Introduction
In recent years, the mortality rate for colorectal
cancer is over 600,000 patients annually (1). Early discovery, diagnosis and treatment
are imperative for patients with colorectal cancer. The occurrence
of colorectal cancer is the result of combined actions of multiple
genes, factors and steps. Epigenetic change is a type of abnormal
gene expression without the occurrence of gene sequence changes,
including histone markers and gene methylation. Gene methylation is
an early tumor event, and the process is reversible, which provides
theoretical evidence for methylation-relevant gene application and
clinical early diagnosis and treatment (2,3).
PAQR3 is a cancer suppressor gene and is
involved in various signal channels that can influence biological
processes such as energy metabolism, cell proliferation,
differentiation and the maturity of reproductive cells, and can
negatively regulate the Ras/Raf/MEK/ERK signal channel (4). In experiments in nude mice with
colorectal cancer, it was verified that PAQR3 plays a role in
cancer inhibition in colorectal cancer tissues, and the deficiency
of PAQR3 expression reduced the survival time of gene-knockout nude
mice (4). In addition, a low
expression of PAQR3 in tumor tissues of gastric cancer, breast
cancer, melanoma and liver cancer has been demonstrated. PAQR3 was
able to enhance the sensitivity of breast cancer SK-BR-3 cells to
epirubicin (5–13). However, studies should be performed on
cancer suppressor gene PAQR3 mRNA and protein levels of
colorectal cancer to assist in the diagnosis and treatment of
colorectal cancer.
PAQR3 is a member of the progesterone and
adiponectin receptor (PAQR) family, which plays a role in cancer
inhibition in colorectal cancer cells, and is involved in
biological processes such as signal transduction, energy
metabolism, cell proliferation, differentiation and the maturity of
reproductive cells. The purpose of adjusting physiological
processes such as proliferation, differentiation, apoptosis,
malignant transformation of cells and the occurrence and
development of cancer is reached through the negative regulation of
the Ras/Raf/MEK/ERK signal channel (5–9). In
addition, the AKT signal channel is negatively regulated by PAQR3
through two mechanisms. After knockout of PAQR3, the expression of
p110α in the cytoplast in combination with the dictyosome is
decreased, PAQR3 combined and changed the subcellular localization
of p110α of PI3K of compounds in the cytoplast, and prevented the
combination with the p85 control site, resulting in the
inactivation of PI3K and the phosphorylation of AKT, a process that
influences the signal channel of insulin. By contrast, PAQR3 is
able to block the Gβ locus of GPCR, and inhibits the function of
AKT in GRCR-activated Gβ/γ locus. In gastric cancer tissues, PAQR3
negatively regulates the phosphorylation of ERK and AKT, which
demonstrates that PAQR3 plays a role in the process of negatively
regulating EMT through inhibition of the ERK and AKT signal channel
(10–13). As the cancer suppressor gene in
colorectal cancer, the silencing mechanism of PAQR3 remains
to be determined.
The aim of the present study was to investigate
PAQR3 gene expression and its methylation level in
colorectal cancer tissues, as well as the association with
colorectal cancer clinical data. The results of the present study
provide theoretical evidence for the diagnosis and treatment of
colorectal cancer.
Patients and methods
Tissue extraction
In total, 54 pairs of colorectal cancer and normal
tissues, respectively, were collected from the Department of
General Surgery of the Affiliated Hospital of Hebei University,
between June, 2013 and July, 2014. Inclusion criteria for the study
were that, the patients did not receive radiotherapy, chemotherapy
or immunotherapy prior to surgery. Any tumor tissues that had
already necrosis on collection of cancer tissue samples, were
excluded. Normal tissues that were >10 cm from the edge of
cancer tissue were collected as the cancer adjacent normal tissues.
Full-thickness of the tissue was selected, and the diagnostic
results were confirmed by the postoperative pathological results.
There were 32 male and 22 female patients, 19 patients with rectal
cancer and 35 patients with colon cancer, with an age range of
36–82 years and an average age of 59.69 years. The samples were
collected during surgery, placed in liquid nitrogen, and preserved
at −80°C for ≤6 months.
Reagents
The Omega E.Z.N.A.™ overall RNA extraction kit,
Vazyme HiScript First Strand cDNA synthesis kit, RT-qPCR
fluorescence quantitation kit, Qiagen EpiTect Fast DNA Bisulfite
kit, proteinase K, RNaseA, RNA enzyme EP tubes, PAQR3 and β-actin
antibodies were all purchased from Shijiazhuang Huiyou Biological
Technology Co., Ltd. (Shijiazhuang, China). β-actin, PAQR3, PAQR3
methylation and non-methylation primers were synthesized according
to the design of Primer Premier 5.0 software, their specificities
were detected through PubMed Blast, and all the primers were
produced by Sangon Biotech Co. Ltd. (Shanghai, China). STE lysate,
SDS, agarose, chloroform, isoamyl, absolute ethanol, 75% absolute
ethanol, Tris-phenol, phenol/chloroform/isoamylol (25:24:1),
chloroform/isoamylol (24:1). TE buffer solution and anhydrous
sodium acetate were used in the process of DNA extraction and
nucleic acid electrophoresis. Protein lysis buffer, PMSF, 1X PBS,
and 5X loading buffer were used for protein isolation. Gel
electrophoresis, separation gel and running buffer, methanol, and
1X transfer membrane buffer were used for western blot analysis.
Coomassie blue staining solution, and Coomassie brilliant blue
bleaching liquid were used for quantification of the isolated
proteins. PVDF membrane was used for immunoblotting. The membrane
was incubated in primary antibody, and the secondary antibody was
added in 5% skim milk in TBST. Materials for western blot analysis
were provided by the Central Laboratory of the Affiliated Hospital
of Hebei University.
Quantitative RT-PCR
Tissue RNA extraction
Tissue (30 mg) was collected and RNA was extracted
according to the instructions of the Omega E.Z.N.A.™ RNA extraction
kit. Product (2 µl) was collected for separation on 1% sepharose
gel for electrophoresis. Reverse transcription was carried out
after the ELISA test, and the content (2,000 ng/µl) and ratio (OD:
1.9–2.0) were determined.
Reverse transcription
RNA content was adjusted according to RNA
quantitative results. RNA (1 µg) was collected, and reverse
transcription was carried out according to the instructions of the
Vazyme HiScript First Strand cDNA synthesis kit. The reaction
conditions were denaturation at 25°C for 5 min, annealing at 42°C
for 30 min, and elongation at 85°C for 5 min.
PCR
PCR primers were designed using Primer Premier 5.0.
The reference gene was β-actin primer:
5′-GTGGACATCCGCAAAGAC-3′ (F), 5′-AAAGGGTGTAACGCAACTAA-3′ (R), and
the length of the product was 302 bp. The PDCD4 primer was
5′-TGGGAGTGACGCCCTTAGAA-3′ (F), 5′-TCCACCTCCTCCACATCATACA-3′ (R),
and the length of the product was 171 bp. The PAQR3 primer was
5′-TCTGTATGCTTTGCTCTGTGGG-3′ (F), 5′-TTTGCCATTGCTGCGTGAG-3′ (R),
and the length of the product was 252 bp. The reaction conditions
were 95°C for 5 min, 95°C for 10 sec, 60°C for 32 sec, for 40
cycles; and 95°C for 15 sec, 60°C for 60 sec, 95°C for 15 sec. The
results were normalized to β-actin.
Western blot analysis
Protein expression levels in tissues were analyzed
using western blot analysis as previously described (14). The membranes were incubated with
primary goat antibody in a dilution of 1:500 (anti-PAQR3; cat. no.
sc-161992; Santa Cruz Biotechnology, Santa Cruz, CA, USA) overnight
at 4°C. The membrane was washed with TBST and incubated with a
peroxidase-conjugated secondary polyclonal rabbit-anti-goat
antibody (dilution: 1:1,000; cat. no. sc-2768; Santa Cruz
Biotechnology) for 1 h. Western blot film was scanned and the
membrane was stripped and reprobed with antibody against β-actin
(dilution: 1:1,500) to confirm equal sample loading.
Methylation-specific PCR (MSP)
The phenol/chloroform extraction method was used to
extract tissue DNA. Hydrosulphite modification was then performed
according to the instructions on the EpiTect Fast DNA Bisulfite
kit. PAQR3 methylation and non-methylation primers were designed
and produced using MethPrimer software (Sangon Biotech Co.,
Ltd.).
The PAQR3 methylation primer was: (M)
5′-TTGTTGAAGAGCGCGTATTATATC-3′ (F), 5′-TAAAAACCCGAAAATCTACTCGTA-3′
(R), and non-methylation primer was: (U)
5′-TTGTTGAAGAGTGTGTATTATATTGA-3′ (F),
5′-TAAAAAACCCAAAAATCTACTCATA-3′ (R). The PAQR4 methylation primer
was: (M) 5′-TTTAGTTTCGGTTTCGTCGTTAC-3′ (F),
5′-GAAAAATCTCTAACCCTTCTCGC-3′ (R), and non-methylation primer was:
(U) 5′-TTTAGTTTTGGTTTTGTTGTTATGA-3′ (F),
5′-CAAAAAATCTCTAACCCTTCTCACT-3′ (R). The reference gene β-actin
primer was: 5′-GTGGACATCCGCAAAGAC-3′ (F),
5′-AAAGGGTGTAACGCAACTAA-3′ (R).
Statistical analysis
Data were analysed using statistical software SPSS
16.0 (Chicago, IL, USA). The χ2 test and Spearman
relevant analysis, and two-tailed test were used for comparisons
between groups. P<0.05 was considered statistically
significant.
Results
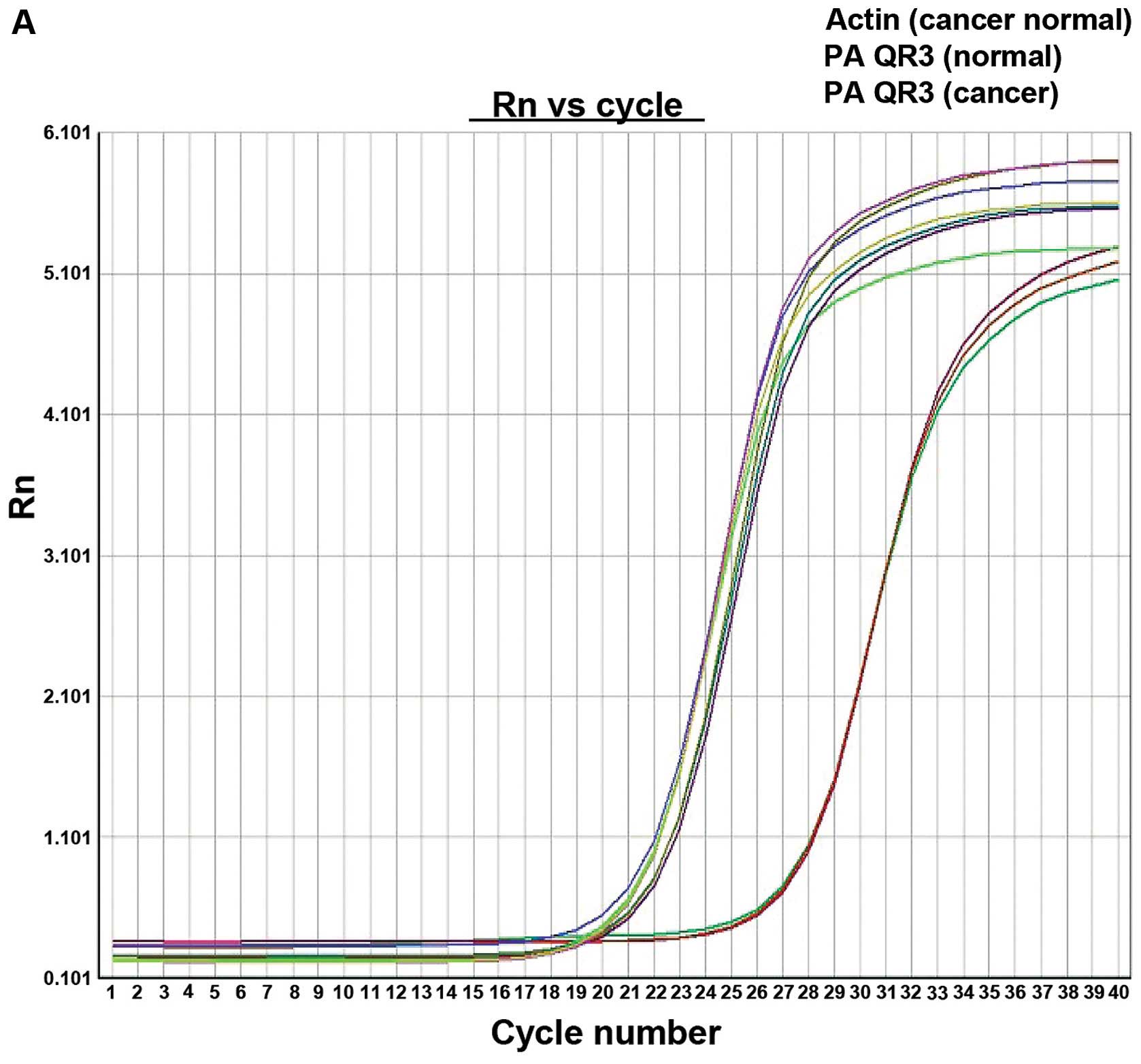
PAQR3 mRNA level in colorectal cancer
tissues
The PAQR3 mRNA expression level in colorectal cancer
tissues was markedly reduced and the expression reduction rate was
57.4% (31/54), with PAQR3 mRNA in cancer tissues being 9.321.97,
while that of normal adjacent tissues was 8.442.42. The difference
was of statistical significance (P=0.001) (Fig. 1).
PAQR3 protein level in colorectal
cancer tissues
The expression level of PAQR3 protein was
significantly low, the protein expression reduction rate in cancer
tissue was 46.3% (25/54), while the protein expression reduction
rate in cancer adjacent tissue was 5.6% (3/54). The difference was
of statistical significance (P<0.05) (Fig. 2).
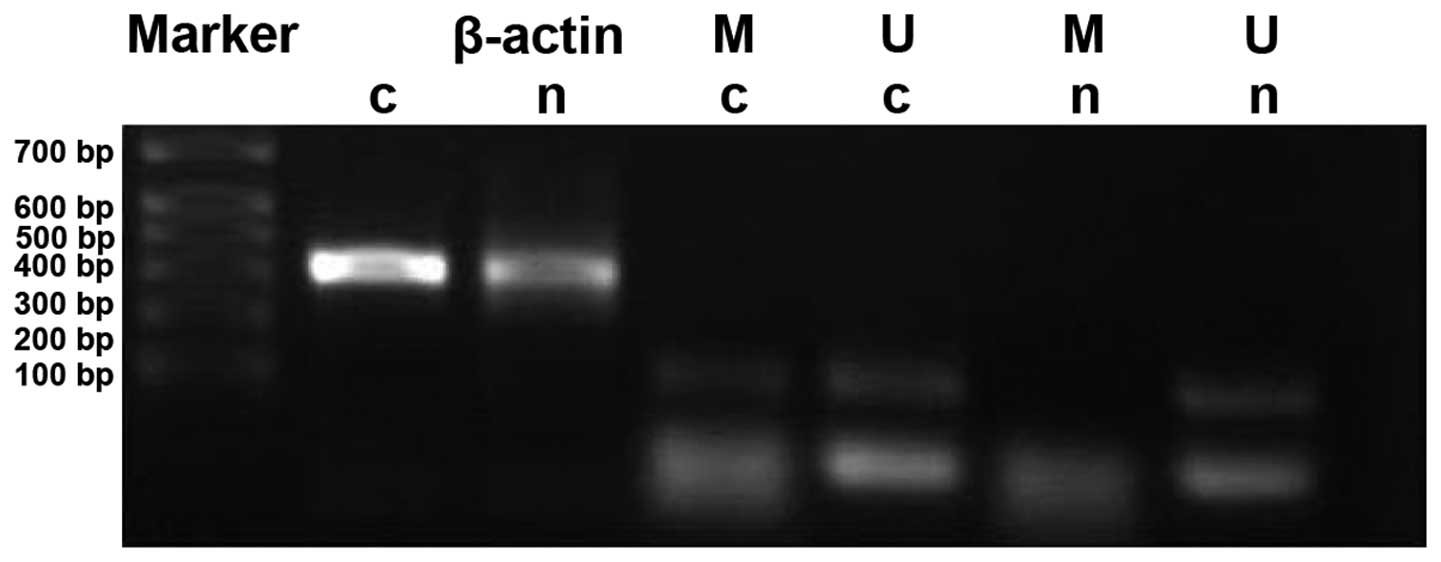
PAQR3 gene methylation level in
colorectal cancer tissues
Methylation rates of PAQR3 in cancer tissues and
cancer adjacent tissues were 33.3% (18/54) and 5.6% (3/54),
respectively. The methylation rate of PAQR3 in cancer tissues was
significantly higher than that in the corresponding normal cancer
adjacent tissues (P<0.05) (Fig.
3).
Association of the PAQR3 mRNA and
protein levels and colorectal cancer clinical parameters
Statistical software was detected using the Fisher's
exact test probability method. The PAQR3 mRNA and protein levels
were not correlated with gender, age or tumor location, but were
statistically significant for differentiation degree, lymphatic
metastasis and tumor infiltration depth (Table I).
 | Table I.Association of the PAQR3 mRNA and
protein levels and colorectal cancer clinical parameters. |
Table I.
Association of the PAQR3 mRNA and
protein levels and colorectal cancer clinical parameters.
|
| Colorectal cancer
tissues |
|---|
|
|
|
|---|
| Clinical
parameters | N=54 | Cases of PAQR3
reduced protein (31) | PAQR3 protein
reduction rate (%) | P-value | PAQR3 mRNA cases,
31 | PAQR3 mRNA rate
(%) | P-value |
|---|
| Gender |
|
| Male | 32 | 18 | 56.2 | 0.836 | 15 | 46.9 | 0.059 |
|
|
| 9 |
|
| 9 |
|
|
|
Female | 22 | 13 | 59.1 |
| 16 |
|
|
|
|
| 4 |
|
| 4 | 72.7 |
|
| Age (years) |
|
|
<60 | 29 | 17 | 68.0 | 0.144 | 19 | 65.5 | 0.194 |
|
|
| 8 |
|
|
|
|
|
| ≥60 | 25 | 14 | 48.3 |
| 12 | 48.0 |
|
|
|
| 5 |
|
| 5 |
|
|
| Location |
|
|
Colon | 35 | 19 | 54.3 | 0.529 | 23 | 65.7 | 0.094 |
|
|
| 6 |
|
| 6 |
|
|
|
Rectum | 19 | 12 | 63.2 |
| 8 | 42.1 |
|
|
|
| 7 |
|
| 7 |
|
|
| Differentiated
degree |
|
| Level
I–II | 30 | 12 | 40.0 | 0.004a | 11 | 36.7 | 0.001a |
|
|
| 9 |
|
| 9 |
|
|
| Level
III | 24 | 19 | 79.2 |
|
|
|
|
|
|
| 4 |
|
| 20 | 83.3 |
|
| Lymphatic
metastasis |
|
| No | 28 | 11 | 39.3 | 0.005a | 12 | 42.9 | 0.025a |
|
|
| 9 |
|
| 9 |
|
|
|
Yes | 26 | 20 | 76.9 |
| 19 | 73.1 |
|
|
|
| 4 |
|
|
|
|
|
| Depth of
infiltration |
|
|
Muscularis mucosae | 31 | 14 | 45.2 | 0.035a | 13 | 41.9 | 0.008a |
| Around
serosa | 23 | 17 | 73.9 |
| 18 | 78.3 |
|
Association between PDCD4 gene
methylation and colorectal cancer clinical data
The methylation level of PAQR3 gene in
colorectal cancer tissues was not correlated with gender or tumor
location, but was associated with age, differentiation degree,
lymphatic metastasis and tumor infiltration depth. The higher the
age of the patient, the lower the differentiation degree observed,
and the deeper the lymphatic metastasis and tumor infiltration the
higher the methylation rate found, and vice versa (Table II).
 | Table II.Association between PDCD4 gene
methylation level and colorectal cancer clinical data. |
Table II.
Association between PDCD4 gene
methylation level and colorectal cancer clinical data.
|
| Colorectal cancer
tissues |
|---|
|
|
|
|---|
| Clinical data | n | Cases of PAQR3
methylation n | PAQR3 methylation
rate (%) | P-value |
|---|
| Gender |
|
|
Male | 32 | 11 | 34.4 | 0.845 |
|
Female | 22 | 7 | 31.8 |
|
| Age (years) |
|
|
<60 | 29 | 6 | 20.7 | 0.034a |
|
≥60 | 25 | 12 | 48.0 |
|
| Location |
|
|
Colon | 35 | 10 | 28.6 | 0.314 |
|
Rectum | 19 | 8 | 42.1 |
|
| Differentiated
degree |
|
| Level
I–II | 30 | 15 | 50.0 | 0.004b |
| Level
III | 24 | 3 | 12.5 |
|
| Lymphatic
metastasis |
|
| No | 28 | 4 | 14.3 | 0.036a |
|
Yes | 26 | 14 | 53.8 |
|
| Depth of
infiltration |
|
|
Muscularis mucosae | 31 | 14 | 45.2 | 0.032a |
| Around
serosa | 23 | 4 | 17.4 |
|
Discussion
The function of the PAQR3 gene and it
inhibitory role in colorectal cancer was identified by Chen and Xie
in 2009 (5). The gene is involved in
various signal channels of cells, which can influence biological
processes such as energy metabolism, cell proliferation,
differentiation and the mature reproductive cells. It is also known
as PKTG, which can encode 7-fold transmembrane protein of 37 kDa,
N-end towards cytoplasm, C-end towards organelle, and is situated
at the Golgi membrane, which can negatively regulate the
Ras/Raf/MEK/ERK signal channel. This signal channel is one of the
typical routes of the crystal-induced mitogen-activated protein
kinase cascade signal. The PAQR3 protein competitively binds to
RAF, and disturbs the combination between RAF and the downstream
signal molecule substrate, which is used to identify extracellular
signals and transfer extracellular stimulating signals into cells.
This can achieve the purpose of physiological processes such as
regulating cell proliferation, differentiation, apoptosis and
malignant transformation of cells, as well as the occurrence and
development of cancer (5–9). Wang et al also verified that
PAQR3 plays a role in cancer inhibition in colorectal cancer
tissues, and the deficiency of PAQR3 expression reduces the
survival time of gene-knockout nude mice (15). In the study on human melanoma A375,
the overexpression of PAQR3 clearly inhibited the amplification and
malignant transformation of cells, and in the nude mouse
transplantation tumor experiment, the re-expression of PAQR3
through siRNA clearly inhibited the effects of subcutaneous tumors,
and the stimulation of abnormally activated ERK downstream was
inhibited (16). In addition, the
expression of PAQR3 in tumor tissues, such as breast cancer, was
reduced, and PAQR3 enhanced the sensitivity of breast cancer
SK-BR-3 cells to epirubicin (17).
The silencing mechanism of PAQR3 is not clear, thus,
the gene promoter mRNA and protein levels in colorectal cancer
tissues were detected to determine their association. In the
present study, through the detection of tissue samples of 54 pairs
of patients with colorectal cancer, it was verified that the
expression of PAQR3 in colorectal cancer is reduced, and this shows
that PAQR3 mRNA and protein levels in colorectal cancer tissues was
not associated with gender, age or tumor location, but were
correlated with differentiation degree, lymphatic metastasis and
tumor infiltration depth. Previous studies only mentioned that the
expression level of PAQR3 protein was reduced; however, the
association between the reduction rate of protein expression and
clinical data in colorectal cancer tissues was not clear (12,15). Due
to the limitation of the number of samples, this result was not
completely consistent.
Wang et al detected that the reduction rate
of PAQR3 mRNA expression in colorectal cancer was 82.3% (51/62),
which is different from our results of 57.4% (31/54) (18). However, compared with normal cancer
adjacent tissues, statistical significance was observed. Authors of
that study mentioned that the reduction rate of PAQR3 mRNA
expression was higher in male compared to female patients, which is
consistent with the epidemiological analysis results that male
patients have higher morbidity in colorectal cancer compared to
female patients (19). In the present
study, there were more male samples collected than female samples.
However, the results showed that, the reduction of PAQR3 expression
was not associated with gender, which was a result based on sample
and regional difference. In addition, Wang et al found that
the PAQR3 mRNA level in colorectal cancer tissues was not
associated with tumor location or age, which is consistent with the
results of the present study. However, its relationship with tumor
differentiation degree was not analyzed. In the present study, the
PAQR3 mRNA level was relevant to lymphatic metastasis, tumor
infiltration depth and tumor differentiation degree, and this was
considered to be relevant to the number of samples collected. These
indexes can be used as prognosis monitoring indicators of patients
and remain to be confirmed by clinical follow-up research.
Through the detection of the methylation level of
PAQR3 gene in colorectal cancer tissues, the present study
evaluated methylation in colorectal cancer, and its association
with clinical data. Gene methylation is an early event of
tumorigenesis that can be used as diagnostic evidence of colorectal
cancer. The methylation rate of PAQR3 was 33.3% (18/54), which was
of statistical significance, compared to the methylation level of
the normal cancer adjacent tissues. To the best of our knowledge,
there are currently no studies on PAQR3 gene methylation,
and the results of our study verified that PAQR3 gene
methylation is involved in the formation of colorectal cancer, and
can be used as the tumor marker thereof. At the same time we found
that PAQR3 methylation level was relevant to age; thus, the older
the patient was, the higher the PAQR3 gene methylation rate.
The analysis of the relevance between gene methylation and age
shows that, with the increase of age, methylation of the organism
was higher. The age-relevant methylation level changing mechanism
remains to be clarified, and this relationship has received
controversion results from different research groups. First of all,
with increasing age, enzymatic activity in the human body is
altered, promoting gene methylation. This may be influenced by
geographical environmental elements, random selection and sample
size, although this difference was not statistically significant
(20). The high methylation level of
PAQR3 gene is associated with differentiated degree,
lymphatic metastasis and tumor infiltration depth. Concerning gene
methylation is the early event of tumorigenesis (21), the lower the differentiated degree the
higher the gene methylation level was, and at a low gene expression
level the ability of cancer inhibition was minimal. With the
increase of differentiation degree, the possibility of methylation
occurrence was less. Therefore, the lower the differentiation
degree was in tumor tissues, the higher the possibility of gene
methylation. PAQR3 methylation was relevant to tumor infiltration
depth of colorectal cancer and lymphatic metastasis, and
considering the consequence of PAQR3 gene silencing, PAQR3
can be used as the monitoring prognostic indicator of colorectal
cancer. The present results verified the role that PAQR3
gene methylation plays in colorectal cancer, which provides
effective theoretical evidences for further in vitro
experiments, reverse gene methylation and re-expression of cancer
suppressor genes.
References
|
1
|
Shaukat A, Mongin SJ, Geisser MS, Lederle
FA, Bond JH, Mandel JS and Church TR: Long-term mortality after
screening for colorectal cancer. N Engl J Med. 369:1106–1114. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Carmona FJ, Azuara D, Berenguer-Llergo A,
Fernández AF, Biondo S, de Oca J, Rodriguez-Moranta F, Salazar R,
Villanueva A, Fraga MF, et al: DNA methylation biomarkers for
noninvasive diagnosis of colorectal cancer. Cancer Prev Res
(Phila). 6:656–665. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Tapp HS, Commane DM, Bradburn DM,
Arasaradnam R, Mathers JC, Johnson IT and Belshaw NJ: Nutritional
factors and gender influence age-related DNA methylation in the
human rectal mucosa. Aging Cell. 12:148–155. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Wu HG, Zhang WJ, Ding Q, Peng G, Zou ZW,
Liu T, Cao RB, Fei SJ, Li PC, Yang KY, Hu JL, Dai XF, Wu G and Li
PD: Identification of PAQR3 as a new candidate tumor suppressor in
hepatocellular carcinoma. Oncol Rep. 32:2687–2695. 2014.PubMed/NCBI
|
|
5
|
Chen Y and Xie XD: The functional research
of RKTG gene. J Cell Biol. 31:9–14. 2009.
|
|
6
|
Tang YT, Hu T, Arterburn M, Boyle B,
Bright JM, Emtage PC and Funk WD: PAQR proteins: a novel membrane
receptor family defined by an ancient 7-transmembrane pass motif. J
Mol Evol. 61:372–380. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Feng L, Xie X, Ding Q, Luo X, He J, Fan F,
Liu W, Wang Z and Chen Y: Spatial regulation of Raf kinase
signaling by RKTG. Proc Natl Acad Sci USA. 104:14348–14353. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Cano E and Mahadevan LC: Parallel signal
processing among mammalian MAPKs. Trends Biochem Sci. 20:117–122.
1995. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Fan F, Feng L, He J, Wang X, Jiang X,
Zhang Y, Wang Z and Chen Y: RKTG sequesters B-Raf to the Golgi
apparatus and inhibits the proliferation and tumorigenicity of
human malignant melanoma cells. Carcinogenesis. 29:1157–1163. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Xie X, Zhang Y, Jiang Y, Liu W, Ma H, Wang
Z and Chen Y: Suppressive function of RKTG on chemical
carcinogen-induced skin carcinogenesis in mouse. Carcinogenesis.
29:1632–1638. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Zhang Y, Jiang X, Qin X, Ye D, Yi Z, Liu
M, Bai O, Liu W, Xie X, Wang Z, et al: RKTG inhibits angiogenesis
by suppressing MAPK-mediated autocrine VEGF signaling and is
downregulated in clear-cell renal cell carcinoma. Oncogene.
29:5404–5415. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Ling ZQ, Guo W, Lu XX, Zhu X, Hong LL,
Wang Z, Wang Z and Chen Y: A Golgi-specific protein PAQR3 is
closely associated with the progression, metastasis and prognosis
of human gastric cancers. Ann Oncol. 25:1363–1372. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Jiang Y, Xie X, Zhang Y, Luo X, Wang X,
Fan F, Zheng D, Wang Z and Chen Y: Regulation of G-protein
signaling by RKTG via sequestration of the G betagamma subunit to
the Golgi apparatus. Mol Cell Biol. 30:78–90. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Padda RS, Gkouvatsos K, Guido M, Mui J,
Vali H and Pantopoulos K: A high-fat diet modulates iron metabolism
but does not promote liver fibrosis in hemochromatotic
Hjv−/− mice. Am J Physiol Gastrointest Liver Physiol.
308:G251–G261. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Wang X, Li X, Fan F, Jiao S, Wang L, Zhu
L, Pan Y, Wu G, Ling ZQ, Fang J, et al: PAQR3 plays a suppressive
role in the tumorigenesis of colorectal cancers. Carcinogenesis.
33:2228–2235. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Ding Q, Huo L, Yang JY, Xia W, Wei Y, Liao
Y, Chang CJ, Yang Y, Lai CC, Lee DF, et al: Down-regulation of
myeloid cell leukemia-1 through inhibiting Erk/Pin 1 pathway by
sorafenib facilitates chemosensitization in breast cancer. Cancer
Res. 68:6109–6117. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Huang JB, Luo XR, Kong LQ, Xiang TX and
Ren GS: The influence of sensitivity of PAQR3 to Epirubicin of
breast cancer cells SK-br-3. J Third Military Medical University.
35:1658–1662. 2013.(In Chinese).
|
|
18
|
Wang Q, Sun Z and Yang HS: Downregulation
of tumor suppressor Pdcd4 promotes invasion and activates both
β-catenin/Tcf and AP-1-dependent transcription in colon carcinoma
cells. Oncogene. 27:1527–1535. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Palamarchuk A, Efanov A, Maximov V,
Aqeilan RI, Croce CM and Pekarsky Y: Akt phosphorylates and
regulates Pdcd4 tumor suppressor protein. Cancer Res.
65:11282–11286. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Lillycrop KA, Hoile SP, Grenfell L and
Burdge GC: DNA methylation, ageing and the influence of early life
nutrition. Proc Nutr Soc. 73:413–421. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Bird A: Perceptions of epigenetics.
Nature. 447:396–398. 2007. View Article : Google Scholar : PubMed/NCBI
|