Introduction
Cancer develops through a multistep process that
involves mutations in certain genes; these include the inactivation
of recessive tumor suppressor genes (TSGs), the activation of
dominant oncogenes, and the inactivation of genes involved in DNA
repair and/or replication (1). Both
copies of TSG alleles have to be inactivated for their function to
be lost (2). While one allele may be
inactivated by point mutation, changes in methylation or a small
deletion, the other allele is frequently inactivated by a large
deletion that involves the whole gene of interest, as well as
adjacent stretches of DNA (3).
Searches for genomic regions that undergo frequent deletion in
cancer samples have aided in the identification and confirmation of
the location of several TSGs.
Minimal deletion region (MDR) analysis of solid
tumors has enabled the delineation of specific regions that are the
likely locations of critical TSGs, and have also provided a
molecular portrait of the pattern of accumulation of genetic
alterations during the multistep progression that leads to cancer
(4–7).
Confirmed TSGs may be examined by MDR analysis, since MDR
consistently identifies the map positions of critical TSGs that are
involved in various different types of cancer (8).
Genome-wide loss of heterozygosity (LOH) data has
previously demonstrated that chromosome regions at 2p23.3, 2p24.3,
2q35, 6p22.2, 7p14.3, 7p22.2, 17q24.3 and 21q22.3 were novel and
frequent LOH regions in non-small-cell lung cancer (NSCLC)
(9). In the present study, refined
mapping data obtained using 9 markers together with expression
analysis, it was demonstrated that LOC51321 may be a
deletion target on 17q24.3 in NSCLC tumors. LOC51321, a
putative gene, named AMZ2, was determined to map at 17q24.2
according to RefSeq. In AceView, it covers 51.26 kb, from 63713640
to 63764901 (NCBI build 35, August 2004), on the direct strand.
Putative homologs of LOC51321 have been identified in
Canis familiaris, Mus musculus and Rattus
norvegicus. Conserved domains from the CDD (conserved protein
domain database) found in protein sequences by rpsblast search
demonstrated that the predicted gene encoded a putative
Zn-dependent protease. Based on the above findings, it could be
hypothesized that LOC51321 may potentially possess metallopeptidase
activity. Therefore, the present study characterized the candidate
TSG, LOC51321 (the AMZ2 gene), by analyzing its mRNA
expression and promoter hypermethylation in lung cancer cell lines
and samples from lung cancer patients.
Materials and methods
Sample preparation and clinical
characterization of the patients
Tissues were collected after obtaining institutional
review board permission from the Human Biobank of the Research
Center of Clinical Medicine, Veteran General Hospital (Taipei,
Taiwan) and informed consent from the recruited patients.
Surgically resected tumor samples from 53 patients admitted to
Veteran General Hospital with NSCLC were collected between 2002 and
2008. Of these patients, 24 had adenocarcinoma (AD), 24 had
squamous cell carcinoma (SCC), and 5 had large-cell carcinoma,
according to the World Health Organization classification (10). Surgically resected tumor samples were
immediately snap-frozen and then subsequently stored in liquid
nitrogen. Information on the gender, age, smoking history and tumor
type of the patients were obtained from the Cancer Data Bank of
Taipei Veteran General Hospital with de-identification of patient
IDs and names performed via the Human Biobank, Research Center of
Clinical Medicine, Taipei Veteran General Hospital.
For the LOH and methylation assay, genomic DNA from
matched pairs of primary tumor samples and nearby normal lung
tissue samples were prepared using proteinase K digestion and
phenol-chloroform extraction, followed by ethanol precipitation
(all reagents from Sigma-Aldrich, St Louis, MO, USA) (9). Serial 5-µm serial sections were cut from
formalin-fixed, paraffin-embedded tumor tissues. All slides were
stained with hemotoxylin and eosin (Sigma-Aldrich), and one of the
slides was coverslipped and used as a guide to localize the tumor
region. Tumor cells were then microdissected from up to three
sections. The cells were pelleted by centrifugation at high speed
(15,115 × g) for 5 min. The DNA was extracted by digesting the
cells in buffer consisting of 50 µM Tris-HCl (pH 8.5), 1 µM
ethylenediamineterraacetic acid (pH 8.0), 0.5% Tween 20, and 200
mg/ml proteinase K, at 55°C for 4–6 h, then at 37°C for 24–48 h,
followed by 10 min of incubation at 95°C to destroy any remaining
proteinase K activity. The cellular DNA was then purified using
phenol/chloroform extraction followed by ethanol precipitation. Any
insoluble material present in the DNA solution was pelleted by
centrifugation and aliquots of supernatant were used directly in
the polymerase chain reactions (PCRs). For the RNA expression
assay, total RNA was prepared from tumors and normal lung tissues,
using Trizol reagent (Invitrogen Life Technologies, Carlsbad, CA,
USA). cDNA was then synthesized using SuperScript™ reverse
transcriptase (Invitrogen Life Technologies) using the protocols
provided by the manufacturer.
Minimal deletion region analysis
Genomic DNA (20 ng) from normal lung cells or from
tumor cells of 48 patients were used for each PCR analysis. PCR
reactions were conducted in a 10 ml volume using 0.05 µM
fluorescently labeled and unlabeled primers in order to detect the
microsatellite markers located in the region of interest. The
microsatellite markers used were as follows: D17S1809, D17S1825,
D17S1882, D17S1816, D17S807, D17S1813, D17S2193, D17S789 and
D17S795. The PCR reaction contained 250 µM each dNTP, 2.5 µM MgCl2,
and 0.5 units AmpliTaq DNA polymerase (PE Applied Biosystems,
Foster City, CA), and the manufacturer's instructions were followed
(PE Applied Biosystems). The primer sequences were obtained from
Research Genetics (Huntsville, AL, USA). Next the PCR products were
mixed with fluorescent molecular weight markers and subjected to
electrophoresis on a MegaBACE 1000 automatic sequencer. Allele
sizes were determined using Genetic Profiler Analysis version 2.0
software. The allelic ratio was calculated as (T1/T2)/(N1/N2),
which represents the ratio of the area values for the tumor (T)
alleles versus the ratio of area values for the normal (N) alleles.
LOH was defined as an allelic ratio >2.0 or <0.5.
Cell lines
The human lung cancer cell lines A549, H23, H226,
H226Br, H1299, H1355 and H1435, and normal lung cell lines MRC5 and
Beas2B were purchased from the American Type Culture Collection
(Manassas, VA, USA). The human lung cancer cell lines CL1-0, CL1-1,
CL1-3, CL1-5-F4, CL2 and CL3 were obtained from a Taiwanese patient
with AD of the lung and was kindly provided by Dr. Pan-Chyr Yang of
the Department of Internal Medicine, National Taiwan University
Hospital (Taipei, Taiwan).
Analysis of LOC51321 mRNA expression
using a multiplex reverse transcriptase-PCR (RT-PCR) assay
LOC51321 (AMZ2 gene) mRNA expression
were assayed by multiplex RT-PCR analysis using the
glyceraldehyde-3-phosphate dehydrogenase (GAPDH) gene
as an internal control. The coding regions of exons 3–7 of the
LOC51321 (AMZ2) gene and of the GAPDH gene
were amplified. Sequences for the primers for the RT-PCR were as
follows: LOC51321F, Forward 5′-CCAGTGCTTGTGTCACAGTATGAG-3′ and
LOC51321R, Reverse 5′-TCTACAATGCTGAAGCCAAC-3′. Reactions were
carried out in a volume of 25 ml with 1 ml cDNA and 0.25
pmol primers using a MyCycler™ thermal cycler (Bio-Rad
Laboratories, Taipei, Taiwan). The PCR was performed for 32 cycles
with an annealing temperature of 61°C. The number of cycles, the
amount of primers and the cDNA used were adjusted to provide
quantitative amplification during multiplex RT-PCR. All PCR
products were separated on 2% agarose gels (100 V/17.5 cm) and
stained with ethidium bromide. To quantify the relative levels of
gene expression in the multiplex RT-PCR assay, the value for the
internal standard (GAPDH) in each test tube was used as the
baseline value for gene expression in that sample, and a relative
value was calculated for each target gene transcript amplified from
each tumor and its matched normal sample. Tumor cells that
exhibited mRNA expression <50% of that of the normal cells were
deemed to have an abnormal pattern of expression.
Methylation-specific PCR (MSP)
assay
The methylation status in the promoter region of the
LOC51321 (AMZ2) gene was determined by chemical
treatment with sodium bisulfite and subsequent MSP analysis. In
total, 500 ng of genomic DNA was denatured at 95°C for 5 min and
then in the presence of NaOH (final concentration, 0.2 M) incubated
at 37°C for 15 min. Next, 10 µM hydroquinone (Sigma-Aldrich), and 3
M sodium bisulfite (Sigma-Aldrich) were added and the mixture was
incubated at 50°C for 18 h. After these treatments, the modified
DNA was purified using a Microcon YM-50 DNA purification column
(Millipore, Bedford, MA, USA). Treatment of the genomic DNA with
sodium bisulfite converted unmethylated but not methylated
cytosines to uracils, which were then converted to thymidines
during the subsequent PCR step. The primers used for the converted
sequence were as follows: LOC51321MF, Forward
5′-CGGAGTCGTCGCGAGCGTAA-3′ and LOC51321MR, Reverse
5′-CGTCGCGCACGCCCTAT-3′; LOC51321UF, Forward
5′-GTTGTGGAGTTGTTGTGAGTGTAA-3′ and LOC51321UR, Reverse
5′-ACCACTACATCACACACACCCTA-3′. The PCR was performed for 35 cycles
with annealing temperatures of 64°C and 50°C for the unmethylated
and methylated reactions, respectively, using 100 ng
bisulfite-modified DNA. All PCR products were separated on 2%
agarose gels and stained with ethidium bromide. Negative and
positive control samples with unmethylated lymphocyte DNA and SssI
methyltransferase treated methylated DNA were also included in each
set of PCR amplifications. The lymphocyte DNA was obtained by
collecting blood samples from a healthy volunteer Mr. Wayren Huang,
one of the investigators. The lymphocyte DNA were isolated using a
Genomic DNA Extraction Miniprep System (Viogene BioTek Corp., New
Taipei City, Taiwan). Hypermethylation was defined as amplification
of more methylated product than unmethylated product from a tumor
sample.
5-Aza-2′-deoxycytidine (5-Aza-dC)
treatment of lung cancer cells
The human lung cancer cell lines CL1-1 and CL1-5-F4
were obtained from a Taiwanese patient with AD of the lung and was
kindly provided by Dr. P-C Yang, Department of Internal Medicine,
National Taiwan University Hospital. Cells were plated at
105 cells/100-mm culture dish on the day before
treatment. The cultures were treated for 3 doubling times with 1 uM
of 5-Aza-dC. The 5-Aza-dC-containing medium was changed after each
cell doubling during the treatment. On the day after the third
doubling, the cells were harvested for an analysis of their
methylation status using an MSP assay as described above, and for
LOC51321 mRNA expression using an RT-PCR assay.
Statistical analysis
Pearson's χ2 test was used to compare the
frequency of LOH in NSCLC patients. All data were each assigned a
number and a color code as follows: the existence of LOH in a
marker of NSCLC was assigned the number −1 and red color; the
retention of a chromosomal region was assigned the number +1 and
green color; and the noninformative data were assigned black color;
no data were assigned gray color. The color-coding system provides
interactive graphic analysis of clustering results in the TreeView
program (http://jtreeview.sourceforge.net) (9). Based on a previously described
clustering algorithm the similarity of LOH profiles of a group of
tumor samples (such as a subgroup of NSCLC features) was calculated
(9). P<0.05 was considered to
indicate a statistically significant difference. SPSS software,
version 19.0 (IBM, Chicago, IL, USA) was used for all statistical
analyses.
Results
Refined mapping of the LOH region on
17q24.3 and expressional validation of LOC51321 using NSCLC cell
lines and patient samples
Since the chromosome region at 17q24.3 has not
previously been identified as a high frequency loss region in NSCLC
samples, the present study further refined the LOH pattern of the
chromosomal region 17q24.2-24.3 using 9 additional microsatellite
markers. This was conducted using a panel of 48 microdissected
NSCLC tumor samples and their matched normal lung tissue samples.
The allelic loss pattern of these tumors indicates that the MDR was
located between markers D17S1882 and D17S789, reaching 50–65% LOH
at this locus (Table I). A total of
32/42 (79%) informative tumors exhibited LOH that was associated
with this region. Markers D17S807 and D17S789 were specifically
associated with patients who smoked (P=0.03 and 0.02, respectively,
Table II). In addition, marker
D17S807 was also demonstrated to be related to SCCs (P=0.003,
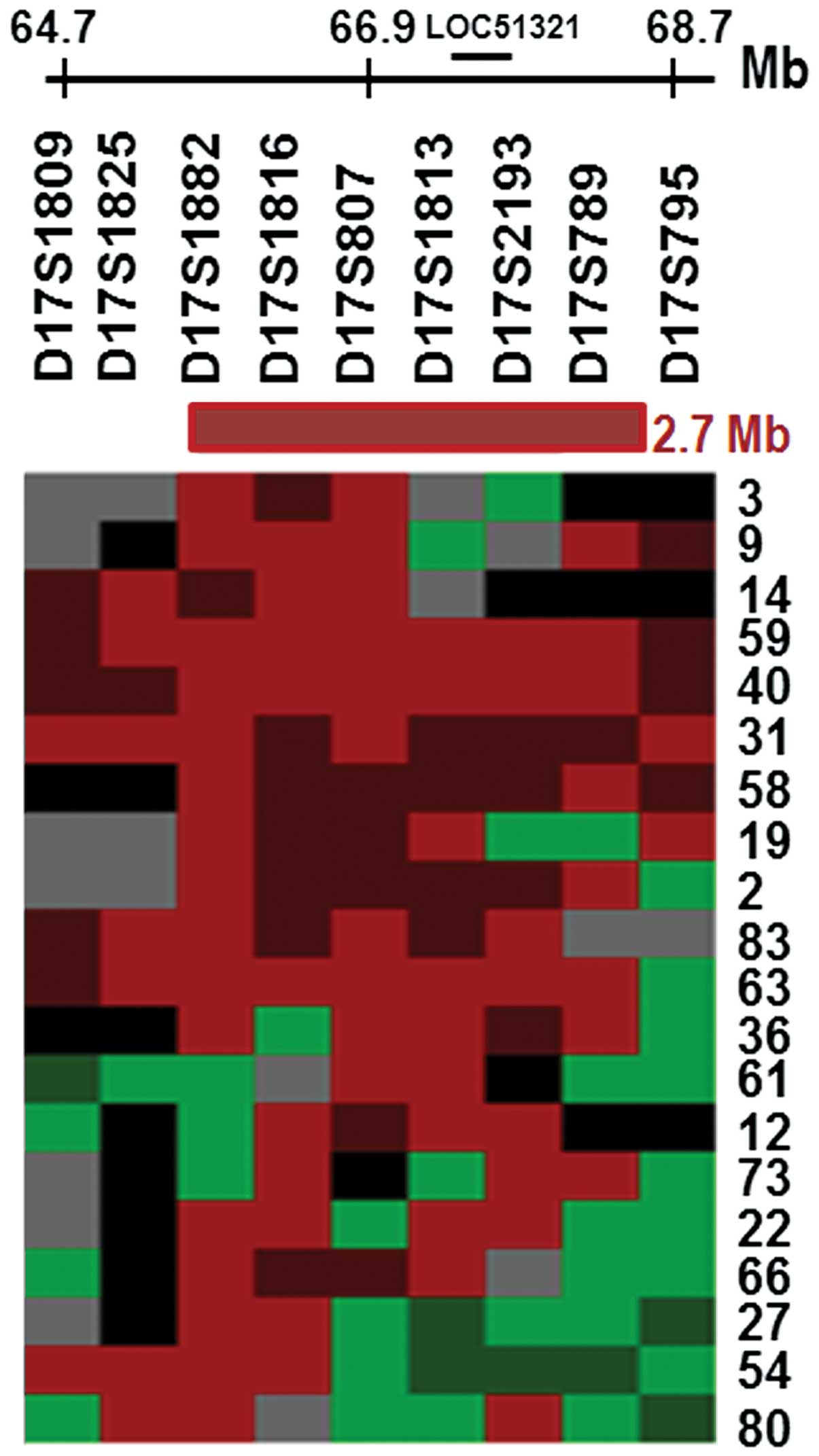
Table II). Fig. 1 summarizes the LOH results for the 20
representative tumors that exhibited allelic imbalance, which
spanned a distance of approximately 2.7 Mb. The smallest commonly
deleted region included markers that are located within
LOC51321, a putative AMZ2 gene that encodes a protein
with a predicted Zn-dependent protease domain. Therefore,
alterations within LOC51321 may occur in NSCLC cell lines
and/or tumor samples.
 | Table I.Refined mapping of 17q24.3 in 48 NSCLC
patients. |
Table I.
Refined mapping of 17q24.3 in 48 NSCLC
patients.
| Markers | STS Map (bp) | LOH% |
|---|
| D17S1809 | 64,701,223 | 40 |
| D17S1882 | 65,916,126 | 58 |
| D17S1816 | 66,424,320 | 65 |
| D17S807 | 66,862,762 | 56 |
| D17S1813 | 67,489,770 | 52 |
| D17S2193 | 68,551,515 | 61 |
| D17S789 | 68,632,310 | 50 |
| D17S795 | 68,681,985 | 19 |
| D17S1826 | 72,538,910 | 39 |
 | Table II.Association between LOH and patient
clinicopathological parametersa. |
Table II.
Association between LOH and patient
clinicopathological parametersa.
| Variable | Markers | Categories | No LOH | LOH | P-value |
|---|
| Smoking |
|
| Marker associated
with smoker patients |
|
|
| D17S1882 | No | 48% | 52% | 0.888 |
|
|
| Yes | 45% | 55% |
|
|
| D17S1816 | No | 29% | 71% | 0.658 |
|
|
| Yes | 38% | 62% |
|
|
| D17S807 | No | 75% | 25% | 0.032 |
|
|
| Yes | 29% | 71% |
|
|
| D17S1813 | No | 35% | 65% | 0.183 |
|
|
| Yes | 67% | 33% |
|
|
| D17S2193 | No | 40% | 60% | 0.907 |
|
|
| Yes | 38% | 62% |
|
|
| D17S789 | No | 75% | 25% | 0.022 |
|
|
| Yes | 31% | 69% |
|
| Tumor
type |
|
| Marker associated
with tumor type |
|
|
| D17S1882 | AD | 52% | 48% | 0.463 |
|
|
| SCC | 40% | 60% |
|
|
| D17S1816 | AD | 22% | 78% | 0.279 |
|
|
| SCC | 45% | 55% |
|
|
| D17S807 | AD | 80% | 20% | 0.003 |
|
|
| SCC | 20% | 80% |
|
|
| D17S1813 | AD | 44% | 56% | 0.305 |
|
|
| SCC | 43% | 57% |
|
|
| D17S2193 | AD | 29% | 71% | 0.940 |
|
|
| SCC | 44% | 56% |
|
|
| D17S789 | AD | 42% | 58% | 0.445 |
|
|
| SCC | 56% | 44% |
|
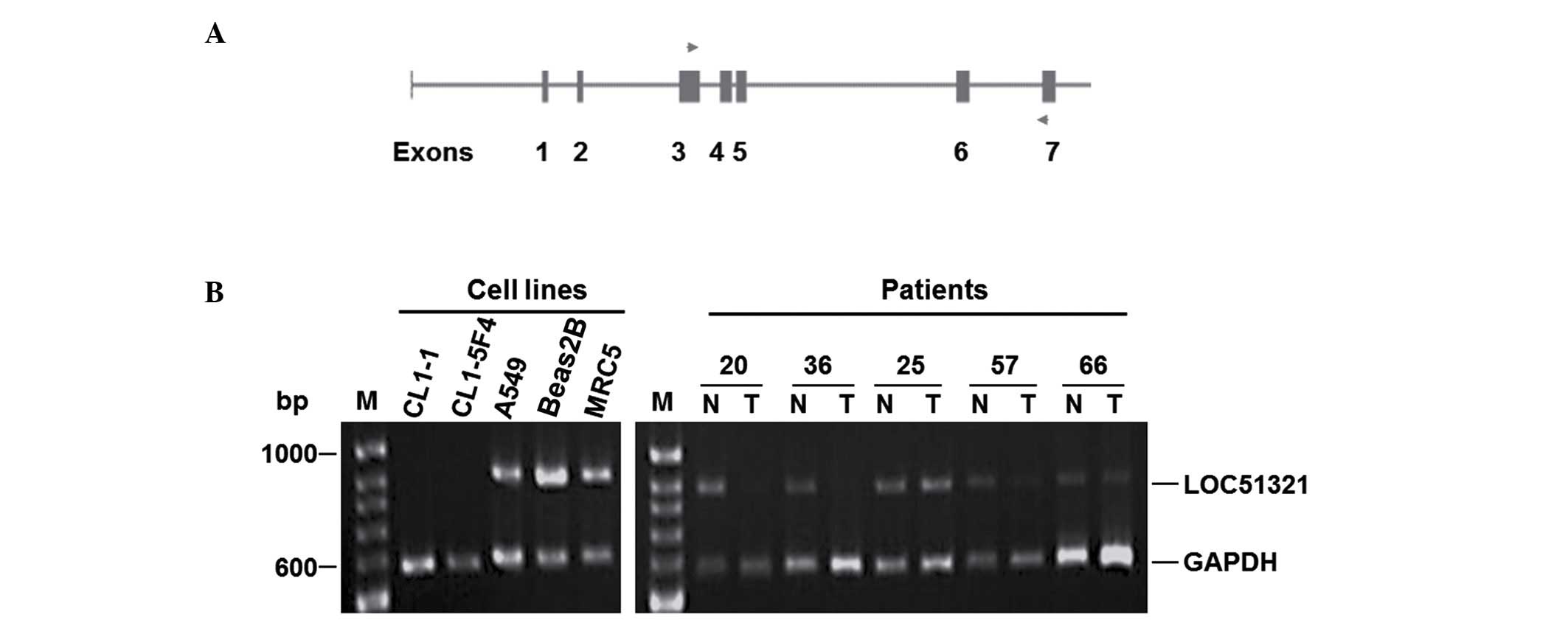
The alteration analysis of LOC1321 in
lung cancer
The expression level of LOC1321 was then
analyzed in two normal lung cancer lines, 13 NSCLC cell lines, and
53 matched normal and tumor tissues by semiquantitative multiplex
RT-PCR. This analysis demonstrated that there was reduced
expression of LOC51321 in 54% (7/13) of the NSCLC cell lines
and 36% (19/53) of the tumor tissues (Fig. 2).
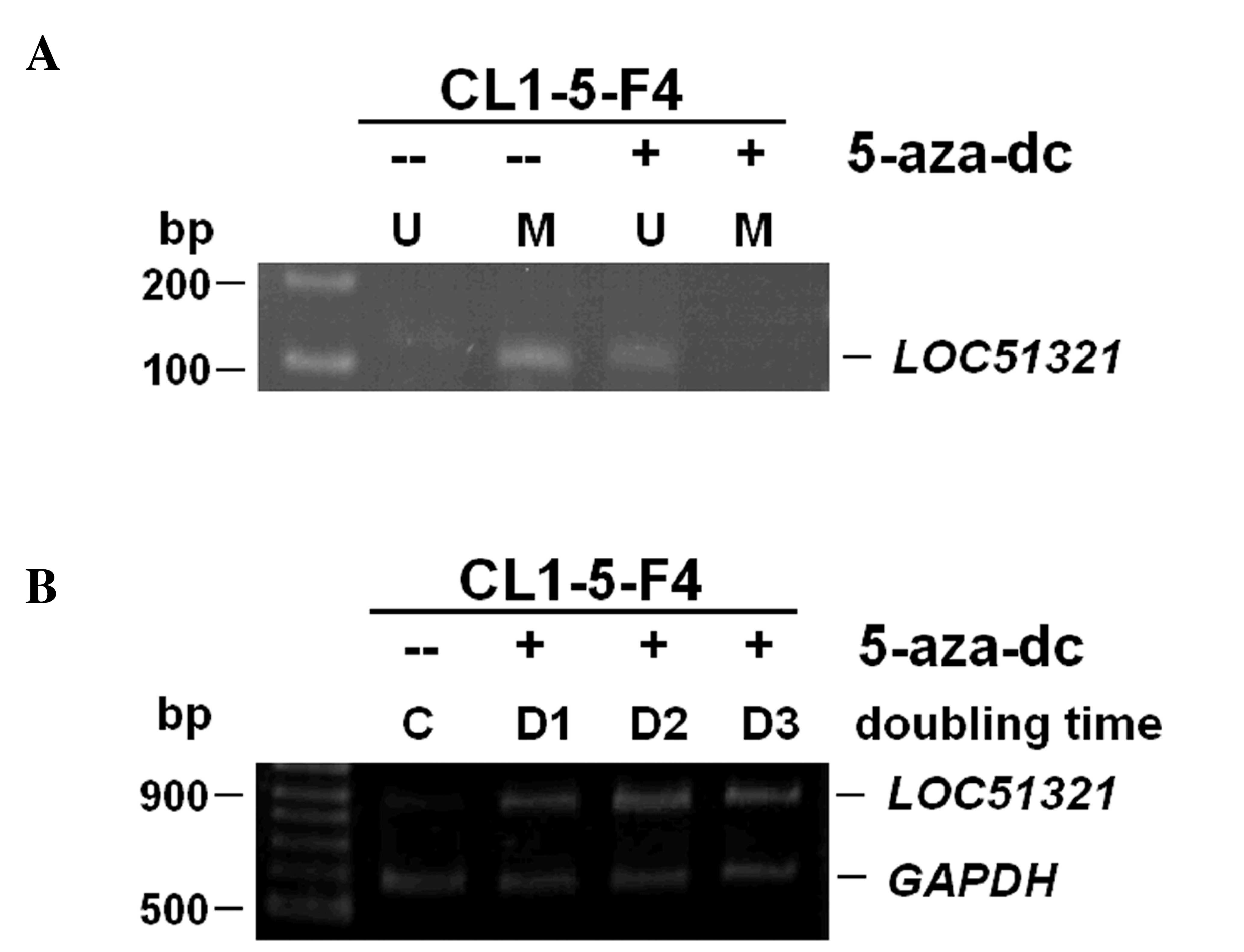
Re-activation of LOC51321 by 5-Aza-dC
treatment
Aberrations in promoter methylation are considered
to be important in the development of lung cancer (11). To determine whether epigenetic change
altered gene expression, promoter hypermethylation of the
LOC51321 was assessed using MSP analysis in lung cancer
cells. One lung cancer cell line CL1-5F4 that demonstrated negative
expression together with promoter hypermethylation of the
LOC51321 gene was treated with the demethylating agent
5-Aza-dC (Fig. 3A). As presented in
Fig. 3B, 5-Aza-dC successfully
restored mRNA and protein expression by de-methylating the putative
promoter region in the cells that lacked LOC51321 expression and
also harbored an appropriately methylated promoter.
Discussion
Both copies of TSGs need to be inactivated for their
function to be lost (2). One allele
may be inactivated by a point mutation, a methylation change, or a
small deletion. Generally, the other allele is often inactivated by
a large deletion that involves the gene of interest as well as
adjacent stretches of DNA (3). Thus,
searches for genomic regions frequently deleted in cancer have
aided in the identification and/or confirmation of the location of
several TSGs. To the best of our knowledge, the present study
reports, for the first time, a high frequency loss region on
chromosomes 17q24.3 in NSCLC samples. Based on refined mapping
using 9 markers together with an expressional study involving
semiquantative RT-PCR, the present study identified a novel gene
LOC51321 that may be one of the main deletion targets on
17q24.3 in NSCLC. LOC51321 was demonstrated to exhibit loss
of mRNA expression in 47 and 36% of examined NSCLC cell lines and
tumor samples, respectively. According to the annotated mRNA
database, LOC51321 may be able to produce a series of
transcripts by alternative splicing. The primers for RT-PCR in the
present study were deliberately designed to cover a large region
and to target sequences not likely to be involved in alternative
splicing. Therefore, if any alternative transcripts occurred, these
should have been detected. However, no aberrant sized cDNA product
was produced by any lung cancer sample, which indicates that
LOC51321 may be a simple gene with a single open reading
frame product. Nevertheless, probable alternatives in terms of
promoters and the possibility of alternative splicing outside the
region examined need to be investigated in the future.
Alterations in mRNA expression associated with
promoter hypermethylation of the LOC51321 gene in CL1-5-F4
cell line were explored and the findings indicate that promoter
hypermethylation may be one possible source of inactivation of the
LOC51321 gene. This conclusion is further strengthened by
the additional findings of the present study that the re-expression
of the mRNA expression together with de-methylation at the promoter
region in the LOC51321 genes using 5-Aza-dC treatment of a
lung cancer cell line.
Based on annotation information from www.genecards.org, LOC31521 (AMZ2) is predicted
to be a Zn-dependent protease. The Zn-dependent and
calcium-dependent family of proteins termed the matrix
metalloproteinases have been demonstrated to be collectively
responsible for the degradation of the extracellular matrix
(12). Members of this family,
including metalloendopeptidase, collagenases, stromelysins and
gelatinases; and are involved in routine tissue remodelling
processes, such as wound healing, embryonic growth and angiogenesis
(12). Loss of expression of some
metalloendopeptidases has been demonstrated to be involved in
tumorigenesis. Shipp et al (13) demonstrated that malignant pulmonary
neuroendocrine cells expressed low levels of the cell surface
metalloendopeptidase CD10/neutral endopeptidase 24.11 (CD10/NEP,
common acute lymphoblastic leukemia antigen) and that this enzyme
hydrolyzes bombesin-like peptides (13). The growth of bombesin-like
peptide-dependent cancer cells is inhibited by CD10/NEP. The
results provide evidence that CD10/NEP is involved in the
regulation of tumor cell proliferation. Since cigarette smoke
inactivates CD10/NEP (13), reduced
cell surface CD10/NEP enzymatic activity may be causally associated
with the development of lung cancer.
Further characterization, including identifying the
various mutations involved in gene inactivation, performing
promoter identification, and conducting a full analysis of
protein(s) associated with LOC51321 will be instrumental in
confirming the mRNA expression findings presented in the current
study. An understanding of the structure, nature, and function of
this chromosomal location will also be necessary to determine its
full role in carcinogenesis.
Acknowledgements
The authors would like to express their thanks to
Dr. Han Shui Hsu (Chief in Division of Thoracic Surgery, Taipei
Veterans General Hospital, Taipei, Taiwan) for providing the
clinical samples. The present study was supported in part by the
Ministry of Science and Technology (No. 101-2320-B-320-008).
References
|
1
|
Fearon ER: Human cancer syndromes: Clues
to the origin and nature of cancer. Science. 278:1043–1050. 1997.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Knudson AG: Hereditary cancer: Two hits
revisited. J Cancer Res Clin Oncol. 122:135–140. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Kohno T and Yokota J: How many tumor
suppressor genes are involved in human lung carcinogenesis?
Carcinogenesis. 20:1403–1410. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Vogelstein B, Fearon ER, Hamilton SR, Kern
SE, Preisinger AC, Leppert M, Nakamura Y, White R, Smits AM and Bos
JL: Genetic alterations during colorectal-tumor development. N Engl
J Med. 319:525–532. 1988. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Lasko D, Cavenee W and Nordenskjöld M:
Loss of constitutional heterozygosity in human cancer. Annu Rev
Genet. 25:281–314. 1991. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Yokota J and Sugimura T: Multiple steps in
carcinogenesis involving alterations of multiple tumor suppressor
genes. FASEB J. 7:920–925. 1993.PubMed/NCBI
|
|
7
|
Gray JW and Collins C: Genome changes and
gene expression in human solid tumors. Carcinogenesis. 21:443–452.
2000. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Thiagalingam S, Foy RL, Cheng KH, Lee HJ,
Thiagalingam A and Ponte JF: Loss of heterozygosity as a predictor
to map tumor suppressor genes in cancer: Molecular basis of its
occurrence. Curr Opin Oncol. 14:65–72. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Tseng RC, Chang JW, Hsien FJ, Chang YH,
Hsiao CF, Chen JT, Chen CY, Jou YS and Wang YC: Genomewide loss of
heterozygosity and its clinical associations in non small cell lung
cancer. Int J Cancer. 117:241–247. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Brambilla E, Travis WD, Colby TV, Corrin B
and Shimosato Y: The new World Health Organization classification
of lung tumours. Eur Respir J. 18:1059–1068. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Zhao Y, Zhou H, Ma K, Sun J, Feng X, Geng
J, Gu J, Wang W, Zhang H, He Y, et al: Abnormal methylation of
seven genes and their associations with clinical characteristics in
early stage non-small cell lung cancer. Oncol Lett. 5:1211–1218.
2013.PubMed/NCBI
|
|
12
|
Borkakoti N: Structural studies of matrix
metalloproteinases. J Mol Med (Berl). 78:261–268. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Shipp MA, Tarr GE, Chen CY, Switzer SN,
Hersh LB, Stein H, Sunday ME and Reinherz EL: CD10/neutral
endopeptidase 24.11 hydrolyzes bombesin-like peptides and regulates
the growth of small cell carcinomas of the lung. Proc Natl Acad Sci
USA. 88:10662–10666. 1991. View Article : Google Scholar : PubMed/NCBI
|