantle cell lymphoma (14), nasopharyngeal carcinoma (15), lung cancer (16), colon cancer (17), breast cancer (18), ovarian cancer (19), pancreatic cancer (20), and HCC (21,22). These
SP cells are typically identified based on their ability to efflux
Hoechst 33342 dye, thought to result from their high expression of
adenosine triphosphate (ATP)-binding cassette (ABC) membrane
transporters (23). These
transporters are also highly expressed in stem cells, suggesting
that cancer-associated SP cells are highly enriched in CSCs
(24).
What remains unclear from studies of SP cells in HCC
is the extent to which these populations show stem cell-like
properties and may therefore be studied in detail as the potential
drivers of HCC tumorigenicity and metastasis. The present study
aimed to determine the proportions of SP cells in four human HCC
cell lines, which are widely used to study the mechanism of HCC
metastasis. The four cell lines increase in metastatic potential in
the following order: HCCLM3>MHCC97-H>MHCC97-L>Huh7
(11,25,26). SP
and MP cells were then prepared from the HCCLM3 cell line and their
tumorigenicity and invasiveness were compared, as well as their
expression of CSC-associated genes.
Materials and methods
Cell culture
The human HCC cell lines HCCLM3, MHCC97-H, MHCC-97
L, and Huh7 were purchased from the Liver Cancer Institute of
Zhongshan Hospital, Fudan University (Shanghai, China). Cells were
cultured in high-glucose Gibco Dulbecco's modified Eagle medium
(DMEM; Thermo Fisher Scientific, Inc., Waltham, MA, USA) containing
Gibco 10% fetal bovine serum (FBS; Thermo Fisher Scientific, Inc.)
and Gibco 1% penicillin/streptomycin (Thermo Fisher Scientific,
Inc.). Cultures were incubated at 37°C in a humidified atmosphere
containing 5% CO2. This study was approved by the
Guangxi Medical Experimental Animal Care Commission.
Analysis of side population (SP)
cells
Cultured cells were detached from culture dishes
using Gibco TrypLE Express Enzyme (Thermo Fisher Scientific, Inc.)
and 1×106 cells/ml were suspended in high-glucose DMEM
supplemented with 5% FBS. The cells were then incubated at 37°C for
90 min with different concentrations of Hoechst 33342 dye (3, 4, 5,
6, 7, 8, 9, or 10 mg/l; Sigma-Aldrich, St. Louis, MO, USA), either
alone or in the presence of 50 µM verapamil (Sigma-Aldrich). Cell
suspensions were washed with cold phosphate-buffered saline (PBS),
then centrifuged and resuspended in 1 ml high-glucose DMEM
supplemented with 5% FBS and 7-amino-actinomycin D (7-aad, 1 mg/l;
Invitrogen, Thermo Fisher Scientific, Inc. to label dead cells.
Cells were filtered through a 40-µm cell strainer (BD Biosciences,
Franklin Lakes, NJ, USA) to obtain single-cell suspensions, which
were maintained at 4°C prior to analysis.
SP and MP cells were analyzed separately by flow
cytometry (FACSComp™; BD Biosciences) based on intracellular levels
of Hoechst 33342 dye (22). The SP
gate was defined as the region where cells disappeared when cells
were exposed to verapamil, which blocks Hoechst 33342 efflux. All
samples were evaluated in 3 independent experiments by the same
flow cytometry expert, who was blinded to the identity of the
samples.
Proliferation assay
Cell proliferation was evaluated in vitro
using the Cell Counting Kit 8 (CCK8; Signalway Antibody LLC.,
College Park, MA, USA) according to the manufacturer's
instructions. Separate SP and MP cultures isolated from the HCCLM3
line were plated at a density of 5×103 cells/well in
96-well microplates with high-glucose DMEM containing 10% FBS and
1% penicillin/streptomycin. At different incubation times (1, 2, 3,
4, 5, 6, or 7 days), the culture medium was replaced with fresh
medium (100 µl/well) containing CCK8 reagent (10 µl per well). The
cultures were incubated for 4 h, then the optical density at 450 nm
was measured using a microplate reader (Multiskan GO; Thermo Fisher
Scientific, Inc.). Experiments were performed 3 times, with each
experiment including 5 replicates of each condition.
Clonogenicity assay
SP and MP cells isolated from HCCLM3 cultures were
plated separately in 6-well dishes at 1×103 cells per
well. Cells were incubated for 2 weeks, and the medium was changed
every 3 days. The cells were fixed with methanol and stained with
Giemsa's solution (ZSGB-BIO, Beijing, China). Clonal populations
containing >50 cells were counted under a microscope (Axiovert
200; Carl Zeiss AG, Oberkochen, Germany) at ×200 magnification from
5 fields and averaged (mean ± standard deviation).
Migration and invasion assays
Cell migration was analyzed using Transwell cell
culture chambers (8 µm pore size; Corning Incorporated, Corning,
NY, USA), and cell invasion was analyzed using the same Transwell
chambers precoated with Matrigel (Corning Incorporated).
For both types of assay, SP and MP cells were
resuspended in FBS-free high-glucose DMEM and added to the upper
chamber of the 24-well Transwell dish at a density of
105 cells/chamber. High-glucose DMEM containing 10% FBS
was added to the well, and the cultures were incubated 24 h. Then
the cells on the upper side of the filters were mechanically
removed by scrubbing with a cotton swab, and the filter was fixed
with methanol for 5 min at room temperature and stained with
Giemsa's solution (ZSGB-BIO) for 10 min. The number of invasive or
migrated cells were counted at 200x magnification (Nikon
Corporation) from 5 fields of each filter and averaged (mean ±
standard deviation).
Tumorigenicity assay
Male nude mice aged 6–8 weeks were purchased from
the Laboratory Animal Centre of Guangxi Medical University
(Nanning, China) and housed in laminar-flow cabinets under specific
pathogen-free conditions. Animal care and experimental protocols
were in accordance with procedures and guidelines established by
the Guangxi Medical Experimental Animal Care Commission.
Varying numbers of SP and MP cells
(5×103-5×105) isolated from the HCCLM3 line
were suspended in a 1:1 (v/v) mixture of 200 µl PBS and Matrigel
(BD Biosciences). The suspensions were injected subcutaneously into
15 mice under anesthesia: Each mouse received an injection of SP
cells on the left side of the back, and an injection of MP cells on
the right side of the back. Tumor growth was monitored weekly for 4
weeks. Subcutaneous tumors were fixed in formalin, embedded in
paraffin, cut into 4 µm sections and subjected to histopathological
examination by staining with hematoxylin-eosin (ZSGB-BIO).
Expression of CSC-associated
genes
Total RNA was isolated from SP or MP cells using
TRIzol reagent (Invitrogen, Thermo Fisher Scientific, Inc.)
according to the manufacturer's instructions. The total RNA
concentration was quantified using the Nanodrop2000 reader (Thermo
Fisher Scientific, Inc.). Total RNA (1 µg) was reverse-transcribed
(RT) using the Prime Script RT Reagent Kit with gDNA Eraser (Takara
Bio, Inc., Otsu, Japan), and the resulting cDNA was used as
template for quantitative polymerase chain reaction with the SYBR
Green/ROX Master Mix on a 7300 Real-Time PCR System (Applied
Biosystems, Thermo Fisher Scientific, Inc.). The thermal cycling
conditions were as follows: 95°C for 1 min, followed by 40 cycles
of 95°C for 5 sec, 60°C for 31 sec and 72°C for 50 sec. Levels of
the mRNA of several CSC-associated genes (ABCG2, CD133, CD90,
EpCAM, CD44, CD13, Nanog, Oct4, Sox2 and Klf4) were determined
using the 2−ΔΔCq method and were normalized to the
transcript levels of the housekeeping gene
glyceraldehyde-3-phosphate dehydrogenase (GAPDH). Primers to
amplify the specific gene sequences are presented in Table I. All reactions were run in
triplicate. Significant differences in gene expression were defined
as those for which normalized transcript levels differed by ≥2-fold
and for which the associated P<0.05.
 | Table I.Primer sequences to assay expression
of cancer stem cell-associated genes by reverse transcription
quantitative-polymerase chain reaction. |
Table I.
Primer sequences to assay expression
of cancer stem cell-associated genes by reverse transcription
quantitative-polymerase chain reaction.
| Gene | Primer sequence
(5′-3′) |
|---|
| ABCG2 | F:
AGGTCTGGATAAAGTGGCA |
|
| R:
GAGGCTGATGAATGGAGAA |
| EpCAM | F:
ATAACCTGCTCTGAGCGAGTG |
|
| R:
TGCAGTCCGCAAACTTTTACTA |
| CD133 | F:
GTCTGACCAGCGTGAAAAC |
|
| R:
GCCATCCAAATCTGTCCTA |
| CD90 | F:
ATGAAGGTCCTCTACTTATCCGC |
|
| R:
GCACTGTGACGTTCTGGGA |
| CD13 | F:
TTCAACATCACGCTTATCCACC |
|
| R:
AGTCGAACTCACTGACAATGAAG |
| CD44 | F:
CTGCCGCTTTGCAGGTGTA |
|
| R:
CATTGTGGGCAAGGTGCTATT |
| Nanog | F:
CCCCAGCCTTTACTCTTCCTA |
|
| R:
CCAGGTTGAATTGTTCCAGGTC |
| Oct4 | F:
GTGTTCAGCCAAAAGACCATCT |
|
| R:
GGCCTGCATGAGGGTTTCT |
| Sox2 | F:
TACAGCATGTCCTACTCGCAG |
|
| R:
GAGGAAGAGGTAACCACAGGG |
| Klf4 | F:
CGGACATCAACGACGTGAG |
|
| R:
GACGCCTTCAGCACGAACT |
| GAPDH | F:
CTGGGCTACACTGAGCACC |
|
| R:
AAGTGGTCGTTGAGGGCAATG |
Statistical analysis
All analyses were performed using SPSS. version 19.0
(IBM SPSS, Armonk, NJ, USA). Data are presented as the mean ±
standard deviation. The difference between means were assessed for
significance using Student's t test or one-way analysis of
variance. The threshold for significance was defined as a two-sided
P<0.05.
Results
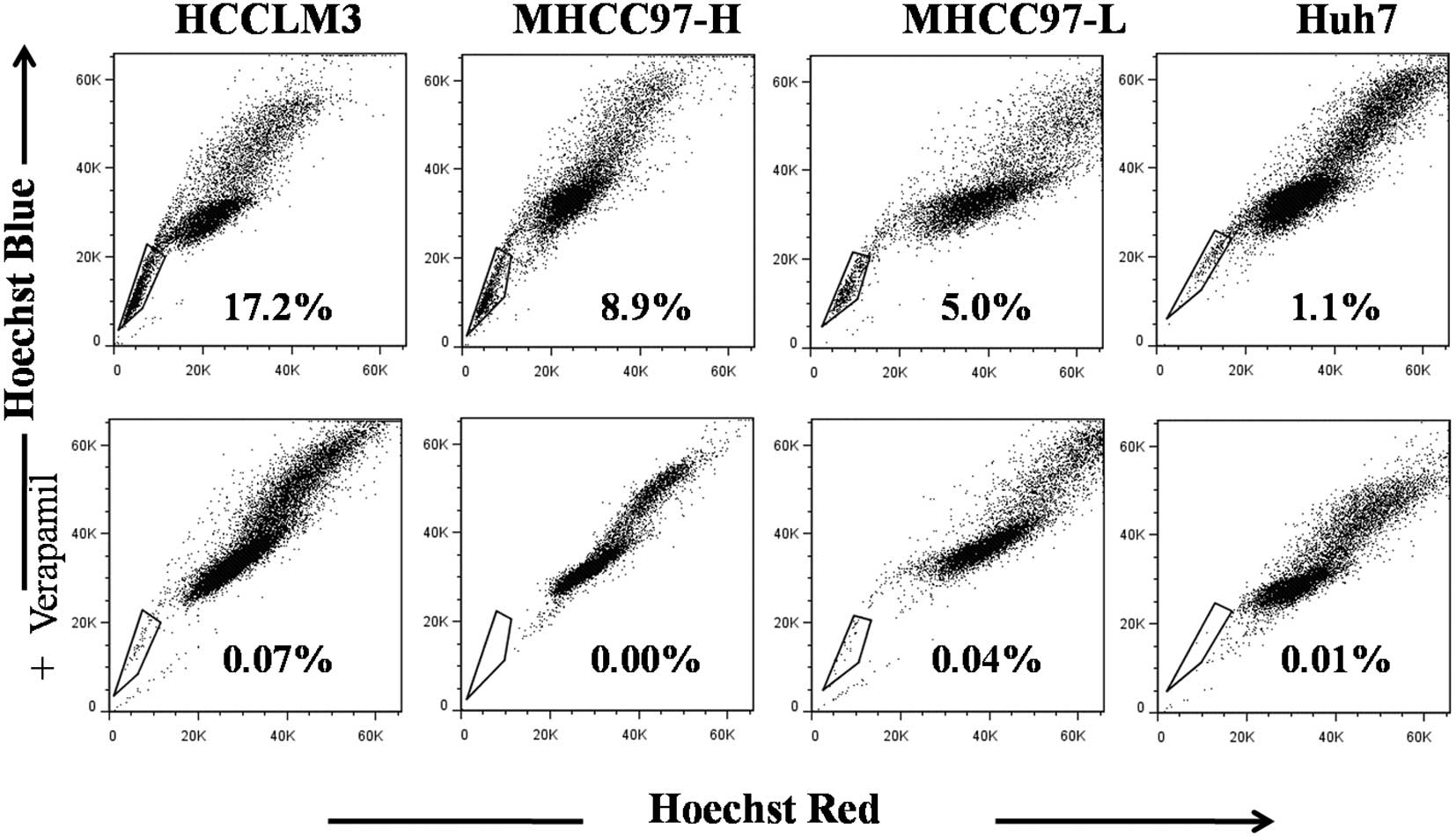
Relative abundance of SP cells
correlated directly with metastatic potential of HCC cell
lines
Flow cytometry was used to differentiate between SP
and MP cells based on retention of Hoechst 33342. In order to be
able to exclude dead cells and debris from the analysis, cells were
also labelled with 7-aad, which causes dead cells to emit intense
red fluorescence. The ability to differentiate SP and MP cells was
dependent on the Hoechst 33342 concentration (Table II). Lower dye concentrations were
associated with higher proportions of SP cells. This proportion
dropped sharply at concentrations between 3 and 5 µg/ml, and this
reduction was not accompanied by increases in the percentage of
dead cells labeled by 7-aad. Thus this drop most likely reflected a
shift in cells from SP to MP regions. Verapamil further lowered the
percentage of cells in the SP region at lower Hoechst 33342
concentrations.
 | Table II.Proportions of total viable cells and
SP cells in HCCLM3 suspensions stained with different
concentrations of H or in the presence of H+V. |
Table II.
Proportions of total viable cells and
SP cells in HCCLM3 suspensions stained with different
concentrations of H or in the presence of H+V.
|
| Total viable cells
(%) | SP cells (%) |
|---|
|
|
|
|
|---|
| Hoechst conc
(mg/l) | H | H+V | H | H+V |
|---|
| 3 | 98.1±0.7 | 96.5±2.2 | 40.5±6.3 | 2.5±1.1 |
| 4 | 96.7±2.4 | 95.7±1.8 | 33.3±4.2 | 0.7±0.3 |
| 5 | 94.8±0.8 | 94.1±1.1 | 21.5±5.1 | 0.4±0.2 |
| 6 | 94.0±1.2 | 92.5±0.9 | 16.3±2.2 | 0 |
| 7 | 88.2±3.1 | 86.3±2.3 | 5.6±2.5 | 0 |
| 8 | 83.5±2.7 | 79.8±3.6 | 2.8±1.3 | 0 |
| 9 | 80.7±4.5 | 75.3±2.8 | 2.3±0.7 | 0 |
| 10 | 76.1±3.3 | 71.6±3.4 | 0.9±0.2 | 0 |
Staining cells with Hoechst dye at concentrations
between 7 and 10 µg/ml caused a large reduction in the SP
proportion, and the residual SP cells disappeared entirely in the
presence of verapamil. These reductions were accompanied by a
marked increase in dead cells, indicating that these dye
concentrations were toxic to the cells. Therefore the dye
concentration for each cell line was optimized in order to identify
a level high enough to ensure a stable proportion of SP cells
without causing appreciable toxicity.
The optimal concentrations (µg/ml) were 6 for HCCLM3
cells, 10 for MHCC97-H, 9 for MHCC97-L, and 7 for Huh7; the
percentage of SP cells at these dye concentrations were 16.3±2.2,
8.4±0.7, 4.7±0.5 and 1.0±0.3% (Fig.
1; P<0.001), resepctively. This relative heirarchy mirrors
the relative metastastic potential of the 4 cell lines.
In order to ensure sufficient numbers of SP cells
for subsequent detailed analysis, SP and MP cells were prepared
from the HCCLM3 cell line.
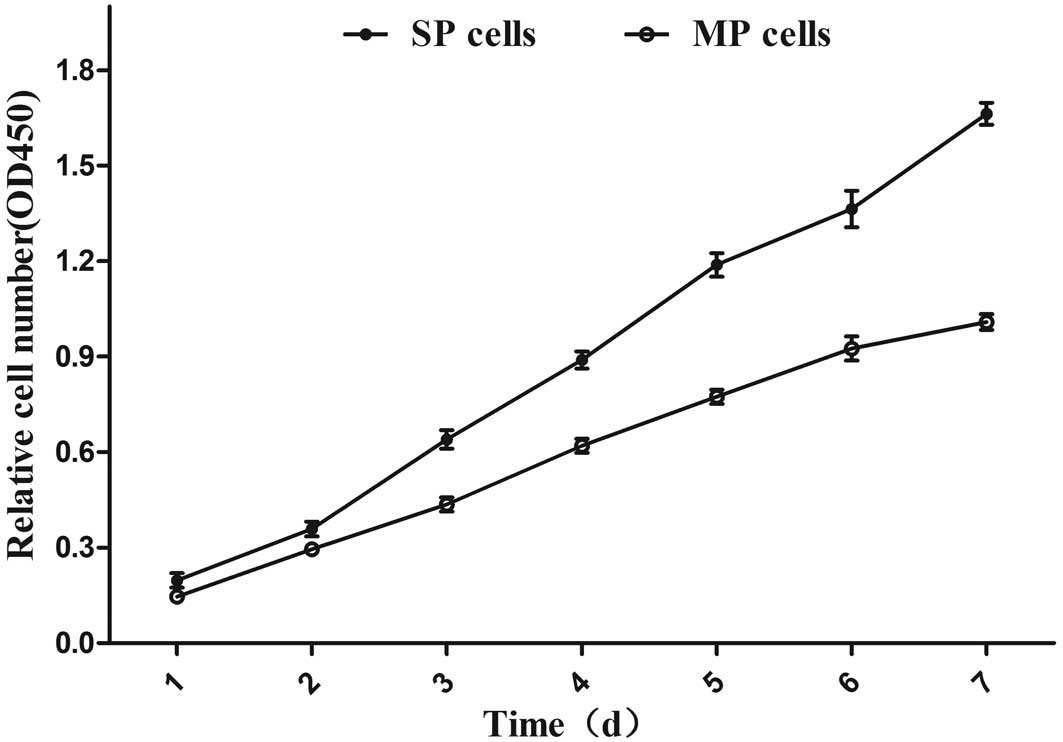
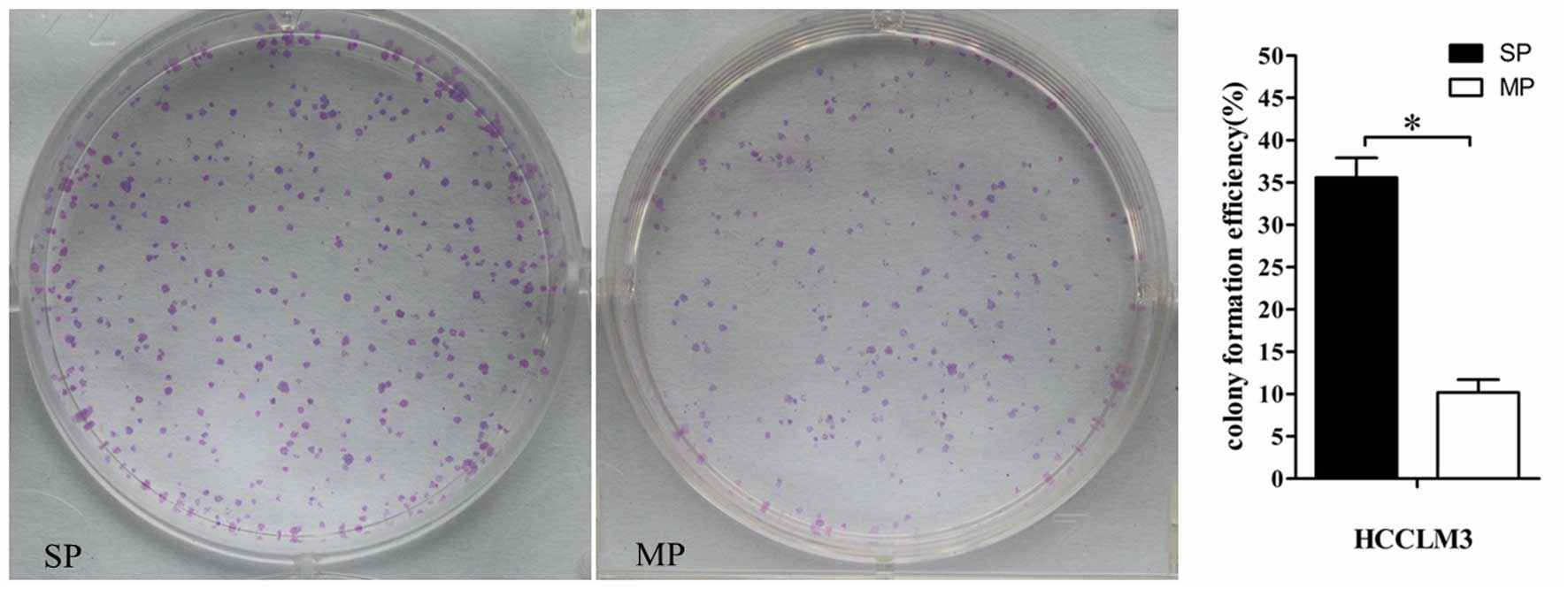
High viability and clonogenicity of SP
cells
The in vitro proliferation of SP and MP cells
was compared by CCK8 assay. The stain in this assay labels only
living cells, so the resulting optical density reflects the number
of viable cells, providing an index of growth. SP cells
demonstrated greater proliferation rates compared with MP cells
(Fig. 2; P<0.05), as well as a
higher colony-forming capacity (Fig.
3; P<0.001).
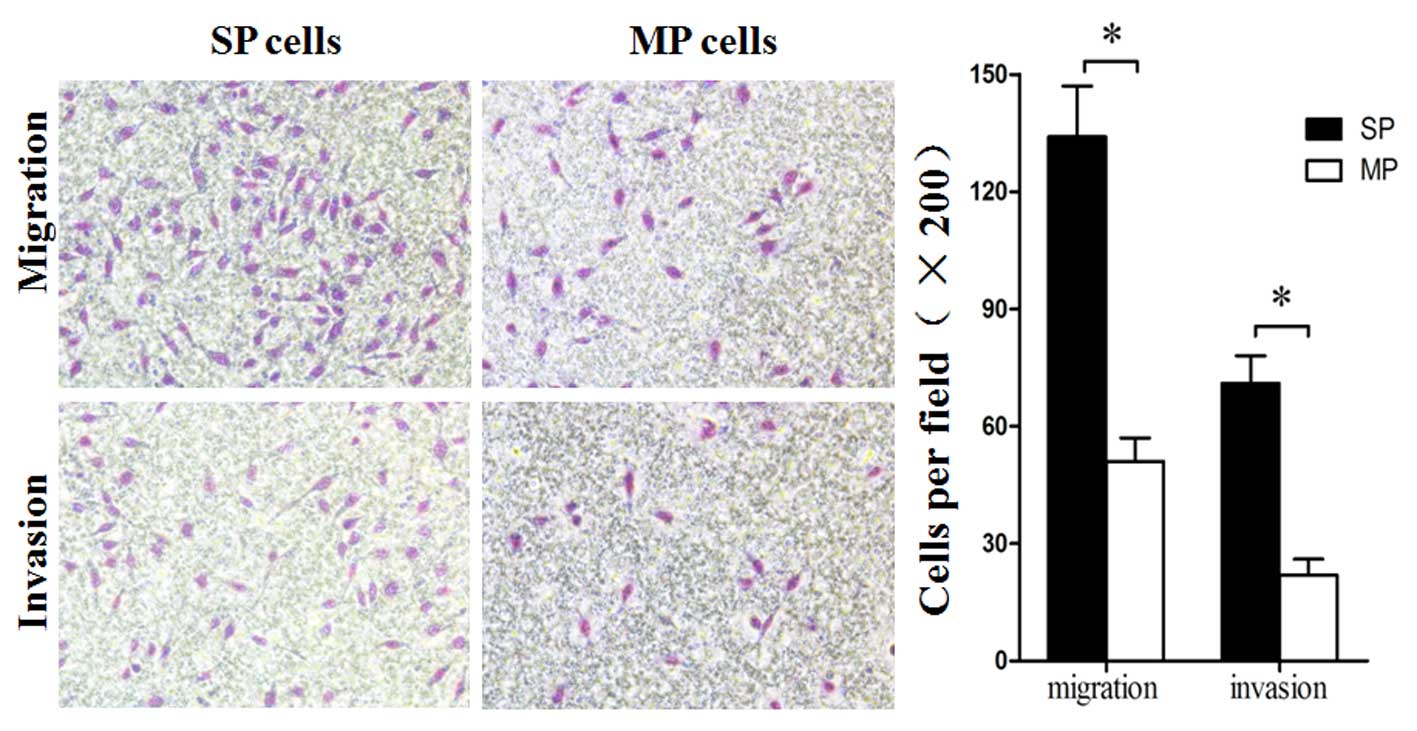
High migration and invasion
capabilities of SP cells
Using a Transwell-based migration and invasion
assay, it was observed that SP cells possessed higher migration and
invasive abilities compared with MP cells (Fig. 4).
High tumorigenicity of SP cells
Nude mice received one injection of SP cells and one
injection of MP cells on opposite sides of the back, after which
tumorigenesis was monitored. Injections of as few as
5×103 SP cells initiated tumor in one of five mice,
while injections of as many as 5×105 MP cells led to
tumors in two of five mice (Table
III). Tumor pathology was confirmed by histopathology. These
results indicate an increase in tumorigenicity in the SP cell
population (Fig. 5).
 | Table III.Tumorigenicity of SP and MP cells
from the HCCLM3 cell line injected into nude mice. |
Table III.
Tumorigenicity of SP and MP cells
from the HCCLM3 cell line injected into nude mice.
|
| No. of animals
showing a tumor at injection sitea |
|---|
|
|
|
|---|
| No. of cells
injected | SP cells | MP cells |
|---|
|
5×103 | 1 | 0 |
|
5×104 | 3 | 0 |
|
5×105 | 5 | 2 |
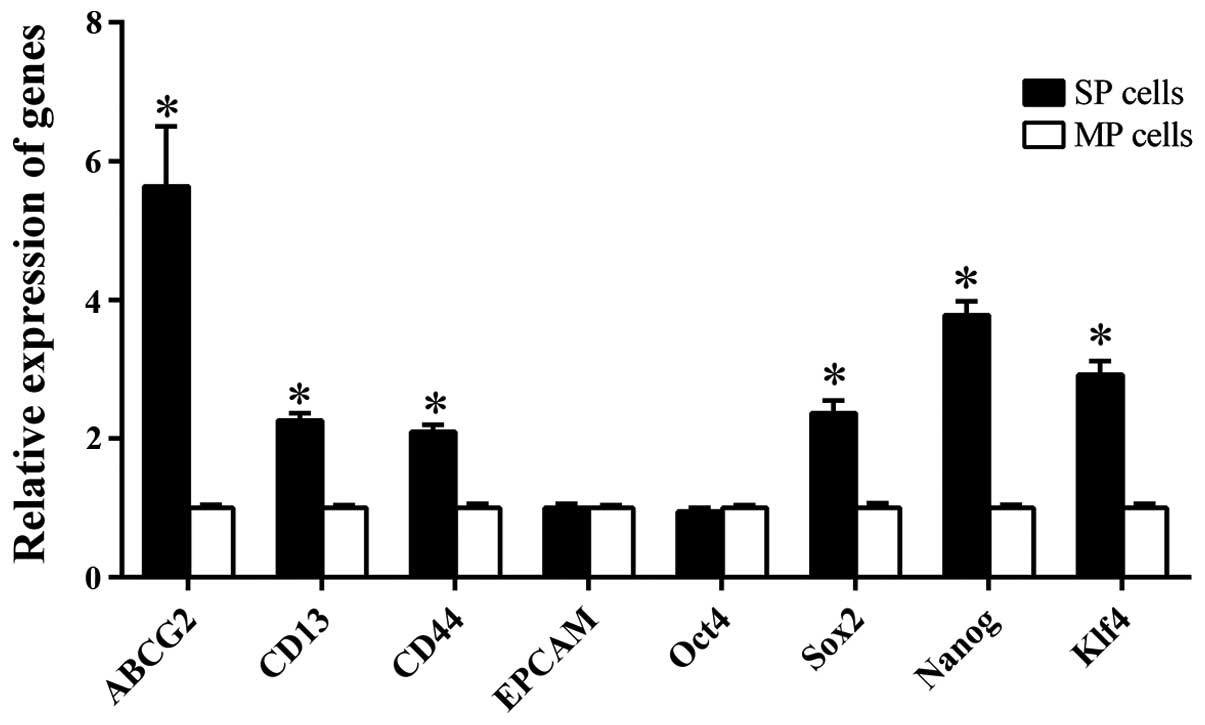
Higher expression of CSC-associated
genes in SP cells
The SP and MP preparations from the HCCLM3 line with
respect to the expression levels of a number of genes linked to the
SP and CSC phenotypes. ABCG2 encodes a protein that can pump a
broad range of cytotoxic substances out of the cell, and it is
essential for maintaining the SP phenotype (27,28). The
genes CD133, CD90, EpCAM, CD44, and CD13 encode cell surface
markers previously used to enrich for CSCs in HCC (9–12). The
expression of Klf4, Oct4, Sox2 and Nanog, were also examined, all
of which encode transcription factors essential for self-renewal
and pluripotency in embryonic stem cells (29–31).
Transcription of the CD133 and CD90
genes were not detected in either SP or MP cells; EpCAM and
Oct4 were expressed at similar levels in the two cell types.
The other genes tested were expressed at significantly higher
levels in SP cells compared with MP cells: for ABCG2, the
difference was 5.64±0.86-fold; CD13, 2.26±0.11; CD44,
2.1±0.1; Sox2, 2.37±0.18; Nanog, 3.78±0.2; and
Klf4, 2.92±0.2 (Fig. 6).
Discussion
CSCs are low-abundance cells with the ability to
proliferate and form tumors, and thought to be responsible for
tumor recurrence and metastasis (5).
Recurrence is a significant obstacle to good prognosis for patients
with HCC, even when it can be treated early, so identifying liver
CSCs and developing a method to isolate them may significantly aid
efforts in studying them as therapeutic targets. SP cells, which
pump out the fluorescrent dye Hoechst33342 through ABC
transporters, display several properties contributable to stem
cell-like cancer cells, and study as a substitute for CSCs
(21,22). IN the present study, SP fractions were
isolated from 4 human HCC cell lines, and this fraction was
demonstrated to contain cells with the phenotypic characteristics
of CSCs. These initial findings lay the groundwork for future
studies in vitro and in vivo aimed at understanding
and ultimately preventing HCC recurrence and metastasis.
In the present study, the proportion of SP cells was
greatly affected by the concentration of Hoechst dye used. At lower
dye concentrations, MP cells may appear to be SP cells in flow
cytometric analysis; at higher concentrations, SP cells may move
into the MP region (Table II).
Oversaturated dye concentrations can also increase cell death.
Therefore, it was necessary to determine the optimal dye
concentrations for each HCC cell line, defined as the concentration
yielding a stable percentage of SP cells without causing
toxicity.
SP cells were successfully identified in suspensions
of HCCLM3, MHCC97-H, MHCC97-L, and Huh7 cells. These cell lines
form a gradient of metastatic potential, in the order
HCCLM3>MHCC97-H>MHCC97-L>Huh7, and these particular cell
lines have been widely used in studies of HCC metastasis (11,25,26). The
proportion of SP cells correlated directly with the metastatic
potential. These findings are similar to those observed in a former
study by Wu et al (32), who
examined the proportion of SP cells in 29 mesenchymal tumors
ranging from benign to high-grade sarcomas, and found that the
highly aggressive tumors contained a higher proportion of SP cells.
Taken together, this evidence indicates that SP cells may be
considered as a substitute for CSCs, and may be involved in
metastasis.
The comparative analysis of SP and MP preparations
from the HCCLM3 cell line performed in the present study, indicated
that SP cells show the characteristics expected of CSCs: Greater
proliferation and colony formation than MP cells, greater migration
and invasion in vitro, and greater tumorigenicity in
vivo. Presumably not all SP cells in the preparations in the
present study were CSCs, yet the strong stem cell-like phenotype of
the SP preparations indicates that they are highly enriched in CSCs
and can be considered a substitute for CSCs in experimental
studies.
A number of studies have used an array of marker
proteins to identify and enrich for liver CSCs, including CD13, and
CD44 (9,12), in addition to the ABC transporter
ABCG2, which pumps Hoechst dye out of stem cells (27,28).
Despite this work, a standardized ‘signature’ of marker proteins
for liver CSCs has not been developed. The comparison of SP and MP
cells prepared under identical conditions from the HCCLM3 line
moves closer to defining a signature by showing that the mRNAs
encoding CD13, CD44 and ABCG2 are expressed at higher levels in SP
cells. These findings provide additional evidence that SP cells are
highly enriched in CSCs, and they suggest that SP cells are
heterogeneous in origin.
In the present study, the expression levels of the
transcription factor genes Klf4, Sox2 and Nanog were
also higher in SP cells compared with MP cells. Therefore these
factors may cooperatively maintain the CSC phenotype, perhaps by
regulating the self-renewal and pluripotency of embryonic stem
cells (30,31). The experimental system described in
the present study may inform future xperiments to test directly
whether and how these genes are involved in the CSC-like phenotype
of SP cells in HCC.
In conclusion, the present study provides evidence
that the proportion of SP cells in human HCC cell lines correlates
with metastatic potential, and the liver SP cells show the
characteristics expected of CSCs, implicating them in HCC
metastasis. Further studies on the identification and
characterization of SP cells using clinical HCC specimens should be
performed to contribute to the understanding of how SP cells are
involved in these disease processes.
Acknowledgements
The authors thank Armando Chapin Rodríguez, PhD for
his language editing, which substantially improved the quality of
the manuscript. This study was funded by the National Natural
Science Foundation of China (grant nos. 81160262, 81260331 and
81360312), National Science and Technology Major Project of the
Ministry of Science and Technology of China (grant no.
2012ZX10002010001009).
References
|
1
|
Jemal A, Bray F, Center MM, Ferlay J, Ward
E and Forman D: Global cancer statistics. CA Cancer J Clin.
61:69–90. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Guo Z, Zhong JH, Jiang JH, Zhang J, Xiang
BD and Li LQ: Comparison of survival of patients with BCLC stage A
hepatocellular carcinoma after hepatic resection or transarterial
chemoembolization: A propensity score-based analysis. Ann Surg
Oncol. 21:3069–3076. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Chun JM, Kwon HJ, Sohn J, Kim SG, Park JY,
Bae HI, Yun YK and Hwang YJ: Prognostic factors after early
recurrence in patients who underwent curative resection for
hepatocellular carcinoma. J Surg Oncol. 103:148–151. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Wu JC, Huang YH, Chau GY, Su CW, Lai CR,
Lee PC, Huo TI, Sheen IJ, Lee SD and Lui WY: Risk factors for early
and late recurrence in hepatitis B-related hepatocellular
carcinoma. J Hepatol. 51:890–897. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Gupta PB, Chaffer CL and Weinberg RA:
Cancer stem cells: Mirage or reality? Nat Med. 15:1010–1012. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Kucia M, Reca R, Miekus K, Wanzeck J,
Wojakowski W, Wieczorek Janowska A, Ratajczak J and Ratajczak MZ:
Trafficking of normal stem cells and metastasis of cancer stem
cells involve similar mechanisms: Pivotal role of the SDF-1-CXCR4
axis. Stem Cells. 23:879–894. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Guo Z, Li LQ, Jiang JH, Ou C, Zeng LX and
Xiang BD: Cancer stem cell markers correlate with early recurrence
and survival in hepatocellular carcinoma. World J Gastroenterol.
20:2098–2106. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Pang RW and Poon RT: Cancer stem cell as a
potential therapeutic target in hepatocellular carcinoma. Curr
Cancer Drug Targets. 12:1081–1094. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Zhu Z, Hao X, Yan M, Yao M, Ge C, Gu J and
Li J: Cancer stem/progenitor cells are highly enriched in
CD133+CD44+ population in hepatocellular carcinoma. Int J Cancer.
126:2067–2078. 2010.PubMed/NCBI
|
|
10
|
Yamashita T, Honda M, Nakamoto Y, Baba M,
Nio K, Hara Y, Zeng SS, Hayashi T, Kondo M, Takatori H, et al:
Discrete nature of EpCAM+ and CD90+ cancer stem cells in human
hepatocellular carcinoma. Hepatology. 57:1484–1497. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Yang ZF, Ho DW, Ng MN, Lau CK, Yu WC, Ngai
P, Chu PW, Lam CT, Poon RT and Fan ST: Significance of CD90+ cancer
stem cells in human liver cancer. Cancer Cell. 13:153–166. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Haraguchi N, Ishii H, Mimori K, Tanaka F,
Ohkuma M, Kim HM, Akita H, Takiuchi D, Hatano H, Nagano H, et al:
CD13 is a therapeutic target in human liver cancer stem cells. J
Clin Invest. 120:3326–3339. 2010. View
Article : Google Scholar : PubMed/NCBI
|
|
13
|
Kondo T, Setoguchi T and Taga T:
Persistence of a small subpopulation of cancer stem-like cells in
the C6 glioma cell line. Proc Natl Acad Sci USA. 101:781–786. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Teshima K, Nara M, Watanabe A, Ito M,
Ikeda S, Hatano Y, Oshima K, Seto M, Sawada K and Tagawa H:
Dysregulation of BMI1 and microRNA-16 collaborate to enhance an
anti-apoptotic potential in the side population of refractory
mantle cell lymphoma. Oncogene. 33:2191–2203. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Wang J, Guo LP, Chen LZ, Zeng YX and Lu
SH: Identification of cancer stem cell-like side population cells
in human nasopharyngeal carcinoma cell line. Cancer Res.
67:3716–3724. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Ho MM, Ng AV, Lam S and Hung JY: Side
population in human lung cancer cell lines and tumors is enriched
with stem-like cancer cells. Cancer Res. 67:4827–4833. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Xu XT, Xu Q, Tong JL, Zhu MM, Nie F, Chen
X, Xiao SD and Ran ZH: MicroRNA expression profiling identifies
miR-328 regulates cancer stem cell-like SP cells in colorectal
cancer. Br J Cancer. 106:1320–1330. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Dubrovska A, Hartung A, Bouchez LC, Walker
JR, Reddy VA, Cho CY and Schultz PG: CXCR4 activation maintains a
stem cell population in tamoxifen-resistant breast cancer cells
through AhR signalling. Br J Cancer. 107:43–52. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Yasuda K, Torigoe T, Morita R, Kuroda T,
Takahashi A, Matsuzaki J, Kochin V, Asanuma H, Hasegawa T, Saito T,
et al: Ovarian cancer stem cells are enriched in side population
and aldehyde dehydrogenase bright overlapping population. PLoS One.
8:e681872013. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Van den Broeck A, Vankelecom H, Van Delm
W, Gremeaux L, Wouters J, Allemeersch J, Govaere O, Roskams T and
Topal B: Human pancreatic cancer contains a side population
expressing cancer stem cell-associated and prognostic genes. PloS
One. 8:e739682013. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Marquardt JU, Raggi C, Andersen JB, Seo D,
Avital I, Geller D, Lee YH, Kitade M, Holczbauer A, Gillen MC, et
al: Human hepatic cancer stem cells are characterized by common
stemness traits and diverse oncogenic pathways. Hepatology.
54:1031–1042. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Chiba T, Kita K, Zheng YW, Yokosuka O,
Saisho H, Iwama A, Nakauchi H and Taniguchi H: Side population
purified from hepatocellular carcinoma cells harbors cancer stem
cell-like properties. Hepatology. 44:240–251. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Goodell MA, Brose K, Paradis G, Conner AS
and Mulligan RC: Isolation and functional properties of murine
hematopoietic stem cells that are replicating in vivo. J Exp Med.
183:1797–1806. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Wu C and Alman BA: Side population cells
in human cancers. Cancer Lett. 268:1–9. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Tang ZY, Ye SL, Liu YK, Qin LX, Sun HC, Ye
QH, Wang L, Zhou J, Qiu SJ, Li Y, et al: A decade's studies on
metastasis of hepatocellular carcinoma. J Cancer Res Clin Oncol.
130:187–196. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Shi GM, Xu Y, Fan J, Zhou J, Yang XR, Qiu
SJ, Liao Y, Wu WZ, Ji Y, Ke AW, et al: Identification of side
population cells in human hepatocellular carcinoma cell lines with
stepwise metastatic potentials. J Cancer Res Clin Oncol.
134:1155–1163. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Huang FF, Zhang L, Wu DS, Yuan XY, Yu YH,
Zhao XL, Chen FP and Zeng H: PTEN regulates BCRP/ABCG2 and the side
population through the PI3K/Akt pathway in chronic myeloid
leukemia. PLoS One. 9:e882982014. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Zhang G, Wang Z, Luo W, Jiao H, Wu J and
Jiang C: Expression of potential cancer stem cell marker ABCG2 is
associated with malignant behaviors of hepatocellular carcinoma.
Gastroenterol Res Pract. 2013:7825812013. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Gagliardi A, Mullin NP, Ying TZ, Colby D,
Kousa AI, Halbritter F, Weiss JT, Felker A, Bezstarosti K, Favaro
R, et al: A direct physical interaction between Nanog and Sox2
regulates embryonic stem cell self-renewal. Embo J. 32:2231–2247.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Wang Z, Oron E, Nelson B, Razis S and
Ivanova N: Distinct lineage specification roles for NANOG, OCT4 and
SOX2 in human embryonic stem cells. Cell Stem Cell. 10:440–454.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Kim MO, Kim S, Cho Y, Nadas J, Jeong C,
Yao K, Kim DJ, Yu D, Keum Y, Lee K, et al: ERK1 and ERK2 regulate
embryonic stem cell self-renewal through phosphorylation of Klf4.
Nat Struct Mol Biol. 19:283–290. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Wu C, Wei Q, Utomo V, Nadesan P, Whetstone
H, Kandel R, Wunder JS and Alman BA: Side population cells isolated
from mesenchymal neoplasms have tumor initiating potential. Cancer
Res. 67:8216–8222. 2007. View Article : Google Scholar : PubMed/NCBI
|