Introduction
Worldwide, esophageal cancer is the eighth most
common type of cancer, and in 2012 there were 456,000 novel cases
and 400,000 mortalities (1).
Esophageal squamous cell carcinoma (ESCC) is the most common type
of esophageal cancer (2); however,
early stage diagnosis and therapeutic outcomes of patients with
ESCC remain poor (3–5). Only 15–25% of patients with ESCC survive
for >5 years following diagnosis (3).
Esophagography, endoscopic ultrasonography,
endoscopy, computed tomography (CT) and 18F-fluorodeoxyglucose
positron emission tomography (FDG-PET) are commonly used in the
diagnosis and staging of ESCC. Endoscopy is the most sensitive
method for the detection and diagnosis of esophageal cancer.
Endoscopic screening has decreased the ESCC-associated mortality,
and esophagography, CT and FDG-PET are used to assess the invasion
and length of tumors, direct invasion to adjacent organs and lymph
node and distant metastasis (6).
There are numerous approaches for the treatment of
patients with ESCC, including surgery, endoscopic therapy,
chemotherapy and radiotherapy. Neoadjuvant chemotherapy and
radiotherapy is performed as a standard treatment for locally
advanced ESCC (7). Radiotherapy is
one of the most common therapeutic methods for the treatment of
patients with ESCC, and leads to the apoptosis of tumor cells by
directly or indirectly destroying the cell DNA (8). A previous study demonstrated that the
radiosensitivity of cells depends on the extent of damage the
radiation exerts on the DNA and the efficiency of the host in
repairing the DNA (9). As DNA damage
accumulates, it may lead to uncontrolled cell proliferation and
differentiation, which may result in tumorigenesis. DNA damage may
be prevented by DNA repair genes; X-ray repair cross complementing
protein 1 (XRCC1) is an important gene for DNA repair (8). It is significant in maintaining
chromosomal stability. An elevated level of sister chromatid
exchange, widely used as an indicator of genetic damage, is
characteristic of XRCC1 functional deficiency, and single
nucleotide polymorphisms (SNPs) of XRCC1 has been associated with a
higher risk of cancer (10). SNP is a
variation in a single nucleotide that may occur at a specific
position in the genome, where each variation is present to a
certain degree within a population. The repairing ability of XRCC1
is negatively associated with the radiosensitivity of cells
(11).
The present study used polymerase chain reaction to
detect SNPs in patients with ESCC. The present study compared the
curative effect of radiotherapy in these patients with various
genotypes and investigated the association between 4 SNPs of XRCC1
and the sensitivity of radiotherapy in patients with ESCC.
Materials and methods
Patients
The present study was approved by the Internal
Review Board of Chaozhou People's Hospital (Chaozhou, China). In
addition, written informed consent was obtained from the patients
involved in the present study. The present study recruited 175
patients with ESCC that were pathologically diagnosed with ESCC and
received radiotherapy as an initial treatment at Chaozhou People's
Hospital between December 2011 and December 2013. Among these
patients, DNA was successfully amplified and 4 SNPs of XRCC1 were
sequenced and analyzed in the blood samples of 112 patients, which
consisted of 81 men and 31 women (average age, 60.6 years; median
age, 60.0 years). The location of tumors in the 112 patients was
cervical and upper thoracic in 10 patients, middle thoracic in 71
patients and lower thoracic in 31 patients. According to the 2010
Clinical Non-Operative Treatment of Esophageal Cancer Staging
Standard (12), there were 28
patients with stage II and 84 patients with stage III ESCC. All the
patients were free from distant metastasis and major organ
dysfunction at diagnosis. The average Karnofsky score of the
patients was ≥70 (13). Written
informed consent was obtained from all patients. The present study
was approved by the Internal Review Board of Chaozhou People's
Hospital.
Radiation treatment
All the patients received radiation treatment, which
was either three-dimensional conformal radiation therapy (3DCRT; 67
patients) or intensity-modulated radiation therapy (IMRT; 45
patients). The radiation treatments were performed on patients in
the supine position with the head extended and using a fixed body
thermoplastic film. The patients underwent enhanced chest CT
(SOMATOM Definition AS; Siemens AG, Munich, Germany) (3DCRT
thickness, 5 mm; IMRT thickness, 3 mm) in order to visualize the
target region for radiation. Following CT, the images were
transferred to the radiotherapy treatment planning system (Eclipse™
version 8.6; Varian Medical Systems, Palo Alto, CA, USA). The
clinical target volume (CTV) comprised the gross tumor volume
visible on the CT scan (GTV), neck metastatic lymph nodes (GTVnd)
and lymphatic drainage region, which required an 0.8 cm extension
in the sagittal and coronal direction and a 5 cm extension upward
and downward from the standard irradiated region of GTV and GTVnd.
Appropriate adjustments were adopted to suit individual patients.
The present study administered a planning target volume (PTV)
extension of 0.5 cm according to the CTV region. The exposure dose
was GTV 60–64 Gy and PTV 50–54 Gy. All the patients were
administered with nedaplatin (20 mg/m2, weekly for 2–4
cycles) as synchronous chemotherapy.
Evaluation
At the end of the radiation treatment and 1 month
following, the 112 patients were administered with a barium swallow
(Qingdao Dongfeng Chemical Co., Ltd., Qingdao, China) and
subsequently underwent enhanced chest CT. According to the Response
Evaluation Criteria In Solid Tumors for curative effect (14), the patients were divided into the
following 4 groups: Complete response (CR), the tumor was not
observed for ≥4 weeks; partial response (PR), the maximal tumor
diameter was reduced by ≥30% for ≥4 weeks; stable disease (SD), the
maximal tumor diameter altered between PR and progressive disease
(PD); and PD, the maximum diameter of the tumor increased ≥20%. CR
and PR were considered as the valid (effective) group and SD and PD
were the null (non-effective) group.
Primer design and DNA
amplification
A Rapid DNA Extraction kit (Yaneng Biotechnology
Shenzhen Co., Ltd., Shenzhen, China) was used to extract genomic
DNA from peripheral blood samples from the patients prior to
radiotherapy. The present study designed 3 pairs of specific
primers using reference strains identified from the
GenBank® gene sequence library (National Institutes of
Health, Bethesda, MA, USA), and optimized them using Primer-BLAST
version 5.0 software (National Institutes of Health; available from
http://www.ncbi.nlm.nih.gov/tools/primer-blast/).
Table I details the primers used in
the present study, which were synthesized by Shanghai Invitrogen
Biotechnology Co., Ltd. (Shanghai, China). Polymerase chain
reaction (PCR) was performed using 3 pairs of specific forward and
reverse primers (DNA polymerase used in the PCR reaction and the
kit used for the master mix of the PCR reaction were provided by
Aidlab Biotechnologies Co., Ltd., Beijing, China) in a total volume
of 50 µl using a PCR machine (Life Express Thermal Cycler
TC-96/G/H(b); Hangzhou Bioer Technology Co., Ltd, Zhejiang, China).
The PCR products (7 µl) were run on a 2% agarose gel (Biowest, Hong
Kong, China) using electrophoresis at 210 volts for 15 minutes
(ethidium bromide, Aidlab Biotechnologies Co., Ltd.). A DNA
Molecular Weight Marker II (200 bp; Aidlab Biotechnologies Co.,
Ltd.) was used as a reference molecular weight ladder. Target gene
fragment amplification bands were observed under UV lamps.
 | Table I.Primers used for restriction fragment
length polymorphism-PCR for 4 SNPs of the X-ray repair cross
complementing protein 1 gene. |
Table I.
Primers used for restriction fragment
length polymorphism-PCR for 4 SNPs of the X-ray repair cross
complementing protein 1 gene.
| SNP | Primer, 5′-3′ | PCR, bp | Temperature, °C |
|---|
|
rs3213245,c.-77C>T | F,
CACTTTAGCCAGCGCAGGG | 392 | 60 |
|
| R,
GGAAGTTCACCTATGGGCTCT |
|
|
|
c26304t,Arg194Trp | F,
TTTGGCTTGAGTTTTGTACGGTT | 595 | 62 |
|
| R,
CGCTGGCTGTGACTATGAAGGGA |
|
|
| G27466A,Arg280His
and | F,
CTCTTTGTCTTCTCCAGTGCCA | 752 | 65 |
|
G28152A,Arg399Gln | R,
CACAGGATAAGGAGCAGGGTT |
|
|
Genotype analysis
Following bidirectional sequencing of the PCR
products, the present study analyzed the sequences using the
following software: BioEdit version 3.0 (Ibis BioSciences,
Carlsbad, CA, USA; available from http://www.mbio.ncsu.edu/bioedit/bioedit.html), Basic
Local Alignment Search Tool (National Institutes of Health;
available from http://blast.ncbi.nlm.nih.gov/Blast.cgi) and
DNASTAR® Primer Premier version 5.0 (DNASTAR, Inc.,
Madison, WI, USA). Subsequently, the present study compared the
coding region of XRCC1 with the GenBank® gene library
reference sequence and performed an analysis on the polymorphisms
of the variations observed in the sequences. In the present study,
the 112 collected identical sequences were merged and divided into
8 genotypes as specified in Table
II.
 | Table II.Association between genotypes and the
curative effect of radiotherapy in patients with esophageal
squamous cell carcinoma. |
Table II.
Association between genotypes and the
curative effect of radiotherapy in patients with esophageal
squamous cell carcinoma.
|
|
|
| Curative effect,
n |
|---|
|
|
|
|
|
|---|
| Genotypes | SNP mutation | Total, n | Null group | Valid group |
|---|
| Total |
| 112 | 17 | 95 |
| T+Arg+Arg+Arg | None | 35 | 9 | 26 |
| C+Arg+Arg+Arg | 1st | 11 | 2 | 9 |
| T+Trp+Arg+Arg | 2nd | 10 | 1 | 9 |
| T+Arg+His+Arg | 3rd | 8 | 1 | 7 |
| T+Arg+Arg+Gln | 4th | 37 | 3 | 34 |
| C+Arg+Arg+Gln | 1st and 4th | 7 | 1 | 6 |
| T+Trp+His+Arg | 2nd and 3rd | 1 | 0 | 1 |
| T+Arg+His+Gln | 3rd and 4th | 3 | 0 | 3 |
Statistics approach
SPSS version 16.0 software (SPSS, Inc., Chicago, IL,
USA) was used for the analysis of data. Differences in sensitivity
to radiotherapy between genotype groups were analyzed using the
χ2 test. P<0.05 was considered to indicate a
statistically significant difference.
Results
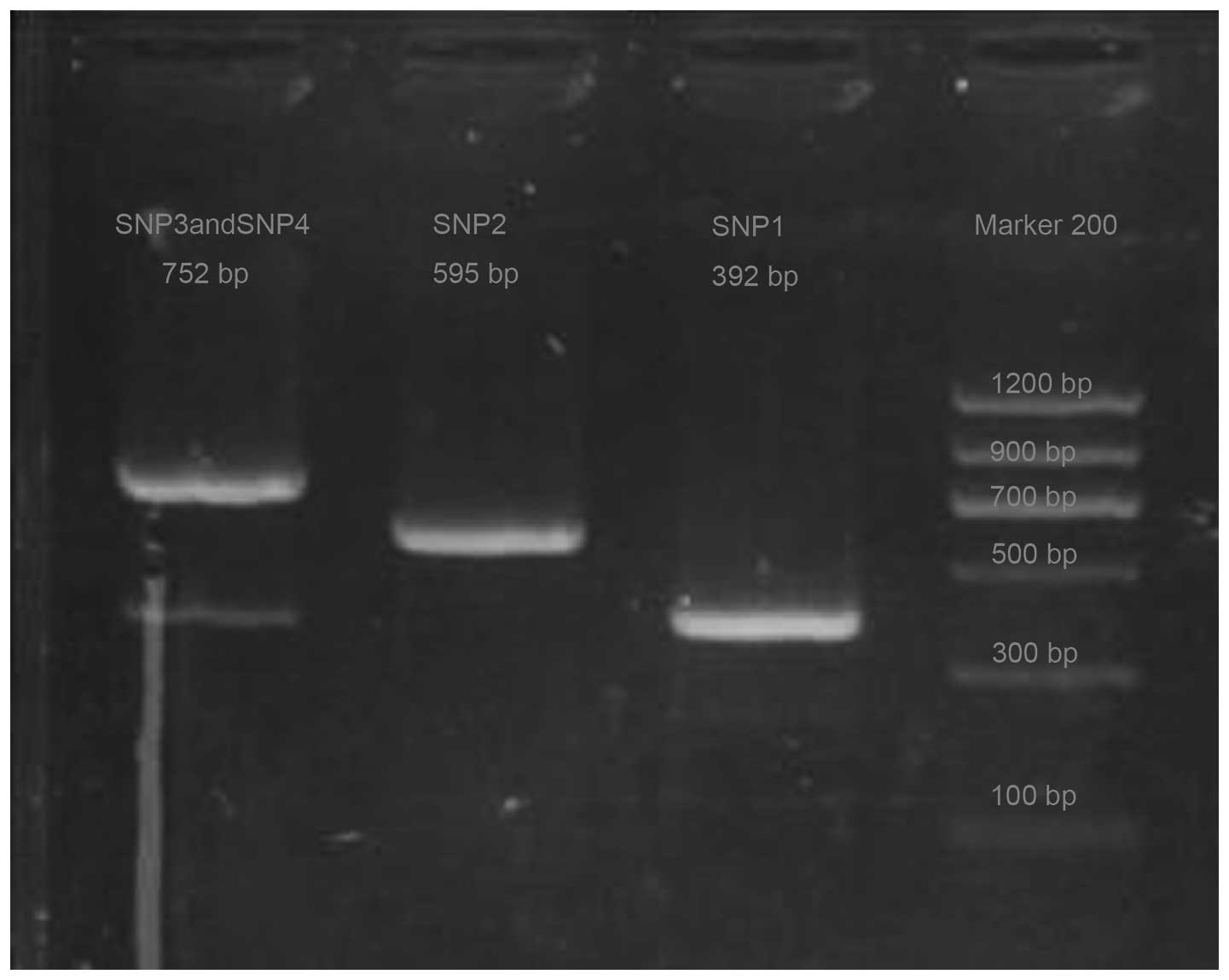
PCR products of 4 SNPs of XRCC1
Following DNA amplification and electrophoresis
analysis, the 112 blood samples of the patients with ESCC exhibited
specific DNA bands at 392, 595 and 752 bp, which corresponded to
the 4 SNPs identified in XRCC1 (Fig.
1).
Mutations in 4 SNPs of XRCC1
An analysis of the base alterations identified in
the 4 SNPS demonstrated that the non-coding base alterations in the
1st SNP locus mutation (rs3213245, C.-77 C>T) indicated the
position of the 77th base ahead of the initiation codon (ATG). In
addition, mutations in the 2nd, 3rd and 4th SNPs resulted in
alterations of the following amino acids: SNP locus mutation
replaced arginine with tryptophan (C26304T, Arg194Trp); 3rd SNP
locus mutation replaced arginine with histidine (G27466A,
Arg280His); 4th SNP locus mutation replaced arginine with glutamine
(G28152A, Arg399Gln).
According to the gene polymorphisms of the samples,
the present study compared the XRCC1 gene sequence with the coding
reference sequence. There were certain identical sequences of XRCC1
in the 112 patient blood samples, which were divided by the present
study into 8 groups. The present study identified that the group
with the majority of patients, in which 35 gene sequences shared
the same sequence in the reference strain and its 4 SNPs, had no
mutations. This group was defined as Chaozhou Local Strains. There
were 11 sequences that possessed the 1st SNP locus mutation
(rs3213245.C.-77 C>T); 10 sequences possessed the 2nd SNP locus
mutation (C26304T, Arg194Trp); 8 gene sequences possessed the 3rd
SNP locus mutation (G27466A, Arg280His); and 37 sequences possessed
the 4th SNP locus mutation (G28152A, Arg399Gln). There were 7 gene
sequences that possessed the 1st and 4th SNP locus, 1 gene sequence
possessed the 2nd and 3rd SNP locus, and 3 gene sequences possessed
the 3rd and 4th SNP locus mutations (Table II).
Follow-up of patients
By the end of February 2014, the follow-up time of
the patients ranged between 2.0 and 26.4 months (median, 16.2
months). The follow-up rate was 95.5%. Survival time was defined as
the time between the beginning of radiotherapy and the last
follow-up or patient mortality. In total, 31 patients succumbed to
ESCC during follow-up, which consisted of 13 patients with local
tumor recurrence, 8 patients with distant metastasis and 10
patients with tumor recurrence and metastasis.
Efficacy of radiotherapy
The effective response rate of radiotherapy in the
112 patients was 84.8%. Table III
summarizes the association between the efficacy of radiotherapy and
patient characteristics. The efficacy rate of radiotherapy in
patients with stage II ESCC was increased compared with patients
with stage III ESCC (100 and 79.8%, respectively;
χ2=6.681; P=0.01). The efficacy rate of radiotherapy had
no significant association with the gender and age of the patients,
tumor location and mode of radiotherapy.
 | Table III.Association between the
characteristics of patients with esophageal squamous cell carcinoma
and the efficacy rate of radiotherapy. |
Table III.
Association between the
characteristics of patients with esophageal squamous cell carcinoma
and the efficacy rate of radiotherapy.
|
| Curative effect,
n |
|
|
|---|
|
|
|
|
|
|---|
| Characteristic | Null group | Valid group | χ2 | P-value |
|---|
| Total | 17 | 95 |
|
|
| Gender |
|
|
|
|
|
Male | 12 | 69 | 0.030 | 0.862 |
|
Female | 5 | 26 |
|
|
| Age, years |
|
|
|
|
|
≥60 | 10 | 48 | 0.398 | 0.528 |
|
﹤60 | 7 | 47 |
|
|
| Tumor location |
|
|
|
|
|
Cervical/upper thoracic | 3 | 7 | 1.963 | 0.375 |
| Middle
thoracic | 10 | 61 |
|
|
| Lower
thoracic | 4 | 27 |
|
|
| Tumor stage |
|
|
|
|
| II | 0 | 28 | 6.681 | 0.010 |
|
III | 17 | 67 |
|
|
| Radiotherapy |
|
|
|
|
|
3DCRT | 9 | 58 | 0.395 | 0.530 |
|
IMRT | 8 | 37 |
|
|
Association between SNP mutations and
ESCC sensitivity to radiotherapy
The efficacy rate of radiotherapy in patients with
no SNP mutations was 74.3%, compared to 89.6% in patients with ≥1
SNP locus mutation (χ2=4.389; P=0.036; Table IV). The efficacy rate of radiotherapy
in patients containing only the 4th SNP locus mutation (G28152 A,
Arg399Gln) and for all patients containing mutations in the 4th and
other SNP locus was 91.9 and 91.5%, respectively (Table II). Compared with the non-mutation
group, the difference was statistically significant
(χ2=4.014 and χ2=4.451, respectively; P=0.045
and P=0.036, respectively; Tables V
and VI). The difference in the
efficacy of radiotherapy in patients with Chaozhou Local Strains
and those with only 1 SNP locus mutation (1st, 2nd or 3rd) was not
significantly different (P>0.05).
 | Table IV.Association between SNP mutations and
the radiotherapy sensitivity of patients with esophageal squamous
cell carcinoma. |
Table IV.
Association between SNP mutations and
the radiotherapy sensitivity of patients with esophageal squamous
cell carcinoma.
|
| Curative effect,
n |
|
|
|---|
|
|
|
|
|
|---|
| SNP | Null group | Valid group | χ2 | P-value |
|---|
| Total | 17 | 95 |
|
|
| None | 9 | 26 | 4.389 | 0.036 |
| ≥1 | 8 | 69 |
|
|
 | Table V.Efficacy rate of radiation in
patients with only the 4th SNP locus mutation. |
Table V.
Efficacy rate of radiation in
patients with only the 4th SNP locus mutation.
|
| Curative effect,
n |
|
|
|---|
|
|
|
|
|
|---|
| SNP | Null group | Valid group | χ2 | P-value |
|---|
| None | 9 | 26 | 4.014 | 0.045 |
| Only 4th | 3 | 34 |
|
|
 | Table VI.Efficacy rate of radiation in all
patients with the 4th SNP locus mutation. |
Table VI.
Efficacy rate of radiation in all
patients with the 4th SNP locus mutation.
|
| Curative effect,
n |
|
|
|---|
|
|
|
|
|
|---|
| SNP | Null group | Valid group | χ2 | P-value |
|---|
| None | 9 | 26 | 4.451 | 0.035 |
| All 4th | 4 | 43 |
|
|
The stratification analysis demonstrated that the
effective rate of radiotherapy in stage III patients with no SNP
mutations was 74.1% and those with ≥1 mutation was 91.2%
(χ2=4.403; P=0.036). The effective rate of radiotherapy
in patients with mutations in the 4th SNP locus and those
containing mutations in the 4th and other SNP locus was 92.3% and
93.5%, respectively, which was not significant compared to the no
mutation group (χ2=3.333 and χ2=3.312,
respectively; P=0.068 and P=0.073, respectively). Table VII summarizes the association
between the SNP genotypes and the sensitivity to radiotherapy in
patients with stage III ESCC.
 | Table VII.Association between genotypes and
radiotherapy sensitivity of patients with stage III esophageal
squamous cell carcinoma. |
Table VII.
Association between genotypes and
radiotherapy sensitivity of patients with stage III esophageal
squamous cell carcinoma.
|
| Curative effect,
n |
|---|
|
|
|
|---|
| SNP | Null group | Valid group |
|---|
| None | 7 | 20 |
| ≥1 | 5 | 52 |
| Only 4th | 2 | 25 |
| All 4th | 3 | 31 |
Discussion
As a radical treatment, radiotherapy is important in
the comprehensive management of esophageal cancer. Radiotherapy
induces the apoptosis of tumor cells by damaging cell DNA directly
or indirectly. However, the 5-year survival rate of patients with
ESCC that are treated solely with radiotherapy is <10% and the
recurrence rate is 60–80%. (12) It
has been reported that an increase in the radiotherapeutic dose
between 50 and 70 Gy does not achieve a corresponding improvement
in efficacy, which suggests that dose is not a key factor for the
efficacy of radiotherapy (15).
Clinically, patients with the same tumor stage often exhibit a
varied response to radiotherapy (15). This difference greatly depends on the
balance between the damage to the DNA and the DNA repair ability.
DNA damage and repair is a complex process that is comprised of
several enzymes and proteins. If a repair gene is mutated, the DNA
repairing capacity of the entire genome decreases, which may lead
to adverse effects, including tumorigenesis (16). The difference in the ability to repair
DNA damage also varies among individuals (17). The balance between DNA damage and
repair ultimately affects the sensitivity of cells to radiotherapy.
Currently, studies that focus on the genes that alter the
sensitivity of tumors to radiotherapy are increasing.
XRCC1 is the first gene that was revealed to affect
cell sensitivity to ionizing radiation (18). It is involved in repairing base
excision and single-strand breaks, which are caused by ionizing
radiation and oxidative damage (18).
The repairing ability of XRCC1 and radiosensitivity is negatively
associated (10). The human genome
mainly manifests itself in multi-nucleotide polymorphisms. In the
process of DNA repair, SNP mutations in XRCC1 alter the encoded
amino acid; therefore altering the structure and activity of the
repair enzymes encoded by XRCC1 (19–21). This
may be the key factor that causes the differences in DNA repairing
ability observed in individuals, and may be responsible for the
various outcomes of radiotherapy.
The identification of SNPs of XRCC1 is valuable to
predict the radiosensitivity of cells. At present, XRCC1 has 3
major SNP locus, which are located in exons 6, 9 and 10. These SNPs
cause alterations at C26304T to Arg194Trp, G27466A to Arg280His and
G28152A to Arg399Gln (22).
The present study identified that 4 SNP locus of
XRCC1, including a locus in the non coding region (rs3213245, C.-77
C>T) that has not been previously reported in radiotherapy, to
the best of our knowledge. This locus is hypothesized to affect the
transcription and mediation of the XRCC1 gene. Hao et al
(23) demonstrated that polymorphisms
of rs3213245, C.-77 C>T increased the binding of the XRCC1
promoter to the transcription inhibitory factor, which lead to a
decrease in the promoter activity and protein expression. The
present study identified that the 1st SNP locus mutation does not
have a significant effect on radiosensitivity of patients compared
with the non-mutation group; however, this may be due to a limited
number of samples. The efficacy of radiotherapy was 83.33% (15/18)
in the mutation group and 74.29% (26/35) in the non-mutation group
(P=0.355).
Previous studies concerning XRCC1 focus on cancer
susceptibility or risk. Sreeja et al (24) identified that 399 homozygous dominant
XRCC1 carriers were more likely to have lung cancer compared with
the heterozygous genotype carriers. Previous studies suggest that
polymorphisms of XRCC1 at Arg399Gln are associated with the
incidence of several types of cancer, including lung, stomach,
esophagal and bladder cancer and nasopharyngeal carcinoma (25–27). Yang
et al (25) demonstrated that
the XRCC1 Gln/Gln genotype was increased in individuals with ESCC
in a population in Taiwan. Therefore, XRCC1 is an important genetic
risk factor for tumor development. A meta-analysis of the XRCC1
gene Arg194Trp by Huang et al (26) indicates that polymorphisms in XRCC1
are a risk factor for cancer in China. Wu et al (27) investigated the polymorphisms of XRCC1
Arg194Trp and Arg399Gln, and the roles these polymorphisms play in
the development of esophageal cancer. The authors demonstrated that
the risk of developing esophageal cancer is associated with
polymorphisms of XRCC1 gene Arg194Trp, and indicate that homozygous
194Trp/Trp carriers with restriction endonuclease Pvu II have the
highest risk of esophageal cancer.
There have been numerous contradictory studies
concerning the association between XRCC1 and the sensitivity of
patients to radiotherapy. Liu et al (28) revealed an association between the
expression of XRCC1 and excision repair cross-complementation group
1 (ERCC1) and the efficacy of radiotherapy and the prognosis of
patients with ESCC. The authors indicated that the expression of
XRCC1 and ERCC1 may play a role in esophageal carcinogenesis.
However, the study did not reveal the association between XRCC1 and
radiosensitivity in esophageal carcinoma. Warnecke-Eberz et
al (29) investigated the
predictive value of ERCC1 and XRCC1 polymorphisms on the effect of
chemotherapy (cisplatin and 5-fluorouracil) in esophageal cancer.
The results demonstrated that ERCC1 may be a predictor for
chemotherapy, and XRCC1 Arg194Trp was not a suitable marker.
However, the authors concluded that SNPs of XRCC1 and ERCC1 may be
applied to treatment strategies for patients in the future.
Cornetta et al (30) used a 2
Gy X-ray irradiation on normal peripheral blood, which revealed
that the DNA loading in homozygous 399Gln/Gln blood was decreased
compared with the DNA loading in wild type and heterozygous DNA.
The authors concluded that polymorphisms in repairing genes may
affect the DNA repairing ability of an individual. Zhang et
al (31) identified SNPs in
hOGG1, XRCC1 and XRCC3 in the peripheral blood of 94 patients with
ESCC using restriction fragment length polymorphism-PCR and
revealed that polymorphisms at Arg399Gln of XRCC1 were clearly
associated with radiosensitivity.
The present study demonstrated that the effective
rate of radiotherapy is associated with patients with the 4th SNP
locus mutation, and the effective rate of radiotherapy in these
patients was significantly increased compared with non-mutation
patients. In addition, the present results indicated that the 4th
SNP locus mutation may be a marker for radiation efficacy in
patients with esophageal cancer. The present study identified that
the efficacy rate of radiation in patients with mutations at ≥1 SNP
locus is increased compared with patients with no mutations.
Although there is no significant difference in the efficacy rate of
radiation between patients with no mutations and patients with
mutations at the 1st, 2nd, 3rd or 4th SNP locus, additional study
is required due to the limited sample size in the present
study.
Based on the stratification analysis of patients
with stage III ESCC, the efficacy rate of radiotherapy in patients
with ≥1 SNP locus mutation was significantly increased compared
with the rate in patients with no mutations. The effective rate of
radiotherapy was not different in patients with mutations only at
the 4th SNP locus or with other locus combining with the 4th SNP
locus.
In conclusion, the sensitivity of radiotherapy to
ESCC is a complex process that involves multiple genes. The present
study suggests that SNP mutations are closely associated with the
sensitivity of radiotherapy in patients with ESCC. Specifically,
mutations in the 4th SNP locus (G28152A, Arg399Gln) of XRCC1
significantly improves the curative effect of radiotherapy.
Therefore, the 4th SNP locus may be a promising marker for the
sensitivity of radiotherapy. Mutations in the 1st SNP locus
(rs3213245, C.-77, C>T), which has never been previously
reported in the literature, is hypothesized to potentially increase
the sensitivity of radiotherapy. Understanding the association
between mutations of various SNPs and the sensitivity of
radiotherapy may aid in the development of individualized
radiotherapy for patients with ESCC.
Acknowledgements
The present study was supported by The Scientific
Research Fund of Guangdong Provincial Health Office (Guangzhou,
China; grant no. A2012838).
References
|
1
|
Torre LA, Bray F, Siegel RL, Ferlay J,
Lortet-Tieulent J and Jemal A: Global cancer statistics, 2012. CA
Cancer J Clin. 65:87–108. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Hongo M, Nagasaki Y and Shoji T:
Epidemiology of esophageal cancer: Orient to Occident. Effects of
chronology, geography and ethnicity. J Gastroenterol Hepatol.
24:729–735. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Gebski V, Burmeister B, Smithers BM, Foo
K, Zalcberg J and Simes J: Australasian Gastro-Intestinal Trials
Group: Survival benefits from neoadjuvant chemoradiotherapy or
chemotherapy in oesophageal carcinoma: A meta-analysis. Lancet
Oncol. 8:226–234. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Cunningham D, Allum WH, Stenning SP,
Thompson JN, Van de Velde CJ, Nicolson M, Scarffe JH, Lofts FJ,
Falk SJ, Iveson TJ, et al: MAGIC Trial Participants: Perioperative
chemotherapy versus surgery alone for resectable gastroesophageal
cancer. N Engl J Med. 355:11–20. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
van Hagen P, Hulshof MC, van Lanschot JJ,
Steyerberg EW, van Berge Henegouwen MI, Wijnhoven BP, Richel DJ,
Nieuwenhuijzen GA, Hospers GA, Bonenkamp JJ, et al: CROSS Group:
Preoperative chemo-radiotherapy for esophageal or junctional
cancer. N Engl J Med. 366:2074–2084. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Ohashi S, Miyamoto S, Kikuchi O, Goto T,
Amanuma Y and Muto M: Recent advances from basic and clinical
studies of esophageal squamous cell carcinoma. Gastroenterology.
149:1700–1715. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Merkow RP, Bilimoria KY, McCarter MD, Chow
WB, Ko CY and Bentrem DJ: Use of multimodality neoadjuvant therapy
for esophageal cancer in the United States: Assessment of 987
hospitals. Ann Surg Oncol. 19:357–364. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Yu X, Xiao H, Zhao B, Zhang X and Wang G:
DNA repair gene ERCC1 C118T polymorphism predicts sensitivity of
recurrent esophageal cancerto radiochemotherapy in a Chinese
population. Thorac Cancer. 6:714–718. 2015. View Article : Google Scholar
|
|
9
|
Zhang Z, Zhang Q, Wu M, Li N and Heng Z:
Effects of 60 Co γ rays on radiosensitivity of down-regulated cell
strain hOGG1 expressed genes. Lin Chuang Fang She Yi Xue Shou Ce.
6:238–241. 2006.(In Chinese).
|
|
10
|
Horton JK, Watson M, Stefanick DF,
Shaughnessy DT, Taylor JA and Wilson SH: XRCC1 and DNA polymerase β
in cellular protection against cytotoxic DNA single-strand breaks.
Cell Res. 18:48–63. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Zhang L, Yao R, Fang S, Wang X and Li X:
Polymorphisms of ERCC1 and XRCC1 predict the overall survival of
advanced gastric cancerpatients receiving oxaliplatin-based
chemotherapy. Int J Clin Exp Med. 8:18375–18382. 2015.PubMed/NCBI
|
|
12
|
Li Q, Wu SG, Gao JM, Xu JJ, Hu LY and Xu
T: Impact of esophageal cancer staging on overall survival and
disease-free survival based on the 2010 AJCC classification by
lymph nodes. J Radiat Res. 54:307–314. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Sachsenheimer W, Piotrowski W and Bimmler
T: Quality of life in patients with intracranial tumors on the
basis of Karnofsky's performance status. J Neurooncol. 13:177–181.
1992. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Kurokawa Y, Shibata T, Ando N, Seki S,
Mukaida H and Fukuda H: Which is the optimal response criteria for
evaluating preoperative treatment in esophagealcancer: RECIST or
histology? Ann Surg Oncol. 20:3009–3014. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Liu S, Zou S, Zhao J, Zhou B, Wan Z, Jia
H, Yuan K and He J: Association between polymorphism of XRCC1 gene
and susceptibility to esophageal carcinoma. Shi Yong Zhong Liu Za
Zhi. 28:253–260. 2013.(In Chinese).
|
|
16
|
Ayiheng Q and Bogela A: Study on laryngeal
cancer related on polymorphism of the Arg399Gln of XRCC1 DNA repair
gene in different nationalities in Xinjiang. Lin Chung Er Bi Yan
Hou Tou Jing Wai Ke Za Zhi. 27:948–951, 954. 2013.(In Chinese).
PubMed/NCBI
|
|
17
|
Kiyohara C, Takayama K and Nakanishi Y:
Association of genetic polymorphisms in the base excision repair
pathway with lung cancer risk: A meta-analysis. Lung Cancer.
54:267–283. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Zhou Y and Che G: Analysis of the
relationship between DNA repair gene polymorphisms and lung cancer
susceptibility. J Environ Occup Med. 40:551–554. 2013.
|
|
19
|
Hoeijmakers JH: DNA damage, aging, and
cancer. N Engl J Med. 361:1475–1485. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Ladiges WC: Mouse models of XRCC1 DNA
repair polymorphisms and cancer. Oncogene. 25:1612–1619. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Almeida KH and Sobol RW: A unified view of
base excision repair: Lesion-dependent protein complexes regulated
by post-translational modification. DNA Repair (Amst). 6:695–711.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Zhou Q, Zou BW, Xu Y, Xue JX, Meng MB, Liu
FJ, Deng L, Ma DY, Ao R and Lu Y: DNA repair gene polymorphisms and
clinical outcome of patients with primary small cell carcinoma of
the esophagus. Tumour Biol. 36:1539–1548. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Hao B, Wang H, Zhou K, Li Y, Chen X, Zhou
G, Zhu Y, Miao X, Tan W and Wei Q: Identification of genetic
variants in base excision repair pathway and their associations
with risk of esophageal squamous cell carcinoma. Cancer Res.
64:4378–4384. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Sreeja L, Syamala VS, Syamala V, Hariharan
S, Raveendran PB, Vijayalekshmi RV, Madhavan J and Ankathil R:
Prognostic importance of DNA repair gene polymorphisms of XRCC1
Arg399Gln and XPD Lys751Gln in lung cancer patients from India. J
Cancer Res Clin Oncol. 134:645–652. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Yang XR, Diehl S, Pfeiffer R, Chen CJ, Hsu
WL, Dosemeci M, Cheng YJ, Sun B, Goldstein AM and Hildesheim A:
Chinese and American Genetic Epidemiology of NPC Study Team:
Evaluation of risk factors for nasopharyngeal carcinoma in
high-risk nasopharyngeal carcinoma families in Taiwan. Cancer
Epidemiol Biomarkers Prev. 14:900–905. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Huang J, Zhang J, Zhao Y, Liao B, Liu J,
Li L, Liao M and Wang L: The Arg194Trp polymorphism in the XRCC1
gene and cancer risk in Chinese Mainland population: A
meta-analysis. Mol Biol Rep. 38:4565–4573. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Wu KS, Hou X, Shao G and Li Y:
Meta-analysis of X-ray repair cross complementing gene 1
polymorphisms and esophageal cancer risk. J Shantou Univ Med Col.
20:206–209, 212. 2007.
|
|
28
|
Liu S, Mu K, Zhang Z, Zeng M and Ge F: A
correlation study of the expression of XRCC1 and ERCC1 to the
effect of radiotherapy and prognosis in esophageal squamous cell
carcinoma. J Xinjiang Med Univ. 33:477–481. 2010.
|
|
29
|
Warnecke-Eberz U, Vallböhmer D, Alakus H,
Kütting F, Lurje G, Bollschweiler E, Wienand-Dorweiler A, Drebber
U, Hölscher AH and Metzger R: ERCC1 and XRCC1 gene polymorphisms
predict response to neoadjuvant radiochemotherapy in esophageal
cancer. J Gastrointest Surg. 13:1411–1421. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Cornetta T, Festa F, Testa A and Cozzi R:
DNA damage repair and genetic polymorphisms: Assessment of
individual sensitivity and repair capacity. Int J Radiat Oncol Biol
Phys. 66:537–545. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Zhang X, Aheli H, Zhang Z, Lv Y and Ge F:
Association study on single nucleotide polymorphism in hOGG1,
XRCC1, XRCC3 and radiosensitivity in esophageal cancer. J Xinjiang
Med Univ. 33:473–476. 2010.
|