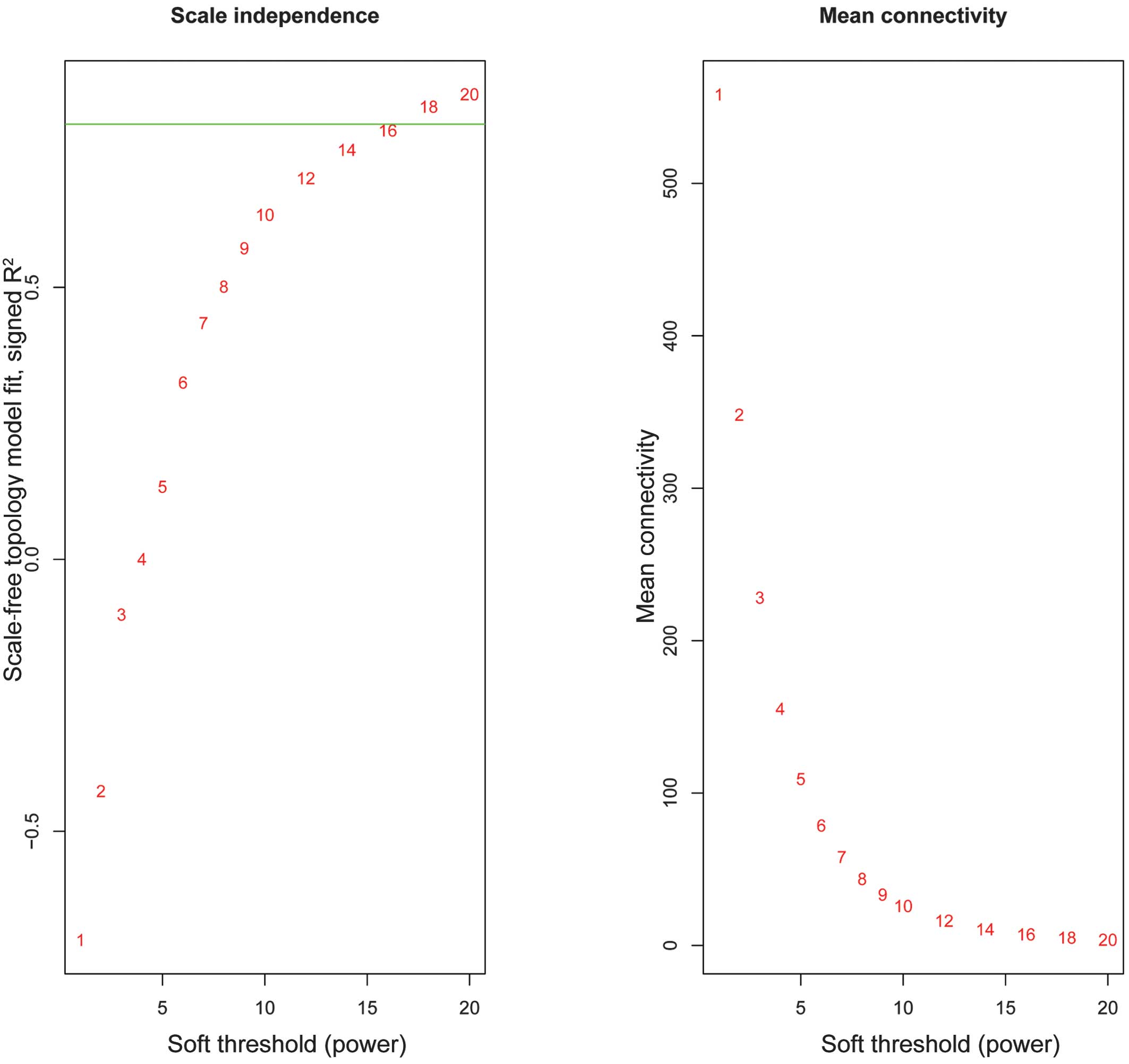
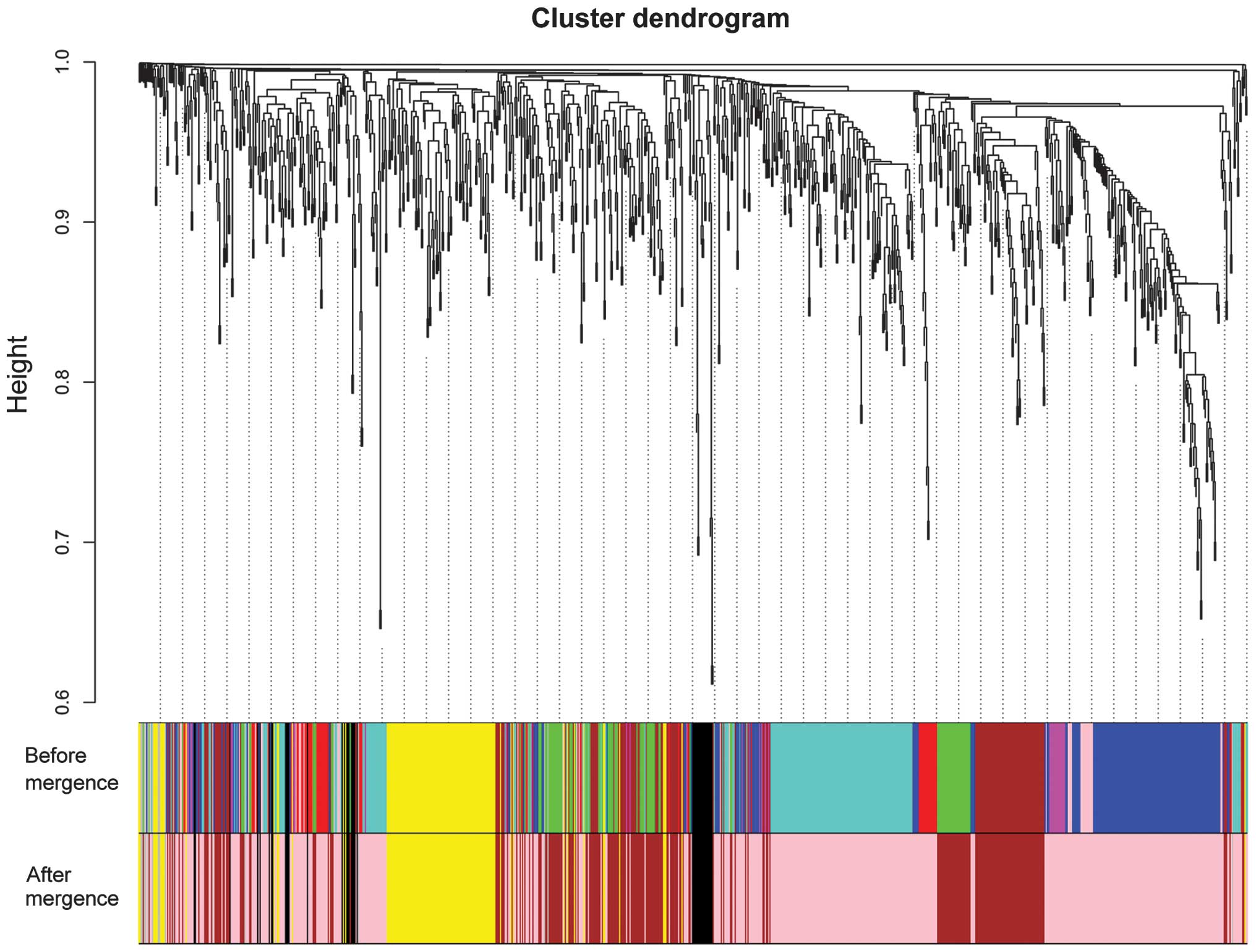
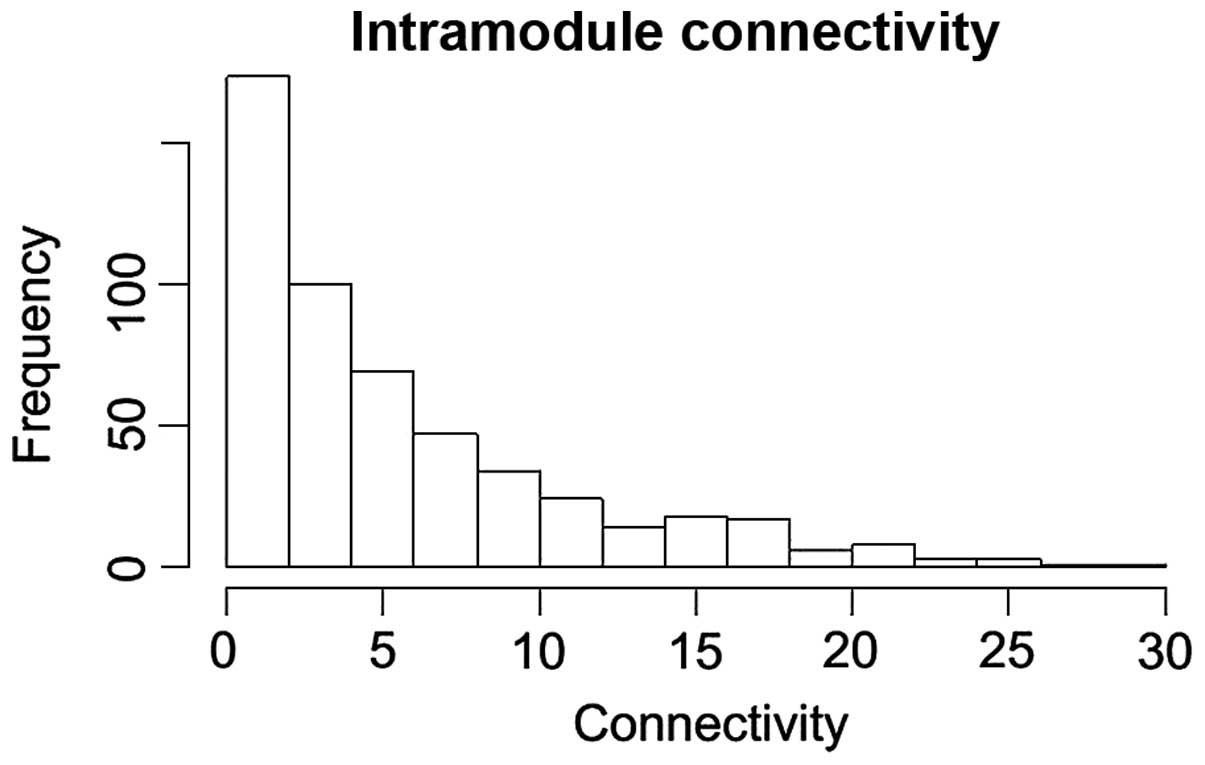
|
1
|
Batsakis JG: Tumors of the head and neck:
Clinical and pathological considerations (2nd). Williams &
Wilkins. Baltimore: 1979. View Article : Google Scholar
|
|
2
|
Million R, Cassisi N and Clark J: Cancer
of the head and neck. Cancer: Principles and Practice of Oncology
(3rd). (JB Lippincott, PA). 488–590. 1989.
|
|
3
|
Hunter KD, Parkinson EK and Harrison PR:
Profiling early head and neck cancer. Nat Rev Cancer. 5:127–135.
2005. View
Article : Google Scholar : PubMed/NCBI
|
|
4
|
Zhang H, Chen X, Jin Y, Liu B and Zhou L:
Overexpression of Aurora-A promotes laryngeal cancer progression by
enhancing invasive ability and chromosomal instability. Eur Arch
Otorhinolaryngol. 269:607–614. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Bazan V, Zanna I, Migliavacca M,
Sanz-Casla MT, Maestro ML, Corsale S, Macaluso M, Dardanoni G,
Restivo S, Quintela PL, et al: Prognostic significance of p16INK4a
alterations and 9p21 loss of heterozygosity in locally advanced
laryngeal squamous cell carcinoma. J Cell Physiol. 192:286–293.
2002. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Rocco JW and Sidransky D: p16
(MTS-1/CDKN2/INK4a) in cancer progression. Exp Cell Res. 264:42–55.
2001. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Ogino T, Shigyo H, Ishii H, Katayama A,
Miyokawa N, Harabuchi Y and Ferrone S: HLA class I antigen
down-regulation in primary laryngeal squamous cell carcinoma
lesions as a poor prognostic marker. Cancer Res. 66:9281–9289.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Dong Y, Sui L, Sugimoto K, Tai Y and
Tokuda M: Cyclin D1-CDK4 complex, a possible critical factor for
cell proliferation and prognosis in laryngeal squamous cell
carcinomas. Int J Cancer. 95:209–215. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Vielba R, Bilbao J, Ispizua A, Zabalza I,
Alfaro J, Rezola R, Moreno E, Elorriaga J, Alonso I, Baroja A and
de la Hoz C: p53 and cyclin D1 as prognostic factors in squamous
cell carcinoma of the larynx. Laryngoscope. 113:167–172. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Lauriola L, Michetti F, Maggiano N, Galli
J, Cadoni G, Schäfer BW, Heizmann CW and Ranelletti FO: Prognostic
significance of the Ca(2+) binding protein S100A2 in laryngeal
squamous-cell carcinoma. Int J Cancer. 89:345–349. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Sun Y, Liu M, Yang B, Li B and Lu J: Role
of siRNA silencing of MMP-2 gene on invasion and growth of
laryngeal squamous cell carcinoma. Eur Arch Otorhinolaryngol.
265:1385–1391. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Liu WW, Zeng ZY, Wu QL, Hou JH and Chen
YY: Overexpression of MMP-2 in laryngeal squamous cell carcinoma: A
potential indicator for poor prognosis. Otolaryngol Head Neck Surg.
132:395–400. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Cao WF, Zhang LY, Liu MB, Tang PZ, Liu ZH
and Sun BC: Prognostic significance of stomatin-like protein 2
overexpression in laryngeal squamous cell carcinoma: Clinical,
histologic and immunohistochemistry analyses with tissue
microarray. Hum Pathol. 38:747–752. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Cao WF, Zhang LY, Zhang B, Liu MB, Liu ZH
and Sun BC: Relationship between SLP-2 expression and prognosis in
laryngeal squamous cell carcinoma and mammary invasive carcinoma.
Zhonghua Bing Li Xue Za Zhi. 39:332–337. 2010.(In Chinese).
PubMed/NCBI
|
|
15
|
Smyth GK Limma: Linear models for
microarray data. Bioinformatics and Computational Biology Solutions
using R and Bioconductor. Gentleman R, Carey V, Dudoit S, Irizarry
R and Huber W: Springer. (New York). 397–420. 2005. View Article : Google Scholar
|
|
16
|
Langfelder P and Horvath S: WGCNA: An R
package for weighted correlation network analysis. BMC
Bioinformatics. 9:5592008. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Janssens F, Zhang L, De Moor B and Glänzel
W: Hybrid clustering for validation and improvement of
subject-classification schemes. Inform Process Manag. 45:683–702.
2009. View Article : Google Scholar
|
|
18
|
Kanehisa M, Goto S, Sato Y, Furumichi M
and Tanabe M: KEGG for integration and interpretation of
large-scale molecular data sets. Nucleic Acids Res. 40(Database
Issue): D109–D114. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Yu G, Wang LG, Han Y and He QY:
ClusterProfiler: An R package for comparing biological themes among
gene clusters. OMICS. 16:284–287. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Kufer TA, Silljé HH, Körner R, Gruss OJ,
Meraldi P and Nigg EA: Human TPX2 is required for targeting
Aurora-A kinase to the spindle. J Cell Biol. 158:617–623. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Asteriti IA, Rensen WM, Lindon C, Lavia P
and Guarguaglini G: The Aurora-A/TPX2 complex: A novel oncogenic
holoenzyme? Biochim Biophys Acta. 1806:230–239. 2010.PubMed/NCBI
|
|
22
|
Reiter R, Gais P, Jütting U, Steuer-Vogt
MK, Pickhard A, Bink K, Rauser S, Lassmann S, Höfler H, Werner M
and Walch A: Aurora kinase A messenger RNA overexpression is
correlated with tumor progression and shortened survival in head
and neck squamous cell carcinoma. Clin Cancer Res. 12:5136–5141.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Guan Z, Wang XR, Zhu XF, Huang XF, Xu J,
Wang LH, Wan XB, Long ZJ, Liu JN, Feng GK, et al: Aurora-A, a
negative prognostic marker, increases migration and decreases
radiosensitivity in cancer cells. Cancer Res. 67:10436–10444. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Chatrath P, Scott IS, Morris LS, Davies
RJ, Rushbrook SM, Bird K, Vowler SL, Grant JW, Saeed IT, Howard D,
et al: Aberrant expression of minichromosome maintenance protein-2
and Ki67 in laryngeal squamous epithelial lesions. Br J Cancer.
89:1048–1054. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Pi JT, Lin Y, Quan Q, Chen LL, Jiang LZ,
Chi W and Chen HY: Overexpression of UHRF1 is significantly
associated with poor prognosis in laryngeal squamous cell
carcinoma. Med Oncol. 30:6132013. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Yang C, Wang Y, Zhang F, Sun G, Li C, Jing
S, Liu Q and Cheng Y: Inhibiting UHRF1 expression enhances
radiosensitivity in human esophageal squamous cell carcinoma. Mol
Biol Rep. 40:5225–5235. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Matsumoto M, Furihata M, Ishikawa T,
Ohtsuki Y and Ogoshi S: Comparison of deregulated expression of
cyclin D1 and cyclin E with that of cyclin-dependent kinase 4
(CDK4) and CDK2 in human oesophageal squamous cell carcinoma. Br J
Cancer. 80:256–261. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Dong Y, Sui L, Tai Y, Sugimoto K and
Tokuda M: The overexpression of cyclin-dependent kinase (CDK) 2 in
laryngeal squamous cell carcinomas. Anticancer Res. 21:103–108.
2000.
|
|
29
|
Dong Y, Sui L, Tai Y, Sugimoto K, Hirao T
and Tokuda M: Prognostic significance of cyclin E overexpression in
laryngeal squamous cell carcinomas. Clin Cancer Res. 6:4253–4258.
2000.PubMed/NCBI
|
|
30
|
Jiang W, Jimenez G, Wells NJ, Hope TJ,
Wahl GM, Hunter T and Fukunaga R: PRC1: A human mitotic
spindle-associated CDK substrate protein required for cytokinesis.
Mol Cell. 2:877–885. 1998. View Article : Google Scholar : PubMed/NCBI
|