Introduction
Hepatocellular carcinoma (HCC) is one of the most
prevalent forms of cancer worldwide, and is associated with chronic
liver diseases, including hepatitis B and/or hepatitis C viral
infection (1,2). The overall survival of HCC patients is
poor due to the high recurrence rate following hepatic resection
(3). Chronic viral inflammation and
reactive oxygen species (ROS), which are increased by chronic
inflammation, are considered as the most important factors in HCC
carcinogenesis (4).
Mitochondria are present in eukaryotic cells and are
ubiquitous organelles that generate energy in the form of ATP
through the process of oxidative phosphorylation (OXPHOS) (5). The human mitochondrial genome is 16,569
bp in length, with double-stranded circular DNA molecules
containing 37 genes encoding 13 peptides for the OXPHOS apparatus,
in addition to 22 transfer RNAs and 2 ribosomal RNAs, which are
required for protein synthesis in the mitochondria. Furthermore,
mitochondrial DNA (mtDNA) contains a non-coding region that
accommodates a unique displacement loop (D-loop), which controls
the transcription and replication of mtDNA. mtDNA is considered to
be more vulnerable to oxidative damage and to have a higher
mutation rate compared with nuclear DNA due to the lack of a repair
system and protective histones within the mitochondria (5,6). A number
of prevalent polymorphisms have been identified in mtDNA, and the
majority accumulate in the D-loop (or regulatory) region (7). Damage to and somatic mutations of mtDNA
may lead to impairment of the OXPHOS system and increased ROS
production, which subsequently enhances the rate of DNA mutations
(8). Numerous researchers have
reported that mtDNA mutations or polymorphisms, particularly in the
D-loop region, were detected in different types of cancer,
including human colorectal (9),
ovarian (10) and thyroid (11) cancer, in addition to HCC (12). Furthermore, the association between
mtDNA polymorphisms or mutations and the prognoses of cancer has
also been investigated, but the results have been ambiguous. In the
present study, the polymorphisms and mutations in the mtDNA D-loop
region of patients with HCC were analysed, and the clinical
significance of these variations was investigated.
Materials and methods
Patients and tissue specimens
A total of 140 patients with HCC who were
hospitalised for radical resection in the Department of
Hepatobiliary Surgery, The First Affiliated Hospital of Guangxi
Medical University (Nanning, China) from January 2003 to August
2012 were studied. Patients who died during the perioperative
period or those who exhibited recurrence within 2 months
post-surgery were excluded. All of the tumour tissues were
confirmed as HCC by histopathology. Of the 140 patients, 123 were
male and 17 were female and the median age was 47 years (range,
25–79 years). All tissues were placed and stored in liquid nitrogen
immediately after surgical resection, according to the guidelines
of the Human Tissue Research Committee of The First Affiliated
Hospital of Guangxi Medical University. Written informed consent
was obtained from each participant prior to enrolment, and this
study was approved by the Medical Ethics Committee of The First
Affiliated Hospital of Guangxi Medical University.
mtDNA extraction and D-loop
amplification
All tumour tissues were removed from liquid nitrogen
and refrigerated at −80°C until DNA extraction. Total cellular DNA
was extracted from the tumour tissue according to the
specifications of the TIANamp Genomic DNA kit (Tiangen Biotech Co.,
Ltd., Beijing, China), and was stored at −20°C until mtDNA
amplification. The mtDNA fragments were amplified using the
following primers: Forward, 5′-ATTCTAACCTGAATCGGAGG-3′; and
reverse, 5′-GATGCTTGCATGTGTAATCT-3′ (Sangon Biotech Co., Ltd.,
Shanghai, China). The fragments were 1, 528 bp in length, including
the complete D-loop region of 1,122 bp, as described previously
(13). Each DNA sample (50 ng) was
amplified using polymerase chain reaction (PCR; MJ Research PTC-200
Thermal Cycler; Bio-Rad Laboratories, Inc., Hercules, CA, USA). PCR
was performed in a final volume of 50 µl with the Taq™ Hot-Start
Version PCR kit (Takara Bio, Inc., Otsu, Japan). The thermal
profile was as follows: Initial incubation at 95°C for 5 min,
followed by 35 cycles of 95°C for 1 min, 55°C for 1 min and 72°C
for 2 min, and a final extension at 72°C for 7 min.
Direct sequencing and analysis of the
D-loop region of mtDNA
Aberrant PCR products were purified with a PCR
purification kit (Beijing Sunbiotech Co., Ltd., Beijing, China) and
were sequenced with an 3730xl DNA Analyzer (Applied Biosystems;
Thermo Fisher Scientific, Inc., Waltham, MA, USA). The direct
sequencing primers were the same as those used for amplification.
As there were numerous structures containing poly-C in the mtDNA
D-loop region, the following two additional primers were used for
PCR experiments (F2: 5′-AATACTTGACCACCTGTAG-′3; and F3:
5-′CCTATGTCGCAGTATCTGTC-′3) to sequence the entire mtDNA D-loop
region (13). Multiple primers
overlapped the sequenced regions. All of the variations were
confirmed by repeated analysis of the mtDNA extracted from the
tissue samples. All of these procedures were performed by Beijing
Sunbiotech Co., Ltd. (Beijing, China).
The sequences of the tumour tissues were compared
with the revised Cambridge Reference Sequence (GenBank®
NC_012920) using DNASTAR Lasergene software version 7.1 (DNASTAR,
Inc., Madison, WI, USA). Subsequently, the variations in the
sequences were considered polymorphisms or mutations according to
the mtDNA database (www.mitomap.org).
Follow-up
The patients with HCC were recommended to receive
postoperative adjuvant transcatheter hepatic arterial
chemoembolisation (TACE). All patients were followed up in the
outpatient clinic every 3 months, and their serum α-fetoprotein
levels were measured. Hepatic ultrasonography was performed every
2–4 months from the date of initial treatment until December 31,
2014, or mortality. If any signs of recurrence were suspected,
further investigation was performed using abdominal computed
tomography or magnetic resonance imaging to confirm the diagnosis.
If recurrence was verified, further treatment was recommended
according to the condition of the patient and included
re-operation, TACE, percutaneous ethanol injection (PEI) and/or
radiofrequency ablation (RFA).
Statistical analysis
SPSS software version 13.0 (SPSS, Inc., Chicago, IL,
USA) was used to perform the analyses. For the univariate analysis,
the χ2 test and Kaplan-Meier method (compared using the
log-rank test) were employed. For the multivariate analysis, a
stepwise logistic regression model and the Cox proportional hazards
regression model were used to identify the most important factors.
P<0.05 was considered to indicate a statistically significant
difference.
Results
Patient follow-up
The median follow-up time was 40.9 months (range,
8–126 months). A total of 100 patients with HCC received
postoperative adjuvant TACE prior to signs of recurrence, while the
remaining 40 patients did not receive any TACE treatment and only
attended follow-up visits at the outpatient clinic. As of December
31, 2014, there were 129 cases with recurrence (median time, 12
months; range, 3–77 months) and 107 mortalities (median time, 37
months; range, 8–126 months). Of the 129 patients with recurrence,
surgery was performed in 42 cases (49 times altogether, twice for 7
cases); additionally, in the same group of patients, TACE was
performed 354 times, RFA was performed 31 times and PEI was
performed 152 times. A total of 73 patients received more than one
treatment following recurrence.
Variations in the mtDNA D-loop
region
A total of 150 point sequence variations were
observed among the 140 cases. Of these variations, there were 13
point mutations, 8 insertions, 20 deletions and 116 polymorphisms
(including a few insertions and deletions according to the mtDNA
database). The variation rate was 13.4% (150/1, 122). The majority
of the variations were located in hypervariable segments I
(NT57-372) and II (NT16024-16383), and they consisted of 46 (30.7%,
46/150) and 63 points (42.0%, 63/150), respectively. No large-scale
deletions were observed.
Clinicopathological characteristics in
patients with early and late recurrence
Early recurrence was characterised by a recurrence
time of <1 year post-radical resection, while recurrences that
occurred after 1 year were characterised as late recurrences. In
the univariate analysis, the clinicopathological characteristics
[including age, tumour size, tumour capsule, surgical margin,
hepatitis B e antigen, hepatitis B virus DNA (HBV DNA),
differentiation degree (Edmondson-Steiner grade), cirrhosis,
microscope embolus, TNM stage, nucleotide 16129 (G/A), nucleotide
16192 (C/T), nucleotide 16297 (T/C), nucleotide 150 (C/T),
nucleotide 199 (T/C) and nucleotide 204 (T/C) of mtDNA] were
significantly different between the early and late recurrence
groups (P<0.05). However, only HBV DNA, differentiation degree,
TNM stage and nucleotide 150 (C/T) of mtDNA were independent
factors in the logistic regression. Nucleotide 150 (C/T) of mtDNA
was a protective factor, while the remainder were risk factors.
Patients with the 150T allele appeared to exhibit later recurrence
(Table I).
 | Table I.Clinicopathological factors for early
recurrent hepatocellular carcinoma in a stepwise logistic
regression model. |
Table I.
Clinicopathological factors for early
recurrent hepatocellular carcinoma in a stepwise logistic
regression model.
| Clinicopathological
variables | B | SE | Exp (B) | 95.0% CI for Exp
(B) | P-value |
|---|
| HBV DNA | 1.152 | 0.446 | 3.164 | 1.320–7.582 |
0.010 |
| Pathological
differentiation degree | 0.841 | 0.402 | 2.318 | 1.054–5.100 |
0.037 |
| TNM | 1.033 | 0.230 | 2.809 | 1.791–4.405 | <0.001 |
| Nucleotide 150
(C/T) | −1.512 | 0.491 | 0.220 | 0.084–0.577 |
0.002 |
Associations between HCC prognosis and
clinicopathological characteristics
Patients with HCC were divided into different groups
based on their clinicopathological characteristics and genotypes at
each variation site. The postoperative tumour-free and overall
survival curves were subsequently plotted using the Kaplan-Meier
method and were compared by the log-rank test. The Cox proportional
hazards regression model was also used to identify the most
important factors.
Among the 140 cases, the median tumour-free survival
time was 12 months, and the recurrence rates for 1, 2, 3, 4 and 5
years were 50, 70, 79, 89 and 92%, respectively. In the univariate
analysis, the clinicopathological characteristics, including tumour
size, tumour capsule, HBV DNA, Child-Pugh class, differentiation
degree, tumour necrosis, microscopic embolism, TNM stage,
nucleotide 16129 (G/A), nucleotide 16263 (T/C), nucleotide 310
(T/CTC/other types) and nucleotide 315 (N/insertion C) were
significantly different between the groups. No significant
differences were observed between the mutation and normal groups,
while in the Cox proportional hazards regression model, only HBV
DNA, Child-Pugh class, differentiation degree, TNM stage,
nucleotide 16263 (T/C) and nucleotide 315 (N/insertion C) were
independent factors for tumour-free survival time. Regarding the
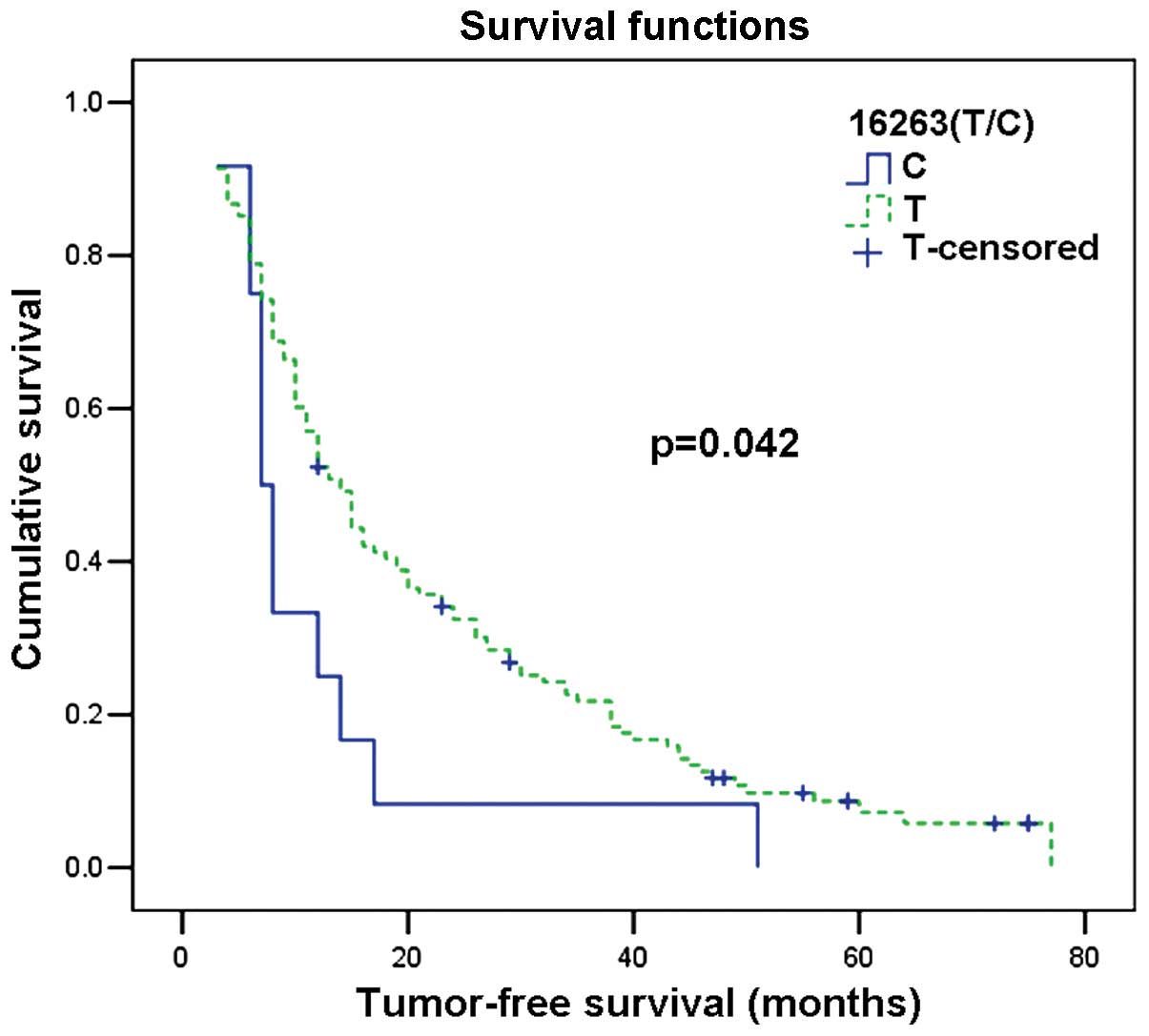
mtDNA variations, patients with the 16263T allele had a greater
tumour-free survival time than patients with the 16263C allele.
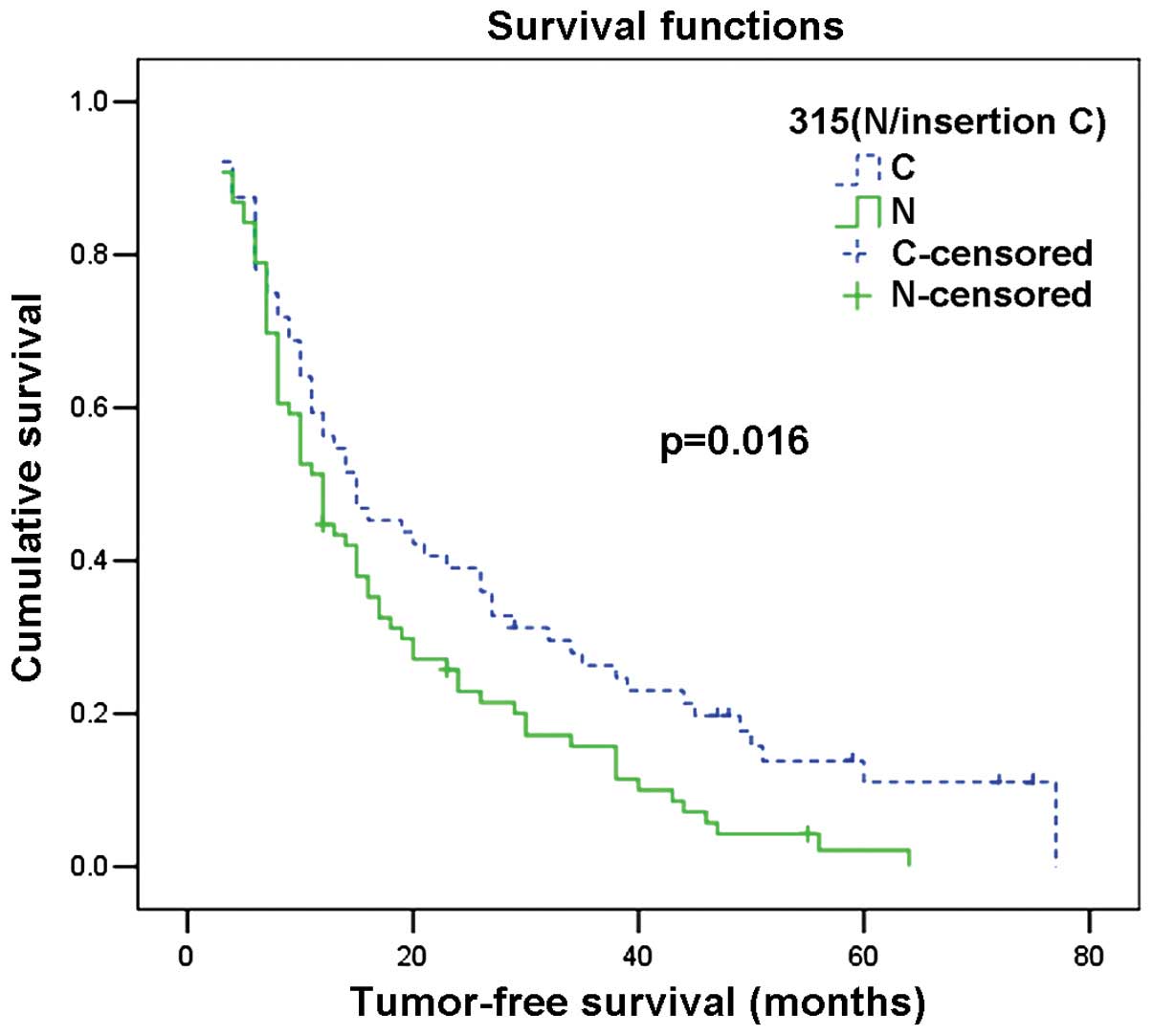
Similarly, patients with 315 insertion C had a superior tumour-free
survival time when compared with patients with the 315 N genotype
(normal) (Table II; Fig. 1 and 2).
 | Table II.Multivariate analysis of the
clinicopathological factors using Cox's proportional hazards
regression model for tumour-free survival. |
Table II.
Multivariate analysis of the
clinicopathological factors using Cox's proportional hazards
regression model for tumour-free survival.
| Clinicopathological
variables | B | SE | Exp (B) | 95.0% CI for Exp
(B) | P-value |
|---|
| HBV DNA | 0.662 | 0.190 | 1.939 | 1.337–2.814 | <0.001 |
| Child-Pugh
class | −0.591 | 0.198 | 0.554 | 0.375–0.817 | 0.003 |
| Pathological
differentiation degree | −0.425 | 0.211 | 0.654 | 0.432–0.989 | 0.044 |
| TNM | 0.517 | 0.093 | 1.677 | 1.399–2.011 | <0.001 |
| Nucleotide 16263
(T/C) | 0.708 | 0.315 | 2.030 | 1.095–3.762 | 0.025 |
| Nucleotide 315
insertion C | −0.462 | 0.186 | 0.630 | 0.437–0.908 | 0.013 |
Among the 140 cases, the median overall survival was
37 months, and the survival rates for 1, 2, 3, 4 and 5 years were
94, 72, 56, 40 and 27%, respectively. In the univariate analysis,
the clinicopathological characteristics, including recurrence type
(early/late), age, tumour size, tumour number, HBV DNA, Child-Pugh
class, differentiation degree, tumour necrosis, microscopic
embolism, TNM stage, adjuvant treatment following tumour recurrence
(none or one/more than one treatment), nucleotide 16263 (T/C),
nucleotide 150 (C/T), nucleotide 249 (insertion A/N), nucleotide
310 (T/CTC/other types), nucleotide 315 (N/insertion C) and
mutations were all independent factors for overall survival, while
in the Cox proportional hazards regression model, only recurrence
type (early/late), Child-Pugh class, TNM stage and adjuvant
treatment following tumour recurrence (none or one/more than one
treatment) were independent factors for overall survival. None of
the mtDNA variations were independent factors. Patients with late
recurrence, Child-Pugh class A, and a low TNM stage and/or those
who received more than one adjuvant treatment following tumour
recurrence had favourable outcomes (Table III).
 | Table III.Multivariate analysis of the
clinicopathological factors using Cox's proportional hazards
regression model for overall survival. |
Table III.
Multivariate analysis of the
clinicopathological factors using Cox's proportional hazards
regression model for overall survival.
| Clinicopathological
variables | B | SE | Exp (B) | 95.0% CI for Exp
(B) | P-value |
|---|
| Recurrence type
(early/late) | 2.017 | 0.271 | 7.513 | 4.413–12.791 | <0.001 |
| Child Pugh
class | −0.669 | 0.206 | 0.512 | 0.342–0.766 | 0.001 |
| TNM | 0.226 | 0.110 | 1.253 | 1.009–1.555 | 0.041 |
| Adjuvant treatment
post-tumour recurrence (none or one/more than one treatment) | −0.431 | 0.207 | 0.650 | 0.433–0.976 | 0.038 |
Discussion
Unlike other forms of cancer, hepatocellular
carcinoma is consistently associated with chronic liver diseases
(14,15). The majority of studies regarding HCC
have focused on the cellular nucleus, with a number of variations
in genes and their expression identified as potential
carcinogenesis factors (16,17). However, the true cause of HCC
carcinogenesis is yet to be elucidated. In population studies for
the detection of a variety of diseases and tumours, polymorphisms
of or alterations in mtDNA have emerged as novel biomarkers
(5). A number of mutations or
polymorphisms in the coding and non-coding regions of the
mitochondrial genome have been associated with an elevated risk of
cancer (6,8). A few studies have reported that mtDNA
mutations were correlated with tumorigenesis (11,18), while
others have focussed on their association with cancer and aetiology
(19,20). The detection of specific mutations in
individual cancer populations has recently become a popular area of
research. In a previous study conducted by the authors, it was
observed that analysing sequence variations in the mtDNA D-loop
region may determine useful biomarkers for identifying the cell
clonal origin of synchronous multinodular HCC (13). In the current study, HCC tissue
samples were screened for mtDNA polymorphisms and mutations, and
their association with HCC prognoses were analysed.
The results demonstrated that mtDNA mutations are
not ubiquitous in HCC, as only a small proportion (13/140, 9.3%) of
the HCC tissue samples had D-loop mutations. Previous studies have
reported that mtDNA mutations were universal in cancer and useful
for predicting the outcomes of patients (21–23). Yin
et al (21) reported that 22%
of HCC tissues studied carried a somatic mutation in the mtDNA
D-loop region. Lee et al (22)
examined mutations in the D-loop region of mtDNA in 61 HCC
specimens, and the results demonstrated that 39.3% carried somatic
mutation(s) in the mtDNA D-loop. Furthermore, Wheelhouse et
al (23) reported that the
percentage of tumour tissue from patients with HCC harbouring
D-loop mutations was 59%. However, in other studies, mtDNA
mutations were not ubiquitous, which is comparable to the present
study (24). In head and neck
squamous cell carcinoma (HNSCC), pooled data from nine studies
investigating D-loop mutations in >400 cases of HNSCC indicated
that approximately one-third of the HNSCC cases harboured D-loop
mutations; however, there was significant variation between
different studies (range, 2–67%) (24). Explanations for these differences may
be include variation in the methods of mutation detection or the
intrinsic genetic heterogeneity of cancer.
In the present study, there were no significant
associations observed between mtDNA mutations and the prognosis of
patients with HCC. mtDNA mutations in the HCC tissues appeared to
be infrequent and lack prognostic utility, consistent with a
previous study (7). However, Liu
et al (25) observed that
patients with oral squamous cell carcinoma with D-loop mutations
had superior survival rates when compared with patients without
such mutations (2-year disease-specific survival rate: 73.4 vs.
45.0%, respectively). In a breast cancer study, Kuo et al
(26) reported that somatic mutations
in the mtDNA D-loop and in tumour protein 53 (TP53) were
independent of each other, indicating that the number of somatic
mtDNA D-loop region mutations may be an indicator of poor prognosis
through a TP53-independent mechanism. However, Challen et al
(24) demonstrated that mtDNA
mutations were not ubiquitous in HNSCC, and that there were no
significant associations between D-loop mutations and determinants
of clinical outcomes. The results were not consistent between
studies, and according to our data, we hypothesised that the
mutations in the mtDNA D-loop region were stochastic events that
may not significantly affect the biology of HCC.
Sequence variations or polymorphisms in the mtDNA
D-loop region are common in HCC. The majority of variations in the
present study were located in hypervariable segments I (NT57-372)
and II (NT16024-16383) (30.7 and 42.0%, respectively). Of these
variations, the nucleotide 150 (C/T) polymorphism of mtDNA was
associated with HCC recurrence. The 150T allele frequency was much
higher in late recurrence of HCC. A number of studies have reported
that the 150 (C/T) polymorphism was correlated with longevity
(27–29). It is considered that the 150 (C/T)
transition functions in remodelling mtDNA replication, thus, if the
150 (C/T) transition is present, replication of the mtDNA heavy
strand would begin at position 149T instead of position 151C, which
would subsequently affect ATP synthesis through the process of
OXPHOS in cell metabolism (30). This
result was similar to that of a previous study of cervical cancer,
in which Zhai et al (31)
reported that a mitochondrial 150 (C/T) polymorphism increased the
risk of cervical cancer and HPV infection. However, based on the
results of the present study, it was not possible to determine the
mechanism of or provide further details regarding this association.
In addition, the current study demonstrated that the 150 (C/T)
polymorphism was not associated with tumour-free or overall
survival. Further research investigating the role of nucleotide 150
(C/T) is required.
In the present study, two polymorphisms, 16263 (T/C)
and insertion 315C, were identified as independent predictive
factors for tumour-free survival time. Patients with the 16263T
allele had a greater tumour-free survival time than patients with
the 16263C allele. Similarly, patients with 315 insertion C had a
superior tumour-free survival time when compared with patients with
315N (normal). However, none of the mtDNA variations were
identified as independent predictors of overall survival. The
association between mtDNA polymorphisms and prognosis has been
analysed in various types of cancer (7,32,33). Wang et al (7) investigated the predictive power of
D-loop single nucleotide polymorphisms (SNPs) in patients with HCC,
and two SNP sites (nucleotides 150 C/T and 146 T/C) were identified
by the log-rank test as statistically significant predictors of HCC
survival. Furthermore, in a multivariate analysis, allele 146 was
identified as an independent predictor, and the survival time of
patients with allele 146C was significantly less than that of
patients with allele 146T. Ding et al (32) also demonstrated that allele 16390 was
an independent predictor of non-small cell lung cancer outcome. The
survival time of patients with allele 16390A was significantly
shorter than that of patients with allele 16390G. Guo et al
(33) identified that nucleotide
16266 C/T was associated with age at onset in patients with
oesophageal squamous cell carcinoma. The age at onset of patients
with the minor allele T genotype was significantly younger than
that of patients with the C genotype at the 16266 site. However,
based on the data from the present study, it was not possible to
determine a specific variation in patients with HCC. Further
investigation is required to determine whether these conclusions
may be extended to other patients with HCC.
Although the mechanisms of mtDNA variations in
tumour development and progression have not yet been elucidated,
studies suggest that variations in the mtDNA D-loop region may
induce significant alterations in mitochondrial function. The mtDNA
D-loop is crucial for regulating the replication and expression of
the mitochondrial genome. It was previously reported that mtDNA
variations had subtle effects on the electron transport chain
(34). However, the accumulation of
numerous subtle changes may result in significant consequences.
Furthermore, mitochondrial respiratory chain activity was affected
notably, which is responsible for the high ROS release and nuclear
genome damage, in addition to cancer initiation and promotion
(5,6,8). High
levels of ROS have been demonstrated to be significant in
activating cell apoptosis and infliction of injury to the genome
(8,35). Thus, the present study hypothesised
that variations in the mtDNA D-loop region may affect the entire
function of mtDNA, further interfering with nuclear genome
expression and function. However, the questions as to whether
variations in the mtDNA D-loop region are the result or cause of
carcinogenesis requires further investigation.
In conclusion, the results of the current study
demonstrated that mtDNA D-loop polymorphisms were associated with
early recurrence and tumour-free survival time, but not with
overall survival. mtDNA D-loop mutations in patients with HCC were
infrequent and lacked prognostic utility. Therefore, the detection
of mtDNA D-loop polymorphisms may aid in identifying risk factors
for HCC prognosis, particularly for the short-term outcome, and
thus aiding the construction of an appropriate therapeutic
strategy. However, mtDNA D-loop variations lacked utility in
predicting long-term outcomes, therefore further biomarkers require
investigation.
Acknowledgements
The present study was supported by a grant from the
National Natural Science Foundation of China (no. 81160262). The
funding body served no role in the study design, data collection
and analysis, decision to publish or manuscript preparation. The
language, grammar, punctuation and spelling of the original
manuscript was edited by the American Journal Experts.
References
|
1
|
Bray F, Ren JS, Masuyer E and Ferlay J:
Global estimates of cancer prevalence for 27 sites in the adult
population in 2008. Int J Cancer. 132:1133–1145. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Jemal A, Bray F, Center MM, Ferlay J, Ward
E and Forman D: Global cancer statistics. CA Cancer J Clin.
61:69–90. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Portolani N, Coniglio A, Ghidoni S, et al:
Early and late recurrence after liver resection for hepatocellular
carcinoma: Prognostic and therapeutic implications. Ann Surg.
243:229–235. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Lim W, Kwon SH, Cho H, et al: HBx
targeting to mitochondria and ROS generation are necessary but
insufficient for HBV-induced cyclooxygenase-2 expression. J Mol Med
Berl. 88:359–369. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Zapico SC and Ubelaker DH: mtDNA mutations
and their role in aging, diseases and forensic sciences. Aging Dis.
4:364–380. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Singh AK, Pandey P, Tewari M, Pandey HP
and Shukla HS: Human mitochondrial genome flaws and risk of cancer.
Mitochondrial DNA. 25:329–334. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Wang C, Zhang F, Fan H, et al: Sequence
polymorphisms of mitochondrial D-loop and hepatocellular carcinoma
outcome. Biochem Biophys Res Commun. 406:493–496. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Fogg VC, Lanning NJ and Mackeigan JP:
Mitochondria in cancer: At the crossroads of life and death. Chin J
Cancer. 30:526–539. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Akouchekian M, Houshmand M, Hemati S, et
al: High rate of mutation in mitochondrial DNA displacement loop
region in human colorectal cancer. Dis Colon Rectum. 52:526–530.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Liu VW, Shi HH, Cheung AN, et al: High
incidence of somatic mitochondrial DNA mutations in human ovarian
carcinomas. Cancer Res. 61:5998–6001. 2001.PubMed/NCBI
|
|
11
|
Ding Z, Ji J, Chen G, et al: Analysis of
mitochondrial DNA mutations in D-loop region in thyroid lesions.
Biochim Biophys Acta. 1800:271–274. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Yin PH, Wu CC, Lin JC, et al: Somatic
mutations of mitochondrial genome in hepatocellular carcinoma.
Mitochondrion. 10:174–182. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Li SL, Su M, Peng T, et al:
Clinicopathologic characteristics and prognoses for multicentric
occurrence and intrahepatic metastasis in synchronous multinodular
hepatocellular carcinoma patients. Asian Pac J Cancer Prev.
14:217–223. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Weinmann A, Koch S, Niederle IM, et al:
Trends in epidemiology, treatment and survival of hepatocellular
carcinoma patients between 1998 and 2009: An analysis of 1066 cases
of a German HCC Registry. J Clin Gastroenterol. 48:279–289. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Nault JC: Pathogenesis of hepatocellular
carcinoma according to aetiology. Best Pract Res Clin
Gastroenterol. 28:937–947. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Kirstein MM and Vogel A: The pathogenesis
of hepatocellular carcinoma. Dig Dis. 32:545–553. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Liu M, Jiang L and Guan XY: The genetic
and epigenetic alterations in human hepatocellular carcinoma: A
recent update. Protein Cell. 5:673–691. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Nishikawa M, Nishiguchi S, Shiomi S, et
al: Somatic mutation of mitochondrial DNA in cancerous and
noncancerous liver tissue in individuals with hepatocellular
carcinoma. Cancer Res. 61:1843–1845. 2001.PubMed/NCBI
|
|
19
|
Zhang R, Zhang F, Wang C, Wang S, Shiao YH
and Guo Z: Identification of sequence polymorphism in the D-Loop
region of mitochondrial DNA as a risk factor for hepatocellular
carcinoma with distinct etiology. J Exp Clin Cancer Res.
29:1302010. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Sharma H, Singh A, Sharma C, Jain SK and
Singh N: Mutations in the mitochondrial DNA D-loop region are
frequent in cervical cancer. Cancer Cell Int. 5:342005. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Yin PH, Lee HC, Chau GY, Wu YT, Li SH, Lui
WY, Wei YH, Liu TY and Chi CW: Alteration of the copy number and
deletion of mitochondrial DNA in human hepatocellular carcinoma. Br
J Cancer. 90:2390–2396. 2004.PubMed/NCBI
|
|
22
|
Lee HC, Li SH, Lin JC, Wu CC, Yeh DC and
Wei YH: Somatic mutations in the D-loop and decrease in the copy
number of mitochondrial DNA in human hepatocellular carcinoma.
Mutat Res. 547:71–78. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Wheelhouse NM, Lai PB, Wigmore SJ, Ross JA
and Harrison DJ: Mitochondrial D-loop mutations and deletion
profiles of cancerous and noncancerous liver tissue in hepatitis B
virus-infected liver. Br J Cancer. 92:1268–1272. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Challen C, Brown H, Cai C, Betts G,
Paterson I, Sloan P, West C, Birch-Machin M and Robinson M:
Mitochondrial DNA mutations in head and neck cancer are infrequent
and lack prognostic utility. Br J Cancer. 104:1319–1324. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Liu SA, Jiang RS, Chen FJ, Wang WY and Lin
JC: Somatic mutations in the D-loop of mitochondrial DNA in oral
squamous cell carcinoma. Eur Arch Otorhinolaryngol. 269:1665–1670.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Kuo SJ, Chen M, Ma GC, Chen ST, Chang SP,
Lin WY, Chen YC, Lee TH, Lin TT and Liu CS: Number of somatic
mutations in the mitochondrial D-loop region indicates poor
prognosis in breast cancer, independent of TP53 mutation. Cancer
Genet Cytogenet. 201:94–101. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Niemi AK, Hervonen A, Hurme M, Karhunen
PJ, Jylhä M and Majamaa K: Mitochondrial DNA polymorphisms
associated with longevity in a Finnish population. Hum Genet.
112:29–33. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Niemi AK, Moilanen JS, Tanaka M, Hervonen
A, Hurme M, Lehtimäki T, Arai Y, Hirose N and Majamaa K: A
combination of three common inherited mitochondrial DNA
polymorphisms promotes longevity in Finnish and Japanese subjects.
Eur J Hum Genet. 13:166–170. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Ren WH, Li XH, Zhang HG, Deng FM, Liao WQ,
Pang Y, Liu YH, Qiu MJ, Zhang GY and Zhang YG: Mitochondrial DNA
haplogroups in a Chinese Uygur population and their potential
association with longevity. Clin Exp Pharmacol Physiol.
35:1477–1481. 2008.PubMed/NCBI
|
|
30
|
Zhang J, Asin-Cayuela J, Fish J, et al:
Strikingly higher frequency in centenarians and twins of mtDNA
mutation causing remodeling of replication origin in leukocytes.
Proc Natl Acad Sci USA. 100:1116–1121. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Zhai K, Chang L, Zhang Q, Liu B and Wu Y:
Mitochondrial C150T polymorphism increases the risk of cervical
cancer and HPV infection. Mitochondrion. 11:559–563. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Ding C, Li R, Wang P, Fan H and Guo Z:
Sequence polymorphisms of the mitochondrial displacement loop and
outcome of non-small cell lung cancer. Exp Ther Med. 3:861–864.
2012.PubMed/NCBI
|
|
33
|
Guo Z, Yang H, Zhang F, Zhang R and Wang
C: Single nucleotide polymorphisms in the mitochondrial
displacement loop and age-at-onset of esophageal squamous cell
carcinoma. Oncol Lett. 3:482–484. 2012.PubMed/NCBI
|
|
34
|
Polyak K, Li Y, Zhu H, Lengauer C, Willson
JK, Markowitz SD, Trush MA, Kinzler KW and Vogelstein B: Somatic
mutations of the mitochondrial genome in human colorectal tumours.
Nat Genet. 20:291–293. 1998. View
Article : Google Scholar : PubMed/NCBI
|
|
35
|
Zamzami N and Kroemer G: The mitochondrion
in apoptosis: How Pandora's box opens. Nat Rev Mol Cell Biol.
2:67–71. 2001. View
Article : Google Scholar : PubMed/NCBI
|