Introduction
Ovarian cancer is the number one cause of
cancer-associated mortality out of all gynecological malignancies
(1). The annual incidence of ovarian
cancer is 12.7/100,000 globally, and the mortality rate is
8.1/100,000 (2). In 2013, there were
22,240 women diagnosed with ovarian cancer in the United States,
and 14,030 of these succumbed to disease (3). It is well-known that cancer antigen 125
(CA125) is used widely for clinical diagnosis and monitoring of the
recurrence of ovarian cancer (4).
However, CA125 is an unsuitable marker. Although it demonstrates
elevated expression in >80% of ovarian cancer patients, its
expression is also observed in various other physiological or
pathological conditions, including menstrual period, early
pregnancy and endometriosis (5).
Thus, the identification of specific biomarkers for early diagnosis
of ovarian cancer is important and urgent.
Phage display is considered to be a powerful
biological technology for screening specific peptides that bind to
the surface of tumor cells (6). It
was initially developed by Smith in 1985 (7). The phage-displayed random peptide
libraries are first incubated on a plate coated with the target
protein or cell. Then, unbound phage with no or low affinity are
washed away. The eluted high-affinity phage are amplified using
host bacteria and submitted to additional binding/amplification
cycles to enrich the pool in favor of binding sequences. After 3–5
rounds, individual clones are characterized by DNA sequencing and
enzyme-linked immunosorbent assay (ELISA) (8) to obtain the corresponding structural and
functional information.
The present study aimed to identify optimal
biomarkers for ovarian cancer by using a Phage Display Peptide
Library to screen for ligands that selectively target ovarian
cancer cells. The results of the present study may assist with the
diagnosis of ovarian cancer and the phages identified may be
utilized as carriers for drug delivery.
Materials and methods
Cells and reagents
The HO-8910 human ovarian cancer cell line, and HeLa
cervical cancer cell line, were purchased from Jilin Baili
Biotechnology Co., Ltd. (Changchun, China). The Chinese hamster
ovary cell line (CHO) was purchased from Sangon Biotech Co., Ltd.
(Shanghai, China). Dulbecco's modified Eagle's medium (DMEM) and
RPMI-1640 medium were purchased from Sangon Biotech Co., Ltd. HeLa
cells were cultured in RPMI-1640, while the other cells were
cultured in DMEM supplemented with 10% (v/v) fetal bovine serum, at
37°C with 5% CO2.
Escherichia coli ER2738 was purchased from
Biovector Co., Ltd. (Beijing, China). The M13K07 phage, anti-M13
mouse monoclonal antibody (#27-9421-01), horseradish peroxidase
(HRP)-conjugated goat anti-mouse antibody (#sc-2005) and
fluorescein isothiocyanate (FITC)-labeled goat anti-mouse antibody
(#sc-2010) were purchased from Sangon Biotech Co., Ltd..
Phage display biopanning
procedures
The Ph.D.-7 Phage Display Peptide Library kit was
purchased from New England BioLabs, Inc. (Ipswich, MA, USA).
Screening procedures were performed according to the manufacturer's
protocol (version 1.0) with some modifications. Firstly, CHO and
HO-8910 cells were digested with trypsin and the number of cells
was adjusted to 1×107/ml. Subsequently, 100 µl CHO cells
were transferred to an Eppendorf tube and 10 µl of the Ph.D.-7
Phage-Display Peptide Library was added, which initially contained
2×1011 plaque-forming units (pfu). The cells were
incubated at 4°C for 2 h. A total of 200 µl organic solvent was
added to the tube, which consisted of 180 µl dibutyl phthalate
(DBP) and 20 µl cyclohexane (Beijing Yiqiangsheng Technology Co.,
Ltd., Beijing China). The tube was subsequently centrifuged at
10,000 × g for 10 min. Following centrifugation, the soluble fluid
upper layer was pipetted into a fresh tube, which contained HO-8910
cells, and was incubated at 4°C for 3 h. The precipitate was
transferred to a fresh tube, and 200 µl Luria-Bertani (LB) with
E. coli ER2738 (mid-log phase) was added and incubated at
37°C for 30 min. Subsequently, phage was titrated and amplified,
according to the manufacturer's instructions (New England BioLabs,
Inc.; www.neb.com/protocols/2014/
05/08/m13-titer-protocol). Finally, 5 rounds of in vitro
reiterative biopanning were performed.
Selection and amplification of
positive clones
Following 5 rounds of biopanning, 60 blue plaques
were randomly selected, and were individually added to E.
coli ER2738 cultures for amplification and titration.
ELISA
HO-8910 cells were plated into 96-well plates at a
density of 104 cells/well. Following 1 h of incubation
at 37°C, the selected positive phage clones (1010
pfu/well), M13K07 phage and phosphate-buffered saline (PBS), were
added individually to the cells and incubated at 37°C for 2 h.
Subsequently, the cells were washed three times with PBS and
cultured at 37°C for 2 h in the presence of anti-M13 mouse
monoclonal antibody (dilution, 1:6,000). Subsequently, the plates
were washed and HRP-conjugated goat anti-mouse antibody (dilution,
1:4,000) was added. Following 2 h of incubation, the plates were
washed and 200 µl fresh substrate solution
(3,3′,5,5′-tetramethylbenzidine; Sigma-Aldrich China, Inc.,
Shanghai, China) was added to each well, and the absorbance values
at 450 nm were recorded using a plate reader.
DNA sequencing
A total of 13 phage clones were selected if their
optical density (OD)450 was >0.6, and the single
stranded DNA from the positive phages was purified using an M13
purification kit (Beijing Sunny Instruments Co., Ltd., Beijing,
China) according to the manufacturer's protocol. The samples were
sent to Sangon Biotech Co., Ltd. for sequencing, and the sequences
were analyzed by using Vector NTI Advance® software
(version 10.3; Thermo Fisher Scientific, Inc., Waltham, MA,
USA).
Immunofluorescence assay
HO-8910, HeLa and CHO cells were seeded onto glass
coverslips at 2×104/ml, and grown to 80% confluence.
Cells were washed gently with PBS, and fixed in 4% paraformaldehyde
for 15 min at room temperature. The selected targeting phage clone
P2 and PBS were added individually to the cells and incubated at
37°C for 2 h, following by washing with PBS three times. Cells were
subsequently incubated with anti-M13 mouse monoclonal antibody
(dilution, 1:500). Following 1 h of incubation at 37°C, the cells
were washed 10 times with PBST, and FITC-labeled goat anti-mouse
antibody (dilution, 1:500) and propidium iodide (dilution, 1:1,000)
were added. Following incubation at 37°C for 30 min, the cells were
visualized using a fluorescence microscope.
Immunohistochemical staining
The selected phage clone was added to ovarian cancer
and normal ovarian tissue samples, which were obtained from the
Department of Gynecology and Obstetrics, The Second Affiliated
Hospital & Yuying Children's Hospital of Wenzhou Medical
University (Wenzhou, China). Following 30 min of incubation at room
temperature, the tissue samples were sequentially incubated with
anti-M13 mouse monoclonal antibody (dilution, 1:300) for 1 h,
followed by incubation with the HRP-conjugated goat anti-mouse
antibody (dilution, 1:100). A total of 1 h later,
3,3′-diaminobenzidine (DAB) solution was added and the samples were
visualized using a light microscope.
Results
Phage display biopanning
In the present study, a 7-mer phage display library
was employed to screen the peptides binding specifically to the
HO-8910 ovarian cancer cell line. The results of this screening
(Table I) revealed that the recovery
rate increased 92-fold following 5 rounds of biopanning, which
demonstrated that the phages binding to HO-8910 cells were
enriched.
 | Table I.Biopanning of the HO-8910 ovarian
cancer cell line using a phage display peptide library. |
Table I.
Biopanning of the HO-8910 ovarian
cancer cell line using a phage display peptide library.
| Round | Input number,
pfu | Output number,
pfu | Recovery
ratea |
|---|
| 1 |
2.0×1011 |
5.0×105 |
2.5×10−6 |
| 2 |
1.5×1011 |
1.0×106 |
6.7×10−6 |
| 3 |
1.8×1011 |
3.0×106 |
1.7×10−5 |
| 4 |
2.5×1011 |
1.0×107 |
4.0×10−5 |
| 5 |
1.6×1011 |
3.6×107 |
2.3×10−4 |
ELISA and DNA sequencing
Following 5 rounds of biopanning, the individual
phages were randomly selected and analyzed using ELISA assay. As
demonstrated in Fig. 1, the
OD490 value of 13 clones was >0.6, and these 13 phage
clones (P2, P3, P9, P12, P15, P30, P35, P37, P48, P54, P57, P58 and
P60) were sequenced. As demonstrated in Table II, 7 distinct sequences from the 13
clones were obtained and the most frequent DNA sequence was AAC CCG
ATG ATT CGC CGC CAG; the corresponding amino acid sequence was
NPMIRRQ.
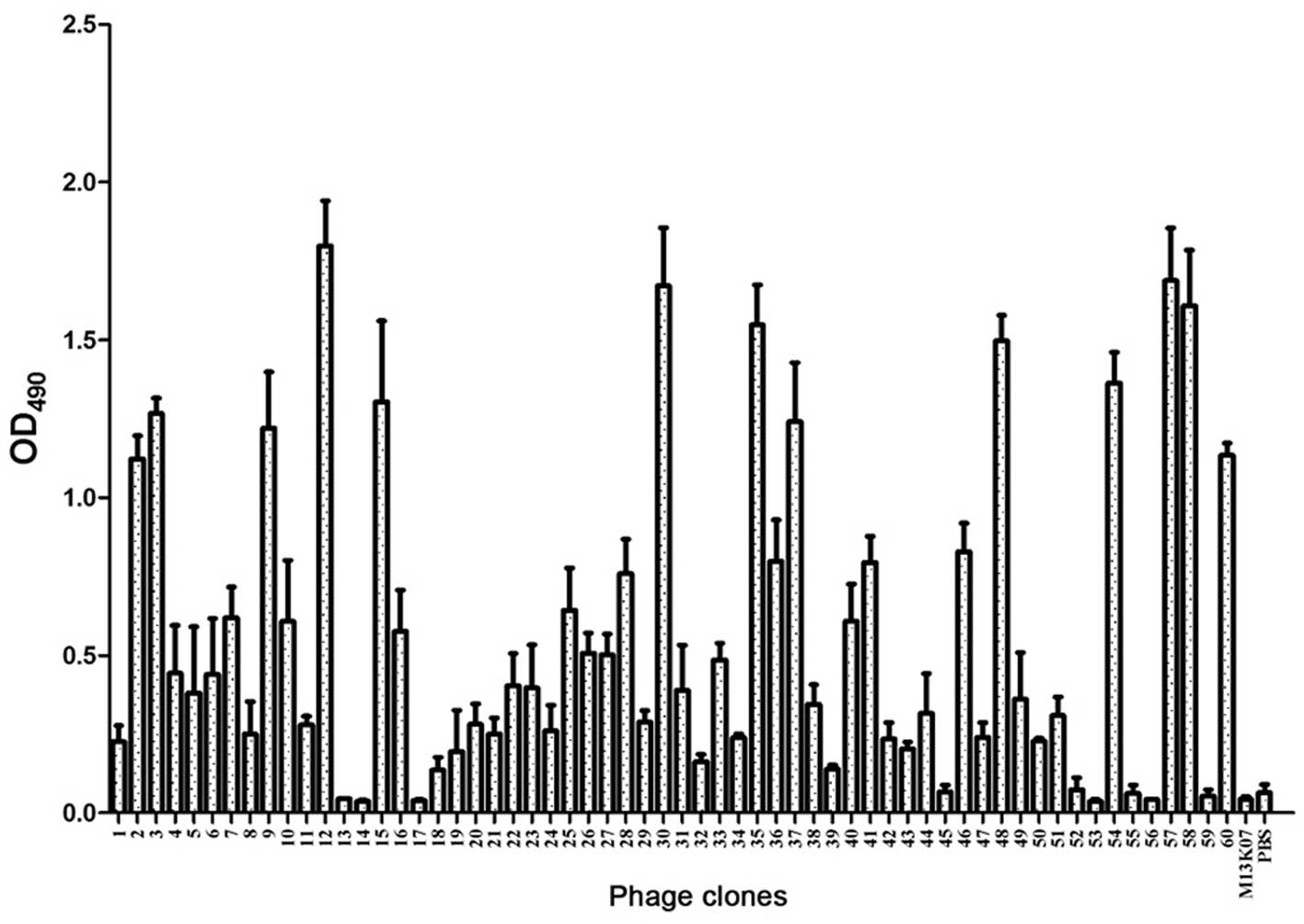 | Figure 1.Enzyme-linked immunosorbent assay
(ELISA) of the selected phage clones binding to HO-8910 cells.
After 5 rounds of biopanning, 60 plaques were randomly selected and
analyzed using ELISA. The OD490 value of 13 clones was
>0.6 (P2, P3, P9, P12, P15, P30, P35, P37, P48, P54, P57, P58
and P60). The M13K07 phage and phosphate-buffered saline were used
as negative controls. OD, optical density. |
 | Table II.DNA sequences of the 13 clones. |
Table II.
DNA sequences of the 13 clones.
| Clone | DNA sequence | Amino acid
sequence | Frequency |
|---|
| P2, P3, P15, P30,
P54 |
AACCCGATGATTCGCCGCCAG | NPMIRRQ | 5 |
| P9, P35, P58 |
ATGCGCATGACCATTATTAAC | MRMTIIN | 3 |
| P57 |
CTGCGCCTGCGCAACACCCGC | LRLRNTR | 1 |
| P12 |
AGCCATCTGCGCCATCGCATT | SHLRHRI | 1 |
| P37 |
AAACTGCCGCTGACCACCAAA | KLPLTTK | 1 |
| P60 |
CCGATTAAAACCAACCGCAAA | PIKTNRK | 1 |
| P48 |
AAACCGACCATTCCGACCAAA | KPTIPTK | 1 |
Immunocytochemical staining and
immunofluorescence assay
According to the results of the phage clones DNA
sequence analysis, the most frequent sequence (NPMIRRQ) was
selected, and as P2, P3, P15, P30 and P54 all contained the same
amino acid sequence, the corresponding phage clone P2 was selected
to perform immunocytochemical staining and immunofluorescence assay
to additionally confirm the specificity of the phage clone binding
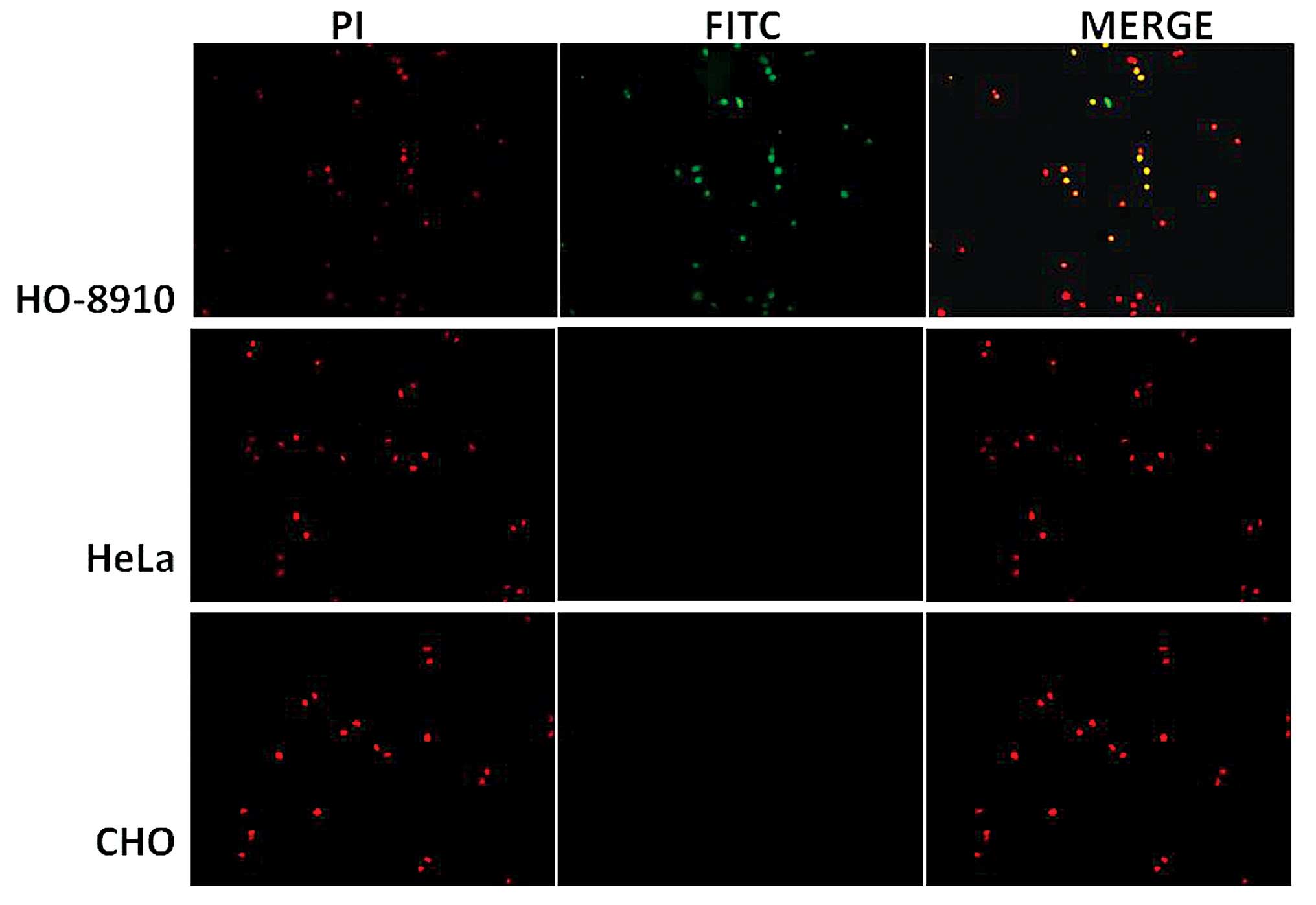
to HO-8910 cells. As demonstrated in Fig.
2, a fluorescent signal was observed in the HO-8910 cells, and
no fluorescent signal was observed in the HeLa and CHO cells.
Immunohistochemical staining
results
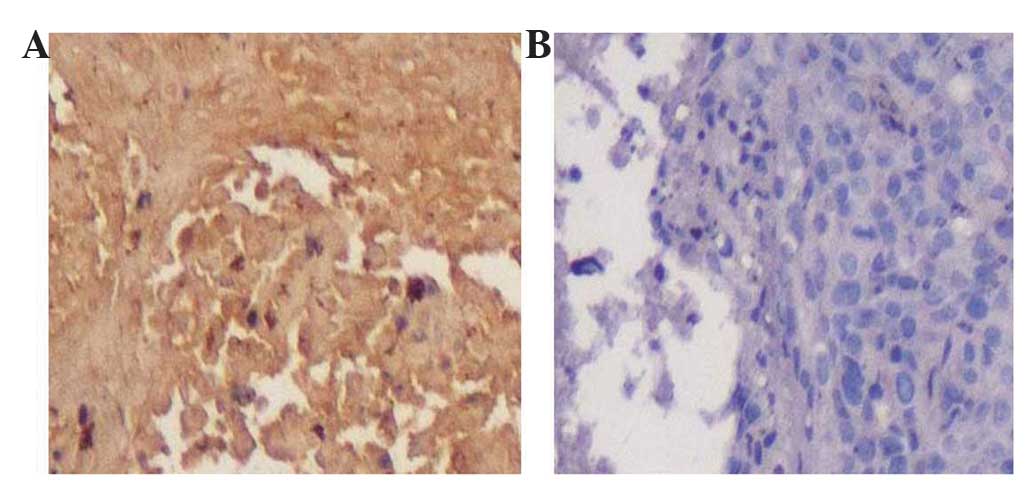
In Fig. 3, positive
DAB staining (brown color) was observed in the ovarian cancer
tissue incubated with phage clone P2, and not observed in normal
ovarian tissue.
Discussion
In recent years, tumor-targeting therapy has become
one of the focuses of clinical cancer treatment, with the aim of
using highly-specific tumor material as a carrier, which will be
able to specifically deliver anti-tumor drugs to the tumor site
(9). Small molecule peptides may be
an ideal carrier of anti-tumor targeted drugs, due to their simple
structure, easy preparation and strong penetration into the tumor
tissue (10). Phage display
technology is able to rapidly screen high-affinity antibodies or
peptide ligands that bind with specific target molecules (11). The advantage of this technology is
that it realizes the linkage between genotype and phenotype
(12), and there is no need to know
the structural information of the target molecule in advance
(13). It is possible to obtain the
amino acid sequence indirectly by sequencing the positive phage
clones following a screening procedure.
Recently, peptides targeting different tumor cells
have been identified using phage display technology (14,15). For
the ovarian cancer cell line SKOV3, the peptides WSGPGVWGASVK
(16) and SVSVGMKPSPRP (17) were identified to have high affinity.
However, there are various pathological types of ovarian cancer,
and there is a limited amount of research focusing on the targeting
of these peptides against the ovarian cancer HO-8910 cell line. In
the present study, a 7-mer phage display peptide library was used
to isolate specific peptides binding specifically to HO-8910 cells.
After 5 rounds of biopanning, the recovery rate of phages
demonstrated a 92-fold increase over the first round, indicating
that the bound phage were amplified. Subsequently, blue plaques
were randomly selected after 5 rounds of biopanning. According to
the results of DNA sequencing, the 13 phages contained 7 distinct
polypeptide sequences, and the amino acid sequence NPMIRRQ was
enriched significantly.
To additionally identify the affinity of the
positive clones, the phage clone P2 was selected for further study.
Immunocytochemical staining and immunofluorescence assay were
performed to confirm the specificity of the phage clone P2 binding
to HO-8910 cells. The results of immunocytochemical staining and
immunofluorescence assay suggested that the phage clone P2 was able
to bind to HO-8910 cells, and not the HeLa cervical cancer cell
line.
In conclusion, the present study identified a novel
peptide, NPMIRRQ, targeting ovarian cancer was isolated using phage
display. The peptide was able to bind to target cells and ovarian
cancer tissues specifically, and therefore potentially be applied
in the diagnostics and treatment of ovarian cancer.
Acknowledgements
The present study was supported by grants from the
Zhejiang Provincal Natural Science Foundation (no. LQ16H160022),
the Zhejiang Provincal Technology Bureau, Zhejiang, China (no.
2014C37003) and Wenzhou Technology Bureau, Wenzhou, China (no.
2015Y0372).
References
|
1
|
Rooth C: Ovarian cancer: Risk factors,
treatment and management. Br J Nurs. 22:S23–S30. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Collins Y, Holcomb K, Chapman-Davis E,
Khabele D and Farley JH: Gynecologic cancer disparities: A report
from the Health Disparities Taskforce of the Society of Gynecologic
Oncology. Gynecol Oncol. 133:353–361. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Siegel R, Naishadham D and Jemal A: Cancer
statistics, 2013. CA Cancer J Clin. 63:11–30. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Bocheva Y, Bochev P and Ivanov S: Ca-125
in diagnosis and monitoring of patients with ovarian cancer. Akush
Ginekol (Sofiia). 54:11–7. 2015.PubMed/NCBI
|
|
5
|
Cannistra SA: Cancer of the ovary. N Engl
J Med. 351:2519–2529. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Jemal A, Siegel R, Xu J and Ward E: Cancer
statistics, 2010. CA Cancer J Clin. 60:277–300. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Smith GP: Filamentous fusion phage: Novel
expression vectors that display cloned antigens on the virion
surface. Science. 228:1315–1317. 1985. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Parmley SF and Smith GP:
Antibody-selectable filamentous fd phage vectors: Affinity
purification of target genes. Gene. 73:305–318. 1988. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Velasco-Velázquez M, Xolalpa W and Pestell
RG: The potential to target CCL5/CCR5 in breast cancer. Expert Opin
Ther Targets. 18:1265–1275. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Bastien JI, McNeill KA and Fine HA:
Molecular characterizations of glioblastoma, targeted therapy, and
clinical results to date. Cancer. 121:502–516. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Henry KA, Arbabi-Ghahroudi M and Scott JK:
Beyond phage display: Non-traditional applications of the
filamentous bacteriophage as a vaccine carrier, therapeutic
biologic, and bioconjugation scaffold. Front Microbiol. 6:7552015.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Kaplan G and Gershoni JM: A general insert
label for peptide display on chimeric filamentous bacteriophages.
Anal Biochem. 420:68–72. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Silacci M, Brack S, Schirru G, Mårlind J,
Ettorre A, Merlo A, Viti F and Neri D: Design, construction, and
characterization of a large synthetic human antibody phage display
library. Proteomics. 5:2340–2350. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Lee KJ, Lee JH, Chung HK, Ju EJ, Song SY,
Jeong SY and Choi EK: Application of peptide displaying phage as a
novel diagnostic probe for human lung adenocarcinoma. Amino Acids.
48:1079–1086. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Che YJ, Wu HW, Hung LY, Liu CA, Chang HY,
Wang K and Lee GB: An integrated microfluidic system for screening
of phage-displayed peptides specific to colon cancer cells and
colon cancer stem cells. Biomicrofluidics. 9:0541212015. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Ma CI, Yin G, Yan D, He X, Zhang L, Wei Y
and Huang Z: A novel peptide specifically targeting ovarian cancer
identified by in vivo phage display. J Pept Sci. 19:730–736. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Zhang L, Yin G, Yan D, Wei Y, Ma C, Huang
Z, Liao X, Yao Y, Chen X and Hao B: In vitro screening of ovarian
tumor specific peptides from a phage display peptide library.
Biotechnol Lett. 33:1729–1735. 2011. View Article : Google Scholar : PubMed/NCBI
|