Introduction
Oral cancer is one of the most common types of
cancer worldwide, and includes malignancies of the oral cavity,
lip, tongue, gums, retromolar region and oropharynx (1). Oral cancer is the sixth leading cause of
cancer-associated mortality in men, with an estimated 263,900 novel
cases and 128,000 associated mortalities annually (2). Oral squamous cell carcinoma (OSCC) is
the most common type of oral cancer, and accounts for >90% of
all oral malignancies, thus being a major cause of
cancer-associated morbidity and mortality worldwide (3). Metastasis is a major consequence of
treatment failure, leading to a poor prognosis in OSCC patients
(4). Therefore, studies on OSCC
metastasis and proliferation are particularly crucial.
MicroRNAs (miRNAs or miRs) are a group of small,
endogenous non-coding RNAs with single strands of ~21–24
nucleotides, and play a crucial role in the regulation of
eukaryotic gene expression by base pairing with their targeted
messenger (m)RNAs at their 3′-untranslated regions (UTRs), which
results in the cleavage or translational inhibition of the target
mRNA (5,6). In human cancer, miRNAs are involved in
multiples biological processes, including cell growth,
differentiation, apoptosis and regulation of the cell cycle
(5,6).
In recent years, numerous studies have identified several
dysfunctional miRNAs associated with the development and
progression of OSCC, including miR-10b, miR-21, miR-31, miR-98,
miR-101, miR-143, miR-184, miR-210, miR-221 and miR-222 (7–16). The
miR-196 family of miRNAs appears to be expressed from intergenic
regions in clusters of homeobox (HOX) genes, where several HOX
genes are regulated by targeting their 3′-UTRs (17,18). A
previous study by the present authors revealed that miRNAs of the
miR-196 family play a potential oncogenic role in gastric cancer,
and their expression may be regulated through DNA methylation
(19–21). Lim et al (22) reported that the expression of miR-196b
positively correlated with HOXA10 expression and poor prognosis in
gastric cancer. Furthermore, miR-196a directly targets annexin A1,
thus inducing cell proliferation and anchorage-independent growth,
and suppressing apoptosis, which suggests that miR-196a has
oncogenic potential in esophageal cancer (23). Collectively, those studies revealed
that miR-196b expression plays an oncogenic role in gastric and
esophageal cancer. In contrast to the aforementioned results, other
studies have reported that miR-196b plays a tumor-suppressive role
in leukemia, as well as breast and cervical cancer (24–27).
Although dysfunctional miR-196b has been reported to be involved in
oral cancer metastasis (13,28), its regulatory mechanism in OSCC
remains unclear. The present study demonstrated that DNA
hypomethylation led to miR-196b overexpression and facilitated OSCC
cell migration and invasion.
Materials and methods
Patients and tissue samples
The protocol for the present study was independently
reviewed and approved by the Institutional Review Board (IRB) of
Kaohsiung Veterans General Hospital (KSVGH; IRB no. VGHKS14-CT6-18,
Kaohsiung, Taiwan). A total of 69 paired tumor tissues and adjacent
normal tissue samples were collected from patients with oral cancer
at the Department of Otolaryngology of Kaohsiung Veterans General
Hospital (Kaohsiung, Taiwan) between 2010 and 2012. The
requirements for written informed consent from the subjects were
waived by the IRB of KSVGH, since all the data and specimens had
been previously collected and analyzed anonymously. Pathological
tumor-node-metastasis classification was determined at the time of
the initial resection of the tumor in accordance with the 2002
guidelines of the American Joint Committee on Cancer staging system
(29). Fresh tissue samples included
tumor tissues and corresponding adjacent normal tissues from 69
OSCC patients. Following surgery, samples were immediately placed
in RNAlater® solution (Qiagen GmbH, Hilden, Germany) and
stored at room temperature overnight prior to be frozen at
−80°C.
DNA and RNA extraction
Total RNA and DNA were extracted from tissues using
TRIzol reagent (Invitrogen; Thermo Fisher Scientific, Inc.,
Waltham, MA, USA), according to the manufacturer's protocol.
Briefly, tissue samples were homogenized in 1 ml TRIzol and mixed
with 0.2 ml chloroform (Avantor Performance Materials, Center
Valley, PA, USA) for extracting protein and DNA, while RNA was
precipitated with 0.6 ml isopropanol (Avantor Performance
Materials). The remaining liquid was mixed with 0.5 ml back
extraction buffer (4 M guanidine thiocyanate, 50 mM sodium citrate
and 1 M Tris; pH 8.0; Avantor Performance Materials), and DNA was
precipitated with 0.4 ml isopropanol. The concentration, purity and
amount of total RNA and DNA were determined with a NanoDrop 1000
spectrophotometer (Nanodrop Technologies; Thermo Fisher Scientific,
Inc.).
Stem-loop
reverse-transcription-quantitative polymerase chain reaction
(RT-qPCR) of mature miRNAs
Total RNA (1 µg) was reverse transcribed using
miR-specific stem-loop miR-196b-RT primer (Gemomics BioSci &
Tech, New Taipei, Taiwan) and SuperScript® III Reverse
Transcriptase (Invitrogen; Thermo Fisher Scientific, Inc.),
according to the manufacturer's protocol. The RT reaction included
1 µg RNA, 1X buffer, 2.5 mM dNTP and 0.5 mM stem-loop miR-196b RT
primer. The reaction was performed using the following incubation
conditions with a PCR thermocycler (TPersonal 20 Thermal Cycler;
Biometra GmbH, Göttingen, Germany): 16°C for 30 min, followed by 50
cycles of 20°C for 30 sec, 42°C for 30 sec and 50°C for 1 sec. The
complementary DNA was used at a dilution of 1:20 in water in the
subsequent qPCR, which was performed using a specific forward
primer and a universal reverse primer, under the following
conditions: Incubation at 94°C for 10 min, followed by 40 cycles of
94°C for 15 sec and 60°C for 30 sec. Gene expression was detected
with SYBR Green I (Applied Biosystems; Thermo Fisher Scientific,
Inc.). The expression levels of miRNAs were normalized to those of
U6 (ΔCq=miR-196b Cq-U6 Cq) as described by Livak and Schmittgen
(30). The individual primers were
synthesized by Qiagen China Co., Ltd. (Shanghai, China) used in the
present study were as follows: miR-196b RT, foward
5′-CTCAACTGGTGTCGTGGAGTCGGCAATTCAGTTGAGCCCAACAA-3′ and gene
specific forward primer, 5′-CGGCGGTAGGTAGTTTCCTGTT-3′;
universal/miR-196b reverse, 5′-CTGGTGTCGTGGAGTCGGCAATTC-3′; and U6
forward, 5′-CTCGCTTCGGCAGCACA-3′ and reverse,
5′-AACGCTTCACGAATTTGCGT-3′.
Genomic DNA bisulfite conversion
Genomic DNA was extracted from oral tissue samples
using TRIzol, and an aliquot (2 µg) was subjected to bisulfite
conversion using the EZ DNA Methylation-Gold™ kit (Zymo Research
Corporation, Irvine, CA, USA). Bisulfite conversion was achieved by
incubating the reaction at 98°C for 10 min, 64°C for 2.5 h and 4°C
for ≤20 h in a PCR thermocycler (Biometra GmbH).
Combined bisulfite restriction
analysis (COBRA) and bisulfite sequencing analysis
Bisulfite-converted genomic DNA was used for
methylation analysis of CpG islands with specific methylation
primers. The PCR conditions were as follows: 94°C for 10 min,
followed by 40 cycles of 94°C for 30 sec, 60°C for 3 sec and 72°C
for 40 sec, with a final extension at 72°C for 10 min. A PCR
thermocycler (Biometra GmbH) and Hot Start Taq DNA polymerase
(Qiagen GmbH) were used for the reaction. The methylation status of
the genomic DNA of individual samples was examined by TaqI
digestion (New England BioLabs, Inc., Ipswich, MA, USA). The
digested PCR fragments were then separated on a 2% agarose gel
(Invitrogen; Thermo Fisher Scientific, Inc.) immersed in 1X
Tris-acetate-ethylenediaminetetraacetic acid buffer (Omics Bio,
Taipei, Taiwan) using Mupid-2plus DNA Gel Electrophoresis system
(Advance Co., Ltd., Tokyo, Japan). Following ethidium bromide
(MDBio, INc., Taipei, Taiwan) staining, the gels were visualized
and anlyzed by a gel documentation system (Uvitec Cambridge,
Cambridge, UK) In addition, the PCR products were cloned into the
pGEM®-T vector (Promega Corporation, Madison, WI, USA),
and several clones were randomly selected for sequencing using a T7
primer (5′-TAATACGACTCACTATAGGG-3′) by Qiagen China Co., Ltd.. The
individual primers used in the COBRA assay were as follows:
miR-196b mF (forward primer), 5′-GAGAGAAAGGTGGATTTTAATAATAGGA-3′
and mR (reverse primer), 5′-AACCTATAACTTCCCCTTCCTTAAC-3′.
Establishment of stable clones
Human oral cancer SAS and Cal-27 cell lines were
kindly provided by Dr Michael Hsiao (Genomics Research Center,
Academia Sinica, Taipei, Taiwan). The oral cancer cell lines, SASA
and CAL27, were cultured in Dulbecco's modified Eagle's medium
(Biological Industries Israel Beit-Haemek Ltd., Kibbutz
Beit-Haemek, Israel) supplemented with 10% fetal bovine serum (GE
Healthcare Life Sciences, Logan, UT, USA) and 1%
penicillin/streptomycin (Sigma-Aldrich, St. Louis, MO, USA). Stable
or transient SAS and CAL-27 cell cultures expressing miR-196b were
generated by transfecting SAS and CAL-27 oral cancer cells with the
pLKO.1 vector (Addgene, Inc., Cambridge, MA, USA) or with
pLKO.1-miRNA-196b plasmid using Lipofectamine® 2000
(Invitrogen; Thermo Fisher Scientific, Inc.). At 24 h
post-transfection, stable cells expressing miR-196b were generated
using puromycin (Sigma-Aldrich) selection for 14 days. The levels
of miR-196b expression were measured by RT-qPCR as described
previously in the ‘Stem-loop reverse-transcription-quantitative
polymerase chain reaction’ section.
Antagomir assay
The sequence complementary to miR-196b,
5′-CCCAACAACAGGAAACUACCUA-3′, and the negative control (NC)
sequence, 5′-GUGUAACACGUCUAUACGCCCA-3′, referred to as
anti-miR-196b and anti-miR-NC, respectively, were synthesized with
2′-O-methyl oligo modified bases exhibiting phosphorothioate on the
first two and final four bases and a 3-cholesterol modification,
which was added through a hydroxyprolinol linkage (Dharmacon; GE
Healthcare Life Sciences, Chalfont, UK). Transfection of these
antagomirs was performed using Lipofectamine® RNAiMAX
(Invitrogen; Thermo Fisher Scientific, Inc.), according to the
manufacturer's protocol. Cells were incubated with 100 nM
anti-miR-196b for 2 days to suppress miR-196b expression.
Cell proliferation, migration and
invasion assays
For the cell proliferation assay, living cells
(3×103) were plated onto 96-well plates, and cell growth
was determined 0–4 days later using WST-1 cell proliferation
reagent (Roche Diagnostics, Basel, Switzerland). The absorbance
value at 450 nm of each well was measured using a plate reader
(Multiskan FC Microplate Photometer; Invitrogen™, Thermo Fisher
Scientific, Inc.). Cells were also tested for their invasion
ability in vitro using Transwell chambers (BD Biosciences,
Franklin Lakes, NJ, USA). The lower side of the polycarbonate
membranes (containing 8-µm pores) of the Transwell chamber was
coated with Matrigel (1 or 0.2 µg/ml/well; BD Biosciences). Cells
(5×104) were then added onto the Matrigel layer in each
well. Following incubation for 48 h at 37°C, the cells that had
migrated through the Matrigel layer were visualized upon fixation
with formaldehyde (BS Chemical, Kretinga, Lithuania) and staining
with crystal violet (EMD Millipore, Billerica, MA, USA). The level
of invasion was determined using a microscope (CKX41; Olympus
Corporation, Tokyo, Japan). For the migration assay, cells
(1.5×106) were plated onto 6-well plates. A straight
line was scratched into the cell monolayer in the middle of the
well using a 200-ml pipette tip. Culture medium containing 10%
fetal bovine serum was replaced with a serum-deprived culture
medium, and the cells were then incubated at 37°C. Wound closure
was monitored and photographed at 0, 6 and 12 h under a
microscope.
Western blotting
Protien was was extracted from 3×106
cells using 200 µl radioimmunoprecipitation assay buffer. A total
of 30 µg of protein lysate was electrophoresed on 10% sodium
dodecyl sulfate polyacrylamide gels (Sigma-Aldrich) and transferred
onto polyvinylidene difluoride membranes (EMD Millipore). The
membranes were blocked with blocking buffer [50 mM Tris-HCl (pH
7.6), 150 mM NaCl, 0.1% Tween 20, 5% non-fat dry milk, 0.05% sodium
azide) for 1 h at room temperature. The membranes were incubated
with the following primary antibodies overnight at 4°C: Polyclonal
rabbit anti-fibronectin (dilution, 1:1,000; catalog no., GTX112794;
GeneTex, Inc., Irvine, CA, USA); monoclonal rabbit anti-Ep-CAM
(dilution, 1:2,000; catalog no., GTX61060; GeneTex, Inc.);
polyclonal rabbit anti-vimentin (dilution, 1:2,000; catalog no.,
GTX100619; GeneTex, Inc.); polyclonal rabbit anti-Twist1/2
(dilution, 1:2,000; catalog no., GTX127310; GeneTex, Inc.);
monoclonal rabbit anti-E-cadherin (dilution, 1:2,000; catalog no.,
GTX61329; GeneTex, Inc.); and monoclonal mouse anti-actin
(dilution, 1:5,000, catalog no., MAB1501). Following washing three
time for 10 min/time with Tris-buffered saline with Tween 20, the
membranes were incubated with the following horseradish
peroxidase-conjugated secondary antibodies for 1 hour at room
temperature: Goat anti-mouse immunoglobulin (Ig)G (dilution,
1:20,000; catalog no., sc-2005; Santa Cruz Biotechnology, Inc.,
Dallas, TX, USA) and goat anti-rabbit IgG (dilution, 1:20,000;
catalog no., sc-2004; Santa Cruz Biotechnology, Inc.). The target
bands were visulized using WesternBright enhanced chemiluminescence
reagent (Advansta, Inc., Menlo Park, CA, USA) and the results of
the immunoreactions were analyzed with a BioSpectrum®
500 Imaging System (Ultra-Violet Products Ltd., Cambridge, UK). The
bands corresponding to individual genes were analyzed using ImageJ
(imagej.nih.gov/ij/index.html) and the
expression levels of these genes were normalized to those of
actin.
Statistical analysis
The expression levels of miR-196b in all of the oral
tissues were determined using stem-loop RT-qPCR. All reactions were
performed in triplicate, and data were analyzed using the paired t
test or Student's t test. The correlation between various
clinicopathological characteristics and miR-196b expression levels
(low vs. high) or methylation status (no vs. yes) was evaluated
using the χ2 test or Fisher's exact test. Cumulative
survival curves were estimated using the Kaplan-Meier method, and
comparisons between the survival curves were performed by using the
log-rank test. P<0.05 was considered to indicate a statistically
significant difference.
Results
DNA hypomethylation results in
miR-196b overexpression in human OSCC
To investigate the biological role of miR-196b in
OSCC, the expression levels in OSCC tissues and adjacent normal
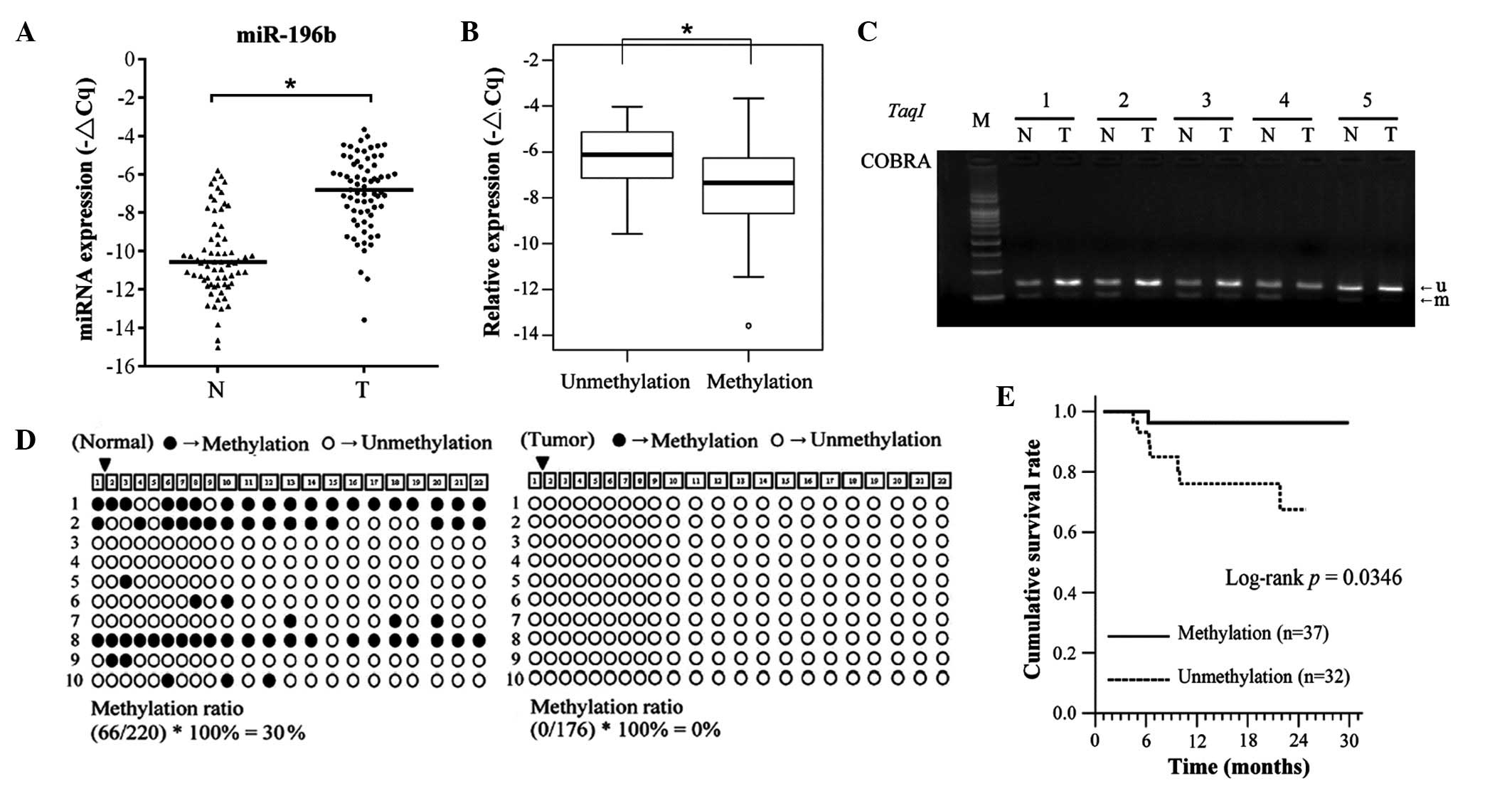
tissues were examined by stem-loop RT-qPCR. As represented in
Fig. 1A, the expression levels of
miR-196b were significantly upregulated in OSCC tissues, compared
with the corresponding adjacent normal tissues (64 of 69, 92.7%,
P<0.001). In addition, the expression levels of miR-196b were
significantly increased in tissues with DNA hypomethylation,
compared with those with DNA hypermethylation in OSCC tissues
(Fig. 1B; P=0.009). A previous study
by the present authors demonstrated that DNA hypomethylation could
lead to miR-196b overexpression in gastric cancer (19). Therefore, the present study examined
whether the DNA methylation status of CpG islands contributed to
miR-196b overexpression in OSCC. Using COBRA, the methylation
status of the miR-196b promoter region was assayed in OSCC samples
and corresponding adjacent normal samples from 69 OSCC patients.
DNA hypomethylation was more frequently observed in oral cancer
tissues than in adjacent normal tissues (32 of 69, 46.3%; Fig. 1C). The methylation status of
individual CpG dinucleotides of the miR-196b promoter region was
verified in selected patients using a bisulfate sequencing
approach. Consistent with the COBRA results, the densities of
methylated CpG dinucleotides were higher in normal tissues
(methylation rate, 30%) than in tumor tissues (methylation rate,
0%; Fig. 1D). The data also indicated
that CpG-rich regions of miR-196b were unmethylated in the oral
cell lines SAS and CAL-27 (data not shown). These results indicated
that abnormal DNA hypomethylation resulted in miR-196b
overexpression in OSCC. The clinicopathological characteristics of
the 69 independent OSCC patients are summarized in Table I. The data indicated that the
expression levels and methylation status of miR-196b were not
associated with any clinicopathological features (Tables II and III). Notably, the disease-specific
survival (DSS) curves of patients with miR-196b promoters
exhibiting unmethylation status were significantly lower than those
displaying methylation status (P=0.035; Fig. 1E).
 | Table I.Clinicopathological data of patients
with oral squamous cell carcinoma. |
Table I.
Clinicopathological data of patients
with oral squamous cell carcinoma.
| Variable | No. of
patients |
|---|
| Age, years |
|
|
≤50 | 18 (26.1) |
|
>50 | 51 (73.9) |
| Cell
differentiation |
|
|
Well | 11 (15.9) |
|
Moderately | 52 (75.4) |
|
Poorly | 6 (8.7) |
| AJCC pathological
stage |
|
| I | 11 (15.9) |
| II | 21 (30.4) |
|
III | 4 (5.8) |
| IV | 33 (47.8) |
| Tumor
classification |
|
| T1 | 11 (15.9) |
| T2 | 26 (37.7) |
| T3 | 6 (8.7) |
| T4 | 26 (37.7) |
| Node
classification |
|
| N0 | 49 (71.0) |
| N1 | 5 (7.2) |
| N2 | 15 (21.7) |
 | Table II.Association between the expression
levels of miR-196b and the clinicopathological data of patients
with oral squamous cell carcinoma. |
Table II.
Association between the expression
levels of miR-196b and the clinicopathological data of patients
with oral squamous cell carcinoma.
|
| miR-196b
(n=69) |
|---|
|
|
|
|---|
|
| No. of patients
(%) |
|
|
|---|
|
|
|
|
|
|---|
| Variable | Low (n=35) | High (n=34) | χ2 |
P-valuea |
|---|
| Gender |
|
| 0.633 | 0.477b |
|
Female | 3
(37.5) | 5
(62.5) |
|
|
|
Male | 32 (52.5) | 29 (47.5) |
|
|
| Age, years |
|
| 4.503 | 0.034 |
|
≤50 | 13 (72.2) | 5
(27.8) |
|
|
|
>50 | 22 (43.1) | 29 (56.9) |
|
|
| Cancer
location |
|
| 2.280 | 0.320 |
|
Buccal | 14 (53.8) | 12 (46.2) |
|
|
|
Tongue | 8
(66.7) | 4
(33.3) |
|
|
| Other
oral mucosal sites | 13 (41.9) | 18 (58.1) |
|
|
| Cell
differentiation |
|
| 2.880 | 0.090 |
|
Well | 3
(27.3) | 8
(72.7) |
|
|
|
Moderate, poor | 32 (55.2) | 26 (44.8) |
|
|
| AJCC pathological
stage |
|
| 0.729 | 0.393 |
| I,
II | 18 (56.3) | 14 (43.8) |
|
|
| III,
IV | 17 (45.9) | 20 (54.1) |
|
|
| Tumor
classification |
|
| 0.354 | 0.552 |
| T1,
T2 | 20 (54.1) | 17 (45.9) |
|
|
| T3,
T4 | 15 (46.9) | 17 (53.1) |
|
|
| Node
classification |
|
| 0.969 | 0.325 |
| N0 | 23 (46.9) | 26 (53.1) |
|
|
| N1,
N2 | 12 (60.0) | 8
(40.0) |
|
|
 | Table III.Association between the DNA
methylation status of miR-196b and the clinicopathological data of
patients with oral squamous cell carcinoma. |
Table III.
Association between the DNA
methylation status of miR-196b and the clinicopathological data of
patients with oral squamous cell carcinoma.
|
| TaqI-DNA
methylation (n=69) |
|---|
|
|
|
|---|
|
| No. of patients
(%) |
|
|
|---|
|
|
|
|
|
|---|
| Variables | Unmethylation
(n=32) | Methylation
(n=37) | χ2 |
P-valuea |
|---|
| Gender |
|
| 1.633 | 0.270b |
|
Female | 2
(25.0) | 6
(75.0) |
|
|
|
Male | 30 (49.2) | 31 (50.8) |
|
|
| Age, years |
|
| 0.129 | 0.720 |
|
≤50 | 9
(50.0) | 9
(50.0) |
|
|
|
>50 | 23 (45.1) | 28 (54.9) |
|
|
| Cancer
location |
|
| 3.256 | 0.196 |
|
Buccal | 9
(34.6) | 17 (65.4) |
|
|
|
Tongue | 5
(41.7) | 7
(58.3) |
|
|
| Other
oral mucosal sites | 18 (58.1) | 13 (41.9) |
|
|
| Cell
differentiation |
|
| 0.528 | 0.468 |
|
Well | 4
(36.4) | 7
(63.6) |
|
|
|
Moderate, poor | 28 (48.3) | 30 (51.7) |
|
|
| AJCC pathological
stage |
|
| 1.891 | 0.169 |
| I,
II | 12 (37.5) | 20 (62.5) |
|
|
| III,
IV | 20 (54.1) | 17 (45.9) |
|
|
| Tumor
classification |
|
| 1.093 | 0.296 |
| T1,
T2 | 15 (40.5) | 22 (59.5) |
|
|
| T3,
T4 | 17 (53.1) | 15 (46.9) |
|
|
| Node
classification |
|
| 0.842 | 0.359 |
| N0 | 21 (42.9) | 28 (57.1) |
|
|
| N1,
N2 | 11 (55.0) | 9
(45.0) |
|
|
miR-196b involvement in the modulation
of the migration and invasion abilities of oral cancer cells
Since miR-196b has been demonstrated to be able to
act as an oncogene or as a tumor suppressor in various types of
human cancer (20,26), stable SAS and CAL-27 clones
overexpressing miRNA-196b were established in the present study by
transfecting SAS and CAL-27 cells with a precursor-miR-196b
construct. As represented in Fig. 2A,
miR-196b was significantly overexpressed in the two stable cell
lines overexpressing miR-196b (P=0.37). The effect of miR-196b on
the proliferation, migration and invasion abilities of SAS and
CAL-27 cells was subsequently investigated. Ectopic overexpression
of miR-196b did not induce a significant proliferation ability in
these two cell lines (Fig. 2B). By
contrast, the wound-healing assay indicated that the overexpression
of miR-196b could significantly promote SAS and CAL-27 migration
(P<0.01; Fig. 2C). Furthermore,
the invasion ability significantly increased in miR-196b-expressing
SAS and CAL-27 cells (SAS cells, P=0.003; CAL-27 cells, P=0.001;
Fig. 2D). Endogenous expression of
miR-196b was knocked down in the SAS and CAL-27 cells by
transfecting the cells with anti-miR-NC or anti-miR-196b. As
indicated in Fig. 3, knockdown of
endogenous miR-196b significantly inhibited the cell migration and
invasion abilities of SAS and CAL-27 cells (P<0.01; Fig. 3A and B).
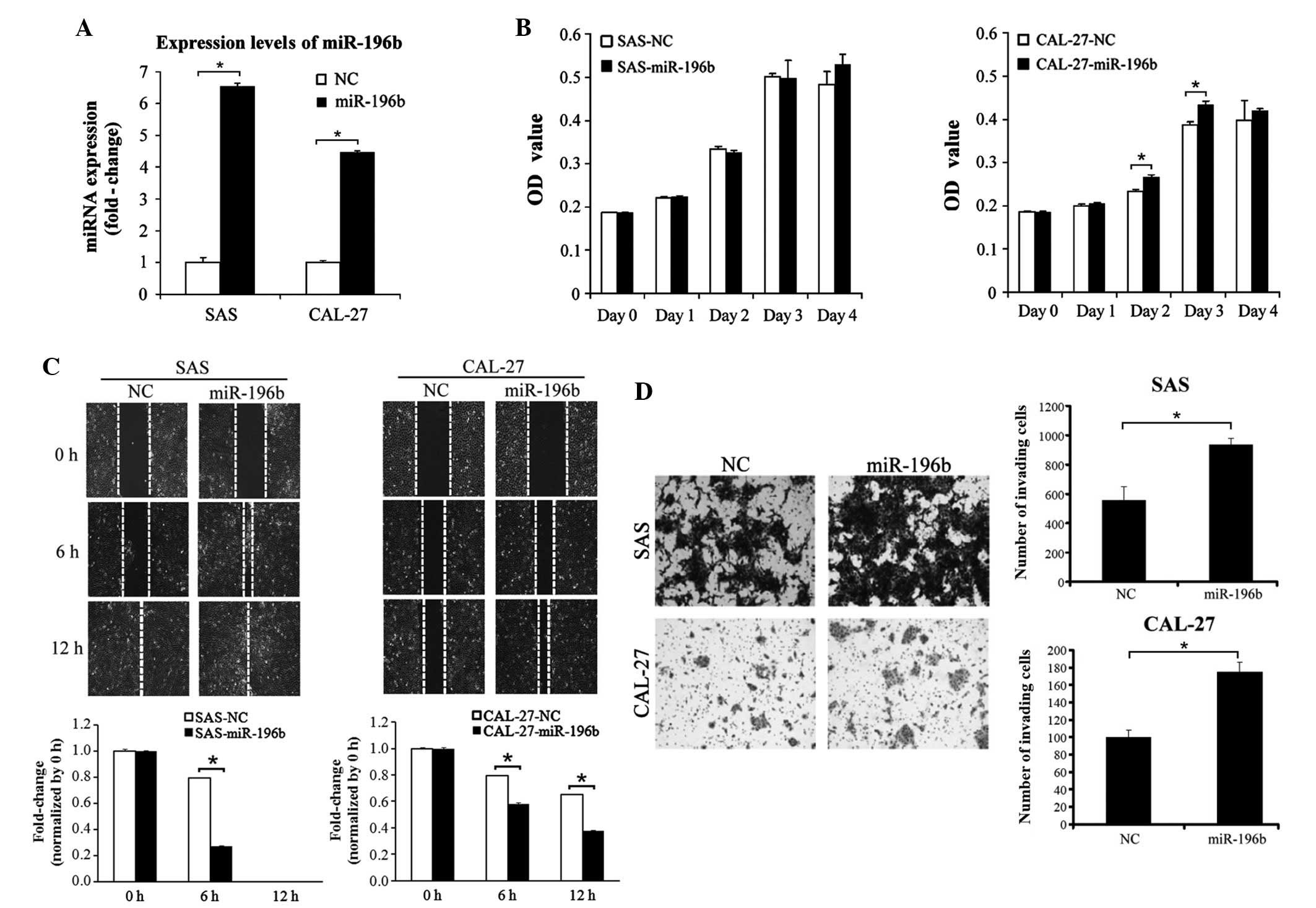 | Figure 2.Overexpression of miR-196b promoted
oral cancer cell migration and invasion abilities. (A) The
expression levels of miR-196b were examined in stable SAS and
CAL-27 cells expressing miR-196b by stem-loop reverse
transcription-quantitative polymerase chain reaction. (B) The
effect of miR-196b overexpression on SAS and CAL-27 cell growth was
examined. Cell growth was assessed using the WST-1 cell
proliferation reagent at days 0, 1, 2, 3 and 4. (C) The effect of
miR-196b overexpression on SAS and CAL-27 cell migration ability
was assessed using a wound-healing assay at 0, 6 and 12 h.
Migration ability was quantified from three independent visible
fields (magnification, ×100). (D) The effect of miR-196b
overexpression on SAS and CAL-27 cell invasion was examined using
an invasion assay with Matrigel concentrations of 1 and 0.2 µg/µl,
respectively. Invasion ability was quantified by counting the
number of invading cells per visible field (magnification, ×100).
Data are presented as the mean ± standard deviation of three
independent experiments (*P<0.05). miR/miRNA, microRNA; OD,
optical density; NC, negative control. |
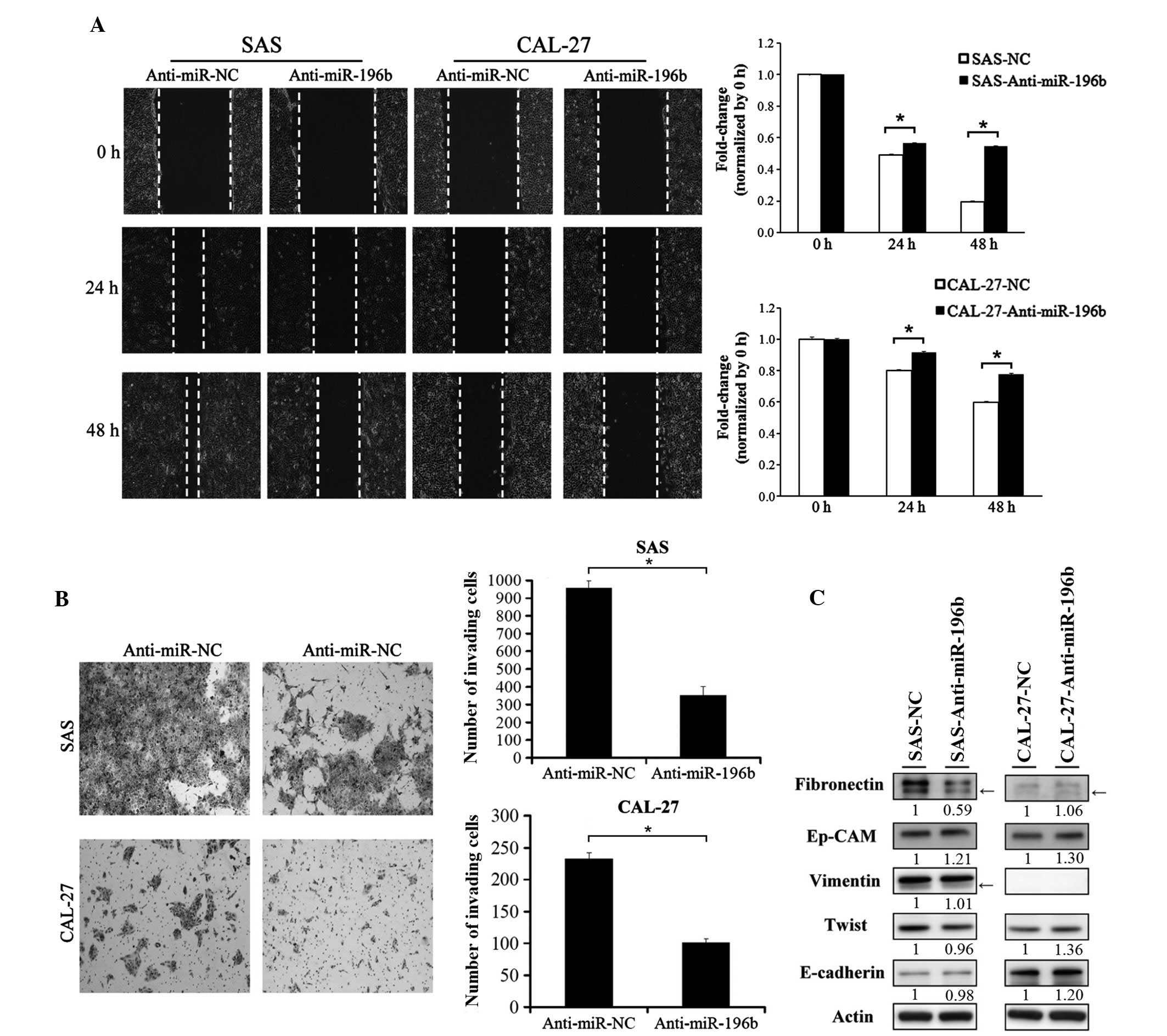 | Figure 3.Knockdown of miR-196b inhibited oral
cancer cell migration and invasion abilities. The expression of
miR-196b was knocked down in SAS and CAL-27 cells by transient
transfection with miR-196b antagomirs (anti-miR-196b). A scrambled
oligonucleotide with a random sequence was used as a negative
control. (A) The effect of miR-196b knockdown on SAS and CAL-27
cell migration was examined. Cancer cell migration ability was
assessed using a wound-healing assay at 0, 24 and 48 h. Migration
ability was quantified from three independent visible fields
(magnification, ×100). (B) The effect of miR-196b knockdown on SAS
and CAL-27 cell invasion was examined. Cancer cell invasion ability
was assessed using an invasion assay, where Matrigel concentrations
of 1 and 0.2 µg/µl were used for SAS and CAL-27 cells,
respectively. Invasion ability was quantified by counting the
number of invading cells per visible field (magnification, ×100).
Data are presented as the mean ± standard deviation of three
independent experiments (*P<0.05). (C) The expression of
epithelial–mesenchymal transition markers, including fibronectin,
epithelial cell adhesion molecule, vimentin, Twist and E-cadherin
was examined by western blotting in SAS and CAL-27 cells with and
without miR-196b knockdown. The bands corresponding to individual
genes were analyzed using ImageJ. Arrows indicate non-specific
bands. Actin was used as an internal control. miR, microRNA; NC,
negative control; Ep-CAM, epithelial cell adhesion molecule. |
Since the epithelial-mesenchymal transition (EMT) is
a crucial process in promoting cancer cell metastasis, the present
study examined whether miR-196b could induce EMT in SAS and CAL-27
cells. As presented in Fig. 3C, the
protein expression of endogenous fibronectin and was clearly
decreased in SAS cells in which miR-196b was downregulated.
However, the expression levels of the other EMT markers were not
clearly altered in SAS cells with miR-196b knockdown.
In summary, DNA hypomethylation resulted in
overexpression of oncogenic miR-196b in OSCC tissues, compared with
corresponding adjacent normal tissues. Furthermore, DNA
hypomethylation may serve as an adverse prognostic marker for
OSCC.
Discussion
The expression pattern and functional role of
miR-196b in various types of cancer remain controversial (19,22–27).
Previous studies have suggested that miR-196b could function as an
oncogene or as a tumor suppressor (19,22–27). In
the present study, miR-196b was demonstrated to be overexpressed in
oral cancer and to perform an oncogenic role in regulating OCSS
cell migration and invasion abilities. In addition, the present
study is the first to report that DNA hypomethylation contributes
to miR-196b overexpression in OSCC. Similar to the results of a
previous study by the present authors (19), the results of the present study
revealed that miR-196b was upregulated in gastric cancer tissue
samples with a hypomethylated promoter region. A high frequency of
miR-196b overexpression was observed in OSCC patients (64 of 69,
92.7%), while hypomethylated promoter regions were observed in
46.4% of the patients (32 of 69). Although DNA methylation is a
critical factor in regulating miR-196b expression, the regulation
of miR-196b expression could also be modulated by other
transcription regulatory factors, including v-Ets erythroblastosis
virus E26 oncogene homolog 2, mixed-lineage leukemia and growth
factor independent 1 (20,31,32).
Using a microarray approach, Lu et al
(13) identified several differences
in miRNA expression between an oral cancer cell line and normal
oral keratinocytes, including increased miR-196b expression in oral
cancer cells. Previous studies have indicated that overexpression
of miR-196b is associated with the biological process of the cell
cycle in glioblastomas (33), and
observed decreased cell viability, proliferation and invasion in
cervical cancer (26). In the present
study, knockdown of miR-196b reduced cell migration and invasion,
whereas overexpression of miR-196b could promote cell migration and
invasion, but not proliferation. The detailed mechanism of miR-196b
involved in regulating OSCC migration ability has not been
thoroughly investigated to date. Therefore, identifying target
genes of miR-196b is essential. Previous studies have identified
various mRNA targets of miR-196b, including breakpoint cluster
region-Abelson murine leukemia viral oncogene homolog 1, HOXA9,
c-Myc, B-cell lymphoma 2, HOXB7, HOXA9, Meis HOX 1, Fas and HOXC8
(26,27,34–36).
Combining a bioinformatic prediction tool with a microarray
approach, 15 putative targets of miR-196b were identified in OSCC
in the present study, including actin-related protein 10 homolog
(Saccharomyces cerevisiae), doublecortin domain-containing
protein 2 (DCDC2), disrupted in renal carcinoma 2, epidermal growth
factor receptor pathway substrate 15, gigaxonin, inhibitor of
growth family, member 5 (ING5), kelch-like family member 23,
ligand-dependent corepressor (LCOR), ovarian tumor
domain-containing protein 6B, Parkin RBR E3 ubiquitin protein
ligase co-regulated like, polymerase (RNA) III (DNA directed)
polypeptide D, 44 kDa, ribonucleoprotein, phosphotyrosine
binding-binding 2, sorting nexin 16, zinc finger DHHC-domain
protein containing 21 and zinc finger, MYND-type containing 11
(ZMYND11; data not shown). Among them, DCDC2, ING5, LCOR and
ZMYND11 have been reported to act as tumor suppressors in different
types of human cancer (36–39). In previous studies, DCDC2 was reported
to be hypermethylated in hepatocellular carcinoma (37), while ING5 could inhibit proliferation
and induce apoptosis in tongue SCC (38), and LCOR could repress prostate cancer
growth in vivo and in vitro, and was observed to be
functionally inactivated by Src, a proto-oncogene tyrosine-protein
kinase (39). In addition, low
expression levels of ZMYND11 in breast cancer patients were
demonstrated to correlate with poor prognosis, and overexpression
of ZMYND11 suppressed cancer cell growth in vitro and tumor
formation in mice (40). At the time
of writing the present manuscript, Lu et al (28)reported that miR-196 expression is
involved in oral cancer metastasis via the non-metastatic cells 4,
protein expressed in-c-Jun N-terminal kinase-tissue inhibitor of
metalloproteinase (MP) 1-matrix MP signaling pathway. Additionally,
assessing the levels of circulating miR-196s in plasma may serve as
an early diagnostic biomarker for oral cancer (41).
Recently, correlations between miR-196b
overexpression and poor prognosis have been identified in several
types of cancer, including osteosarcoma, gastric cancer and
glioblastoma (22,33,38). Qi
and Zhang (38) observed that high
serum expression of miR-196a and/or miR-196b, and joint expression
of miR-196a and miR-196b, are independent prognostic factors for
overall and disease-free survival of osteosarcoma patients. Lim
et al (22) reported that
overexpression of miR-196b and HOXA10 is a biomarker of poor
prognosis in patients with gastric cancer. Ma et al
(33) demonstrated that the
upregulation of miR-196b is an independent biomarker of poor
prognosis that promotes cellular proliferation in glioblastoma.
Although Lu et al (28)
reported that the expression levels of miR-196b were associated
with clinical N stage. However, no clinicopathological features
were significantly associated with miR-196b expression in oral
cancer in the present study. Notably, the present data revealed
that the unmethylation status of miR-196b in OSCC patients
significantly correlated with poor survival. In conclusion, DNA
hypomethylation contributed to regulating the expression of the
oncogenic miR-196b, and the methylation status of miR-196b could be
used as a prognostic biomarker for OSCC.
Acknowledgments
The present authors would like to acknowledge Dr
Michael Hsiao (Genomic Research Center, Academia Sinica, Taipei,
Taiwan) for kindly providing the SAS and CAL-27 cell lines. The
present study was supported by grants from the KSVGH (Kaohsiung,
Taiwan; grants no. KSVGH 103-049 and KSVGH 103-106).
References
|
1
|
Trotta BM, Pease CS, Rasamny JJ, Raghavan
P and Mukherjee S: Oral cavity and oropharyngeal squamous cell
cancer: Key imaging findings for staging and treatment planning.
Radiographics. 31:339–354. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Jemal A, Bray F, Center MM, Ferlay J, Ward
E and Forman D: Global cancer statistics. CA Cancer J Clin.
61:69–90. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Nair S and Pillai MR: Human papillomavirus
and disease mechanisms: Relevance to oral and cervical cancers.
Oral Dis. 11:350–359. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Kamangar F, Dores GM and Anderson WF:
Patterns of cancer incidence, mortality, and prevalence across five
continents: Defining priorities to reduce cancer disparities in
different geographic regions of the world. J Clin Oncol.
24:2137–2150. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Esteller M: Non-coding RNAs in human
disease. Nat Rev Genet. 12:861–874. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Wu HH, Lin WC and Tsai KW: Advances in
molecular biomarkers for gastric cancer: miRNAs as emerging novel
cancer markers. Expert Rev Mol Med. 16:e12014. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Avissar M, McClean MD, Kelsey KT and
Marsit CJ: MicroRNA expression in head and neck cancer associates
with alcohol consumption and survival. Carcinogenesis.
30:2059–2063. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Banerjee R, Mani RS, Russo N, Scanlon CS,
Tsodikov A, Jing X, Cao Q, Palanisamy N, Metwally T, Inglehart RC,
et al: The tumor suppressor gene rap1GAP is silenced by
miR-101-mediated EZH2 overexpression in invasive squamous cell
carcinoma. Oncogene. 30:4339–4349. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Gee HE, Camps C, Buffa FM, Patiar S,
Winter SC, Betts G, Homer J, Corbridge R, Cox G, West CM, et al:
Hsa-mir-210 is a marker of tumor hypoxia and a prognostic factor in
head and neck cancer. Cancer. 116:2148–2158. 2010.PubMed/NCBI
|
|
10
|
Hebert C, Norris K, Scheper MA, Nikitakis
N and Sauk JJ: High mobility group A2 is a target for miRNA-98 in
head and neck squamous cell carcinoma. Mol Cancer. 6:52007.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Liu CJ, Tsai MM, Hung PS, Kao SY, Liu TY,
Wu KJ, Chiou SH, Lin SC and Chang KW: miR-31 ablates expression of
the HIF regulatory factor FIH to activate the HIF pathway in head
and neck carcinoma. Cancer Res. 70:1635–1644. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Liu X, Jiang L, Wang A, Yu J, Shi F and
Zhou X: MicroRNA-138 suppresses invasion and promotes apoptosis in
head and neck squamous cell carcinoma cell lines. Cancer Lett.
286:217–222. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Lu YC, Chen YJ, Wang HM, Tsai CY, Chen WH,
Huang YC, Fan KH, Tsai CN, Huang SF, Kang CJ, et al: Oncogenic
function and early detection potential of miRNA-10b in oral cancer
as identified by microRNA profiling. Cancer Prev Res (Phila).
5:665–674. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Peschiaroli A, Giacobbe A, Formosa A,
Markert EK, Bongiorno-Borbone L, Levine AJ, Candi E, D'Alessandro
A, Zolla L, Finazzi Agrò A and Melino G: miR-143 regulates
hexokinase 2 expression in cancer cells. Oncogene. 32:797–802.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Wong TS, Liu XB, Wong BY, Ng RW, Yuen AP
and Wei WI: Mature miR-184 as potential oncogenic microRNA of
squamous cell carcinoma of tongue. Clin Cancer Res. 14:2588–2592.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Yang CJ, Shen WG, Liu CJ, Chen YW, Lu HH,
Tsai MM and Lin SC: miR-221 and miR-222 expression increased the
growth and tumorigenesis of oral carcinoma cells. J Oral Pathol
Med. 40:560–566. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Yekta S, Shih IH and Bartel DP:
MicroRNA-directed cleavage of HOXB8 mRNA. Science. 304:594–596.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Hornstein E, Mansfield JH, Yekta S, Hu JK,
Harfe BD, McManus MT, Baskerville S, Bartel DP and Tabin CJ: The
microRNA miR-196 acts upstream of Hoxb8 and Shh in limb
development. Nature. 438:671–674. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Tsai KW, Hu LY, Wu CW, Li SC, Lai CH, Kao
HW, Fang WL and Lin WC: Epigenetic regulation of miR-196b
expression in gastric cancer. Genes Chromosomes Cancer. 49:969–980.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Liao YL, Hu LY, Tsai KW, Wu CW, Chan WC,
Li SC, Lai CH, Ho MR, Fang WL, Huang KH and Lin WC: Transcriptional
regulation of miR-196b by ETS2 in gastric cancer cells.
Carcinogenesis. 33:760–769. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Tsai KW, Liao YL, Wu CW, Hu LY, Li SC,
Chan WC, Ho MR, Lai CH, Kao HW, Fang WL, et al: Aberrant expression
of miR-196a in gastric cancers and correlation with recurrence.
Genes Chromosomes Cancer. 51:394–401. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Lim JY, Yoon SO, Seol SY, Hong SW, Kim JW,
Choi SH, Lee JS and Cho JY: Overexpression of miR-196b and HOXA10
characterize a poor-prognosis gastric cancer subtype. World J
Gastroenterol. 19:7078–7088. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Luthra R, Singh RR, Luthra MG, Li YX,
Hannah C, Romans AM, Barkoh BA, Chen SS, Ensor J, Maru DM, et al:
MicroRNA-196a targets annexin A1: A microRNA-mediated mechanism of
annexin A1 downregulation in cancers. Oncogene. 27:6667–6678. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Bhatia S, Kaul D and Varma N: Functional
genomics of tumor suppressor miR-196b in T-cell acute lymphoblastic
leukemia. Mol Cell Biochem. 346:103–116. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Coskun E, von der Heide EK, Schlee C,
Kühnl A, Gökbuget N, Hoelzer D, Hofmann WK, Thiel E and Baldus CD:
The role of microRNA-196a and microRNA-196b as ERG regulators in
acute myeloid leukemia and acute T-lymphoblastic leukemia. Leuk
Res. 35:208–213. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
How C, Hui AB, Alajez NM, Shi W, Boutros
PC, Clarke BA, Yan R, Pintilie M, Fyles A, Hedley DW, et al:
MicroRNA-196b regulates the homeobox B7-vascular endothelial growth
factor axis in cervical cancer. PLoS One. 8:e678462013. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Li Y, Zhang M, Chen H, Dong Z, Ganapathy
V, Thangaraju M and Huang S: Ratio of miR-196s to HOXC8 messenger
RNA correlates with breast cancer cell migration and metastasis.
Cancer Res. 70:7894–7904. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Lu YC, Chang JT, Liao CT, Kang CJ, Huang
SF, Chen IH, Huang CC, Huang YC, Chen WH, Tsai CY, et al:
OncomiR-196 promotes an invasive phenotype in oral cancer through
the NME4-JNK-TIMP1-MMP signaling pathway. Mol Cancer. 13:2182014.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Greene FL, Page DL, Fleming ID, Fritz AG,
Balch CM, Haller DG and Morrow M: AJCC Cancer Staging Manual (6th).
Springer. New York, NY: 2002. View Article : Google Scholar
|
|
30
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) Method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Popovic R, Riesbeck LE, Velu CS, Chaubey
A, Zhang J, Achille NJ, Erfurth FE, Eaton K, Lu J, Grimes HL, et
al: Regulation of mir-196b by MLL and its overexpression by MLL
fusions contributes to immortalization. Blood. 113:3314–3322. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Velu CS, Baktula AM and Grimes HL: Gfi1
regulates miR-21 and miR-196b to control myelopoiesis. Blood.
113:4720–4728. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Ma R, Yan W, Zhang G, Lv H, Liu Z, Fang F,
Zhang W, Zhang J, Tao T, You Y, et al: Upregulation of miR-196b
confers a poor prognosis in glioblastoma patients via inducing a
proliferative phenotype. PLoS One. 7:e380962012. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Liu Y, Zheng W, Song Y, Ma W and Yin H:
Low expression of miR-196b enhances the expression of BCR-ABL1 and
HOXA9 oncogenes in chronic myeloid leukemogenesis. PLoS One.
8:e684422013. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Abe W, Nasu K, Nakada C, Kawano Y,
Moriyama M and Narahara H: miR-196b targets c-myc and Bcl-2
expression, inhibits proliferation and induces apoptosis in
endometriotic stromal cells. Hum Reprod. 28:750–761. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Li Z, Huang H, Chen P, He M, Li Y,
Arnovitz S, Jiang X, He C, Hyjek E, Zhang J, et al: miR-196b
directly targets both HOXA9/MEIS1 oncogenes and FAS tumour
suppressor in MLL-rearranged leukaemia. Nat Commun. 3:6882012.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Inokawa Y, Nomoto S, Hishida M, Hayashi M,
Kanda M, Nishikawa Y, Takeda S, Sugimoto H, Fujii T, Yamada S and
Kodera Y: Detection of doublecortin domain-containing 2 (DCDC2), a
new candidate tumor suppressor gene of hepatocellular carcinoma, by
triple combination array analysis. J Exp Clin Cancer Res.
32:652013. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Qi L and Zhang Y: Truncation of inhibitor
of growth family protein 5 effectively induces senescence, but not
apoptosis in human tongue squamous cell carcinoma cell line. Tumour
Biol. 35:3139–3144. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Asim M, Hafeez BB, Siddiqui IA, Gerlach C,
Patz M, Mukhtar H and Baniahmad A: Ligand-dependent corepressor
acts as a novel androgen receptor corepressor, inhibits prostate
cancer growth, and is functionally inactivated by the Src protein
kinase. J Biol Chem. 286:37108–37117. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Wen H, Li Y, Xi Y, Jiang S, Stratton S,
Peng D, Tanaka K, Ren Y, Xia Z, Wu J, et al: ZMYND11 links histone
H3.3K36me3 to transcription elongation and tumour suppression.
Nature. 508:263–268. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Lu YC, Chang JT, Huang YC, Huang CC, Chen
WH, Lee LY, Huang BS, Chen YJ, Li HF and Cheng AJ: Combined
determination of circulating miR-196a and miR-196b levels produces
high sensitivity and specificity for early detection of oral
cancer. Clin Biochem. 48:115–121. 2015. View Article : Google Scholar : PubMed/NCBI
|