Introduction
Epigenetic modifications affect gene expression
without altering the DNA structure. DNA methylation and histone
acetylation, or methylation, are two of the most important
mechanisms. Epigenetic gene expression modulation is an aspect of
physiological development, and its dysregulation is involved in
carcinogenesis (1). In
myelodysplastic syndromes and acute myeloid leukemia, epigenetic
modifiers are used in routine therapy (2,3), and such
approaches are becoming popular treatment options for solid tumours
(4). Epidermal growth factor receptor
is downregulated by epigenetic modifications, and the re-expressed
receptor after treatment with, for example, hypomethylating agents
is a potential therapeutic target leading to cell death (5,6). Several
studies have demonstrated the growth-inhibiting effect of
Trichostatin A (TSA) on carcinomas (7,8). Vigushin
et al (7) described histone
hyperacetylation of histone H4, thereby inhibiting proliferation in
breast cancer cell lines following TSA treatment. Ailenberg and
Silverman (8) described apoptosis
induction in tumour cells following TSA treatment, which restored
the expression of cell-cycle-controlling genes.
The compound 5-aza-2′-deoxycytidine (aza) is a
cytosin analogue, which can be integrated into newly synthesised
DNA strands. Aza irreversibly binds and inhibits DNA
(cytosine-5)-methyltransferase 1 (DNMT1), which transfers
methylation patterns to newly synthesised DNA. Loss of DNMT1
activity therefore leads to a loss of methylation during the next
cell cycles and the re-expression of specific genes (9). Methylation inhibition of CpG islands of
the estrogen receptor leads to its downregulation, and treatment
with aza restores estrogen receptor expression (10). This principle has been demonstrated
for numerous other genes, including e-cadherin in ovarian cancer
(11) or tissue inhibitor of
matrixmetalloproteinase (TIMP)-3 in gastric cancer (12).
Urothelial cancer develops from a preinvasive stage
into an invasive cancer capable of developing metastasis. Matrix
metalloproteinases (MMPs) are key molecules in extracellular
remodelling, and are most likely to be important for the step from
non-invasive to invasive urothelial cancer (13). MMP-9 was previously shown to be
upregulated in invasive cancer compared with superficial bladder
carcinomas (13). Furthermore,
certain studies have shown that the increased expression of MMP-9
and TIMP-2 is associated with an increased recurrence rate of
superficial bladder cancer (14).
Depending on the grade of differentiation and stage, urothelial
cancer may be treated by local chemo- or immunotherapy (15). However, intravesical treatment, either
by instillation of Mitomycin C or intravesical immunotherapy by
induction of an inflammation with Bacillus Calmette-Guérin (BCG),
holds the potential of severe adverse effects, including severe
urocystitis or even systemic BCG infections (16,17).
Epigenetic approaches may offer potential therapeutic options for
urothelial cancer. Nevertheless, such modifiers also indirectly
affect the expression of genes, which are not under the influence
of CpG islands in their promoter regions.
The present study analyzed the effect of the
epigenetic modifiers aza and TSA on the proliferation, migration
and invasion of four biologically different human urinary bladder
cancer cell lines (RT-4, RT-112, VMCUB-1, T-24). In addition, the
mRNA expression of various MMPs, TIMPs and extracellular matrix
metalloproteinase inducer (EMMPRIN) was analyzed in the four cell
lines following aza and TSA treatment.
Materials and methods
Cell culture
Human urinary bladder transitional cell papilloma
RT-4 and human urinary bladder transitional cell carcinoma RT-112
(low-grade), VMCUB-1 and T-24 (high grade) cell lines were obtained
from the German Collection of Cell Cultures and Microorganisms
(Braunschweig, Germany). They were cultivated in Dulbecco's
modified Eagle's medium (GE Healthcare, Chalfont, UK), supplemented
with 10% foetal calf serum, 1% penicillin/streptomycin and 1%
L-glutamine (all Sigma-Aldrich Chemie Gmbh, Munich, Germany) in a
humidified incubator at 37°C with 5% CO2.
Tumour doubling time
To calculate the tumour doubling time,
105 cells were seeded in a 25-cm2 flask and
cell density was counted after 24 and 48 h.
Cell proliferation and treatment with
epigenetic modifiers
All measurements were performed in triplicate in
three independent experiments. In total, 5,000 tumour cells were
seeded per well in 200 µl cell culture medium, and cultivated for
24 h. For sole treatment with aza, cells were treated with 10 µMol
aza for an additional 48 h. For sole treatment with TSA, cells were
stimulated with 200 nMol TSA for 24 h. For combined treatment,
cells were sequentially stimulated with 10 µMol aza for 24 h, after
which, 200 nMol TSA was added for an additional 24 h. After
stimulation, cell cultures were labelled for a further 14 h with
bromodeoxyuridine (BrdU) at a final concentration of 10 µMol.
Proliferation analysis was performed using a BrdU-Cell
Proliferation ELISA (Roche Diagnostics GmbH, Penzberg, Germany),
according to the manufacturer's protocol. Absorbance was measured
using a microplate reader (BioRad Laboratories, Inc., Hercules, CA,
USA) at 450 nm (control wavelength 655 nm).
Cell migration and invasion
Cell migration was analyzed using microwell inserts
(pore size, 8-µm; Nunc™ Cell Culture Inserts; Thermo Fisher
Scientific, Inc., Waltham, MA, USA), while cell invasion was
analyzed using Corning® BioCoat™ Growth Factor Reduced
Matrigel Invasion Chambers (pore size, 8-µm; BD Biosciences,
Franklin Lakes, NsJ, USA). For the two assays, culture medium
supplemented with 20% heat-inactivated foetal calf serum was used
as a chemoattractant in the lower reservoir. All measurements were
performed in three independent experiments using sequential
treatment with aza and TSA. Negative controls were performed using
acetic acid and dimethyl sulfoxide alone. Subsequent to 72 h,
pre-treated cells were harvested, washed and seeded in the upper
reservoir. After 24 h, cells on the lower side of the membrane were
counted following staining with Giemsa. All values were corrected
against the proliferation ratio of BrdU (aza + TSA) / BrdU
(control).
Quantitative polymerase chain reaction
(qPCR) analysis of MMP, TIMP and EMMPRIN expression
Total RNA was extracted from untreated cells and
cells that were treated using the aforementioned method with the
RNeasy Mini kit® (Qiagen GmbH, Hilden, Germany),
according to the manufacturer's protocol. Since preliminary
experiments did not reveal genomic DNA contamination, no RNAse
treatment was performed. RNA integrity and concentration was
determined using an Agilent 2100 Bioanalyzer (Agilent Technologies
Deutschland GmbH & Co., KG, Waldbronn, Germany). In total, 500
ng total RNA were reverse transcribed into cDNA (Omniscript Reverse
Transcriptase kit; Qiagen GmbH). For specific gene expression, 125
ng cDNA was subjected to qPCR using an iCycler® (BioRad
Laboratories, Inc.) with SYBR®-Green I as an
intercalating dye (SYBR Green I Supermix; BioRad Laboratories,
Inc.). For qPCR, 50 cycles were performed under the following
conditions: Denaturation for 30 sec at 95°C; annealing for 30 sec
(for temperatures see Table I);
followed by elongation for 30 sec at 95°C. Primer sequences were
synthesized by Eurofins Genomics (Ebersberg, Germany) and are
listed in Table I. All experiments
were performed in triplicate, and specific reverse
transcription-PCR measurements were performed in duplicate. The
specificity of the PCR reaction was proven by melting point
analysis and agarose gel electrophoresis. Expression values were
calculated with reference to porphobilinogen deaminase gene
expression using the ∆∆Cq method (∆∆Cq = ∆∆Cq treated - ∆∆Cq
untreated control) and the equation y = 2−∆∆Cq (18).
 | Table I.Primers used for quantitative
polymerase chain reaction and their annealing temperatures. |
Table I.
Primers used for quantitative
polymerase chain reaction and their annealing temperatures.
| MMP type | Sense primer | Antisense primer | Temperature, °C |
|---|
| MMP-1 |
CTGGGAGCAAACACATCTGA |
AAGGAGAGTTGTCCCGATGA | 63 |
| MMP-2 |
ACAGTGGACATGGCGGTCTCAG |
AGCCAAGTGGTCCGTGTGAA | 62 |
| MMP-3 |
CCTTTTGATGGACCTGGAAA |
TGAAAGAGACCCAGGGAGTG | 56 |
| MMP-7 Ex4/5 |
TGCTCACTTCGATGAGGATG |
TGGGGATCTCCATTTCCATA | 59 |
| MMP-8 |
CTTTCAGGGAAACCAGCAAC |
TCCACGGAGTGTGGTGATAG | 56 |
| MMP-9 |
GCCACTTGTCGGCGATAGG |
CACTGTCCACCCCTCAGAGC | 63 |
| MMP-10 |
TGGGTTTTCCTCCAACCATA |
AGGCTCAACTCCTGGAAAGTC | 59 |
| MMP-11 |
TGTGACGCCACTCACCTTTA |
ATCCCCTTCTCGGTGAGTCT | 56 |
| MMP-12 |
TTCCCCTGAACAGCTCTACAAGCCTGGAAA |
GATCCAGGTCCAAAAGCATGGGCTAGGATT | 65 |
| MMP-13 |
AACATCCAAAAACGCCAGAC |
GGAAGTTCTGGCCAAAATGA | 53 |
| MMP-14 |
CGGTCATCATCGGGCAGCACAAAA |
CGCTACGCCATCCAGGGTCTCAAA | 63 |
| MMP-15 |
GGAATTCCCCCTCATGTAT |
GGGATCCCTTTCCAGACTGT | 63 |
| MMP-16 |
GGAATTCCCCCTCATGGTAT |
GGGATCCCTTTCCAGACTGT | 63 |
| MMP-17 |
GTGTGCGGGAGTCTGTGTC |
AAAGCTTCACCCCGGATCT | 68 |
| MMP-19 |
CACAATATGGGTACCTACAGAAGC |
GATCCTCTAGGCCACAACGA | 59 |
| MMP-20 |
GCACGTGCAGCAAATAGATG |
TCGATTTGGCCATTTACTCC | 56 |
| MMP-21 Ex5/6 |
ATGGGGACCCTATCCAAATC |
GGTCATAAAACGCCGTGTCT | 59 |
| MMP-23 |
GATCAACCACACGGACTGC |
CGTGTTGTGAGTGCATCAGG | 56 |
| MMP-24 |
CCTATGACTCACGGGCATCT |
GCCTCCACTTCTGTCCAGTC | 59 |
| MMP-25 |
CCCAAACCCCATATGACAAG |
AGGGGCCTTTGAAGAAGAAA | 56 |
| MMP-26 |
GATATGAAGCCATCCGCAGT |
AGGCATGGCCTAAGATACCA | 63 |
| MMP-27 |
GCCAGATTATCCCAAATCC |
TTACCACTCTCTGCGGGAAC | 59 |
| MMP-28 |
GAGACCTGGGACTCCTACAGC |
CTCTGAGACGTTGCCATCAG | 61 |
| TIMP-1 |
ACCAGACCACCTTATACCAGCG |
GGACTGGAAGCCCTTTTCAGAG | 65 |
| TIMP-2 |
ATGCAGATGTAGTGATCAGGGC |
GATGAAGTCACAGAGGGTGATG | 63 |
| TIMP-3 |
GGGGAAGAAGCTGGTAAAG |
AAGTCACAAAGCAAGGCAG | 57 |
| TIMP-4 |
CACCCTCAGCAGCACATCT |
TTTGATTTCATACCGGAGCA | 59 |
| EMMPRIN |
CCGGCACAGTCTTCACTACC |
TACTCTCCCCACTGGTCGTC | 60 |
| PBGD |
TCAATGTTGCCACCACACTGTCCGTCT |
TGTCTGGTAACGGCAATGCGGCTGCAAC | 70 |
Statistical analysis
Statistical significance was calculated using
Student's t-test. Post hoc comparisons were made by the
Bonferroni test for repeated measurements. P<0.05 was considered
to indicate a statistically significant difference. All tests were
performed using GraphPad Prism version 4 software (GraphPad
Software Inc., La Jolla, CA, USA).
Results
Antiproliferative effects of
epigenetic modifiers
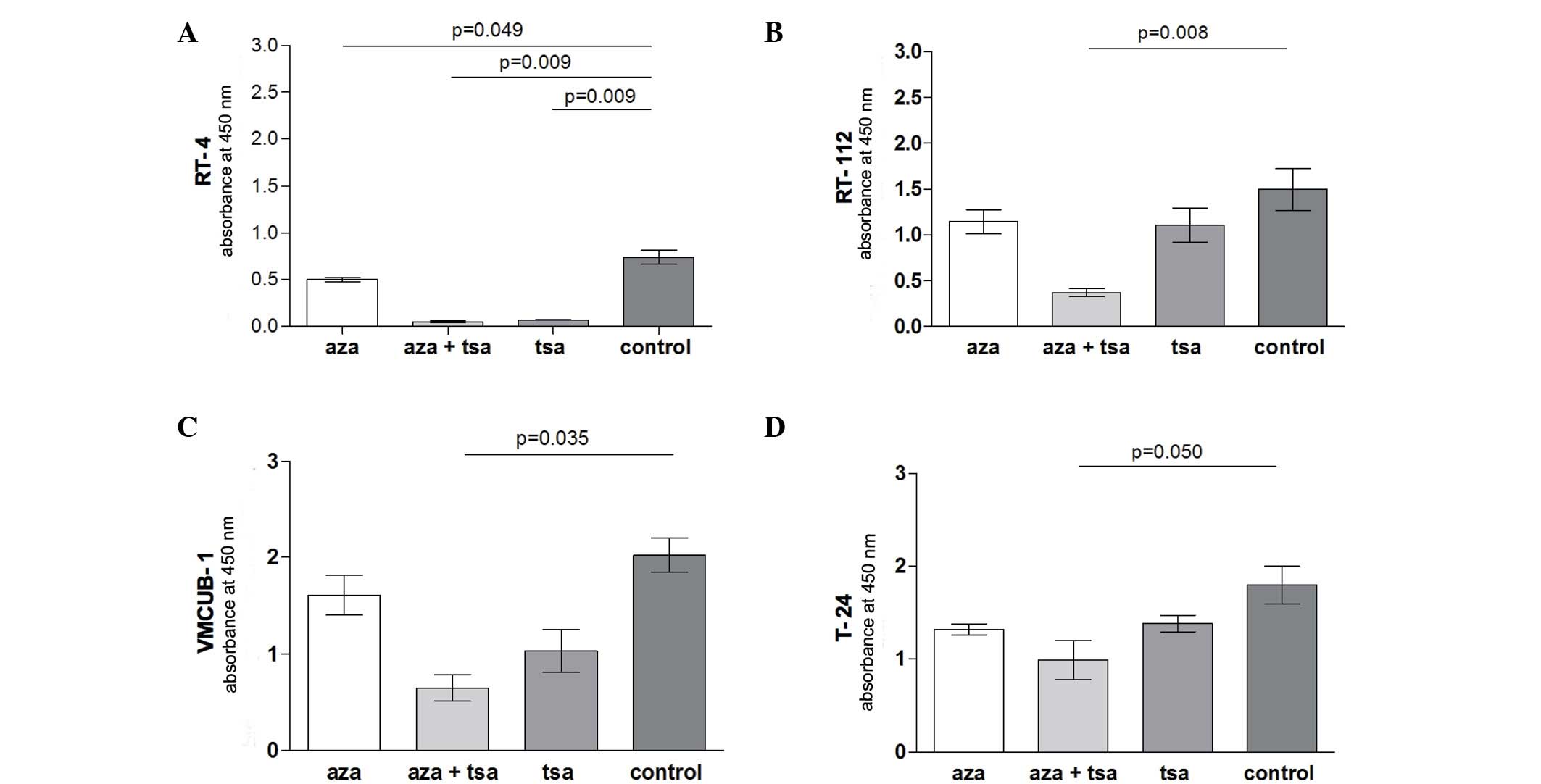
The calculation of the tumour doubling time of
unstimulated cell lines revealed a doubling time of 41.6 h for cell
line RT-4, 15.9 h for RT-112, 11.3 h for VMCUB-1 and 9 h for
T-24.
In cell lines RT-112 (P=0.008), VMCUB-1 (P=0.035)
and T-24 (P=0.050) only combined aza and TSA treatment resulted in
a significantly reduced proliferation compared with untreated
controls. By contrast, the proliferation of RT-4 cells was
significantly suppressed by treatment with TSA (P=0.009) or aza
treatment alone (P=0.049), and the TSA and aza combination
treatment (P=0.009) (Fig. 1).
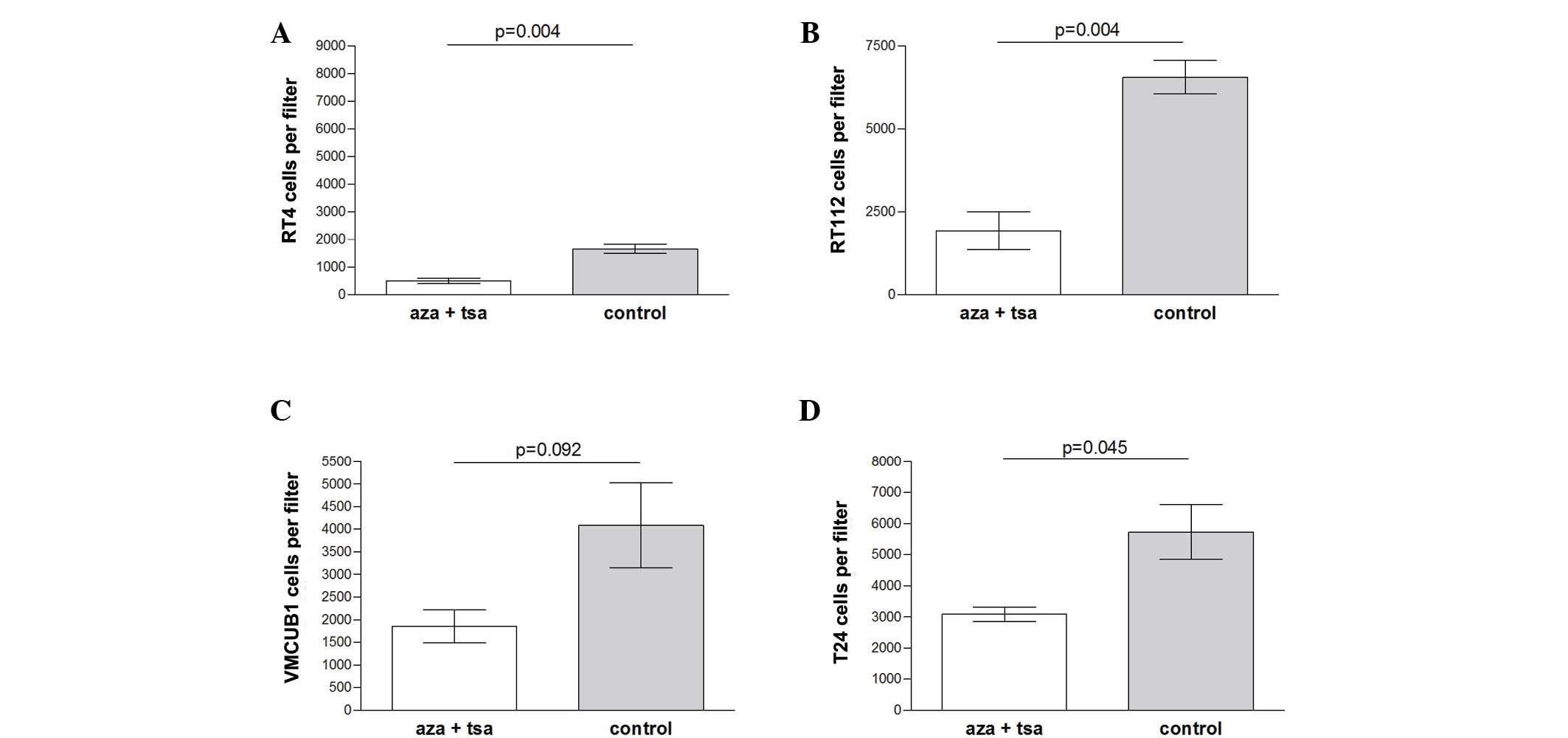
Migration inhibition by epigenetic
modifiers
Migration was significantly inhibited by combined
treatment of aza and TSA in the low grade RT-4 (P=0.004) and RT-112
(P=0.004) cell lines, and slightly less inhibited in the high grade
T-24 cell line (P=0.045). No significant difference was observed
for VMCUB-1 cells (P=0.092) (Fig.
2).
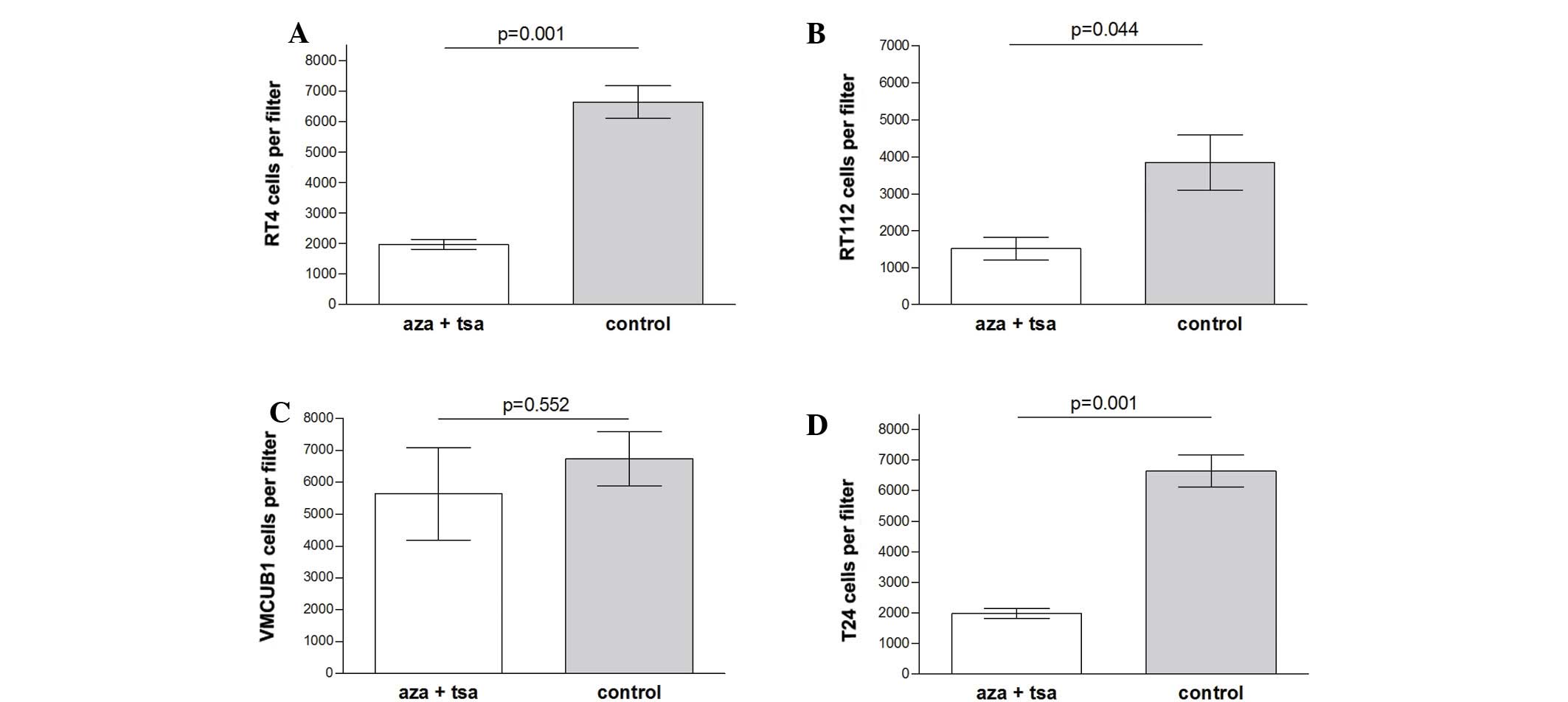
Invasion inhibition by epigenetic
modifiers
Cell invasion was significantly inhibited by
combined treatment of aza and TSA in the low grade RT-4 (P=0.001)
and RT-112 (P=0.044) cell lines, and the high grade T-24 cell line
(P=0.013). No significant difference was observed in VMCUB-1 cells
(P=0.552) (Fig. 3).
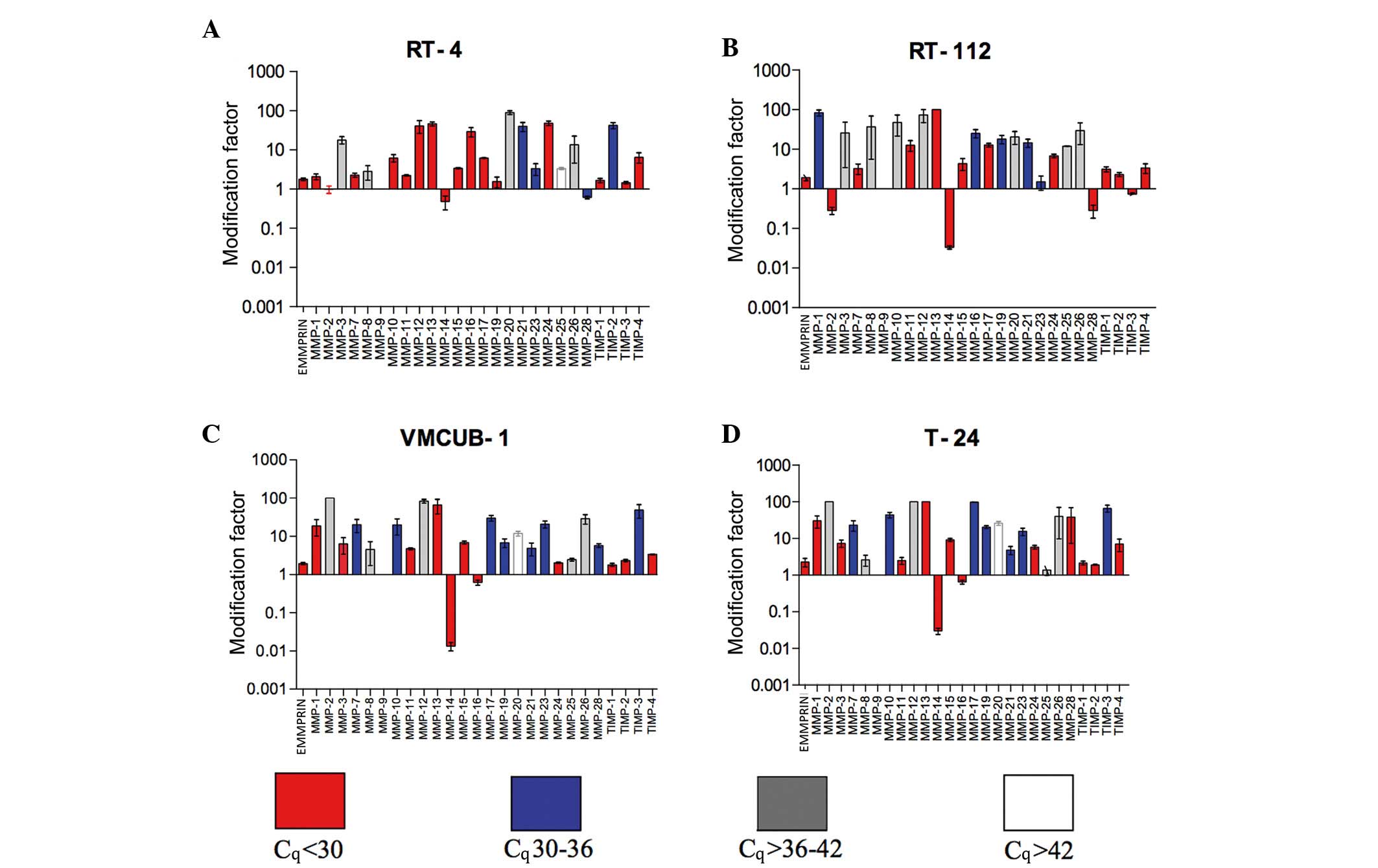
Effects of epigenetic modifiers on
MMP, TIMP and EMMPRIN mRNA expression
Overall
All data regarding smRNA expression levels and the
effect of epigenetic modifiers are summarized in Fig. 4 and Table
II.
 | Table II.P-values for mRNA expression of MMPs,
TIMs and EMMPRIN in human urinary bladder cancer cell lines agaisnt
untreated cells. |
Table II.
P-values for mRNA expression of MMPs,
TIMs and EMMPRIN in human urinary bladder cancer cell lines agaisnt
untreated cells.
|
| Cell lines,
P-value |
|---|
|
|
|
|---|
|
| RT-4 | RT-112 | VMCUB-1 | T-24 |
|---|
| EMMPRIN |
0.0254 |
0.0193 |
0.0618 |
0.0518 |
| MMP-1 |
0.0104 |
0.0010 |
0.0199 |
0.0015 |
| MMP-2 |
0.7801 |
0.0206 |
0.0002a |
<0.0001a |
| MMP-3 |
0.0001 |
0.1097a |
0.3396 |
0.0006 |
| MMP-7 |
0.0009 |
0.0123 |
0.0027 |
0.0004 |
| MMP-8 |
0.0611 |
0.0879 |
0.1041 |
0.0566 |
| MMP-9 | NE | NE | NE | NE |
| MMP-10 |
0.0108 |
0.0076 |
0.0068 |
0.0015 |
| MMP-11 |
0.0099 |
0.0274 |
0.0048 |
0.0602 |
| MMP-12 |
0.0003 |
0.0036a |
0.0023a |
<0.0001a |
| MMP-13 | <0.0001 |
0.0002 |
0.0017 | <0.0001 |
| MMP-14
(MT1-MMP) |
0.2083 | <0.0001 |
0.0005 |
0.0003 |
| MMP-15
(MT2-MMP) | <0.0001 |
0.0036 |
0.0004 |
0.0002 |
| MMP-16
(MT3-MMP) |
0.0011 |
0.0002 |
0.0201 |
0.0875 |
| MMP-17
(MT4-MMP) | <0.0001 | <0.0001 | <0.0001 |
0.0130 |
| MMP-19 |
0.8661 |
0.0002 |
0.0011 | <0.0001 |
| MMP-20 |
<0.0001a |
0.0035a |
0.0008a |
0.0065a |
| MMP-21 | <0.0001 |
0.0210 |
0.0127 |
0.0083 |
| MMP-23 |
0.0534 |
0.6542 |
0.0009 |
0.0013 |
| MMP-24
(MT5-MMP) | <0.0001 |
0.0054 |
0.0014 | <0.0001 |
| MMP-25
(MT6-MMP) |
<0.0001a | <0.0001 |
0.0325a |
0.6737a |
| MMP-26 |
0.1823 |
0.2333 |
0.1719 |
0.4009 |
| MMP-28 |
0.0130 |
0.0307 |
0.0161 |
0.1305 |
| TIMP-1 |
0.0506 |
0.0004 |
0.0026 |
0.1179 |
| TIMP-2 | <0.0001 |
0.0051 |
0.0006 |
0.0025 |
| TIMP-3 |
0.0106 |
0.1888 |
0.0002 |
0.0002 |
| TIMP-4 |
0.0073 |
0.0797 |
0.0028 |
0.0094 |
EMMPRIN expression
TSA and aza induced a significant mRNA increase in
the low-grade RT-4 (P=0.0254) and RT-112 (P=0.0193) cell lines.
This induction was nearly significant in VMCUB-1 (P=0.0618) and
T-24 (P=0.0518) cells.
Membrane-type (MT) MMPs
With the exception of RT-4 cells (P=0.2083), MMP-14
mRNA expression was significantly suppressed by epigenetic modifier
treatment in RT-112 (P<0.0001), VMCUB-1 (P=0.0005) and T-24
(P=0.0003) cell lines. The mRNA expression of all other MT-MMPs was
increased in RT-4, RT-112, VMCUB-1 and T-24 cell lines, the latter
exhibited an alteration in MMP-16 and MMP-25 mRNA expression.
However, MMP 25 mRNA was expressed only at a very low level. All
P-values are provided in Table
II.
Gelatinases
MMP-9 mRNA was not detected in the cell lines. By
contrast to a significant mRNA suppression in RT-112 cells
(P=0.0206), MMP-2 mRNA expression was not altered in RT-4 cells
(P=0.7801). MMP-2 mRNA expression was clearly induced in the
high-grade cell lines at a low level (VMCUB-1, P=0.0002; T-24,
P<0.0001).
Collagenases
MMP-1 was induced significantly in all cell lines.
MMP-8 mRNA increased from a low level. In all four cell lines,
MMP-13 mRNA expression increased significantly from a high base
level. All P-values are provided in Table II.
Stromelysins
MMP-3 mRNA was expressed in high-grade VMCUB-1 and
T-24 cell lines at a high level, in contrast to RT-4 and RT-112
cell lines. However, only RT-4 and T-24 cells upregulated MMP-3
mRNA significantly (P=0.0001 and P=0.0006, respectively). The same
result was observed for MMP-10 and MMP-11 mRNA, with the exception
of MMP-11 in the T-24 cell line (Table
II).
Other MMPs
Notably, mRNA of MMP-12, which is typically
expressed in macrophages, exhibited a high expression level in RT-4
cells and was significantly induced in other cell lines. MMP-28
mRNA was suppressed by epigenetic modifier treatment in RT-4 and
RT-112 cells. The mRNA expression levels of MMP-19, −20, −21, −23
and −27 exhibited individual patterns, which were either stable or
induced following treatment with epigenetic modifiers. P-values are
provided in Table II.
TIMPs
TIMP-3 was upregulated following treatment with TSA
and aza combination, which was at higher level in RT-4 and RT-112
cells compared to VMCUB-1 and T-24 cells. TIMP-1 and TIMP-2 mRNA
was scantly expressed, but significantly upregulated in all four
cell lines, with the exception of TIMP-2 mRNA in T-24 cells.
Treatment with epigenetic modifiers significantly induced the
expression of TIMP-4 mRNA in RT-4, VCUMB-1 and T-24 cell lines,
whereas in RT-112 cells this appeared to be a statistical trend.
All P-values are provided in Table
II.
Discussion
Epigenetics may offer additional treatment options
for solid malignancies. The present study analyzed the effects of
the histone deacetylase inhibitor Trichostatin A (TSA) and the DNA
methyltransferase inhibitor 5-aza-2′-deoxycytidine (aza) on
proliferation, migration and invasion of four urothelial cancer
cell lines with various proliferation characteristics. In addition,
the present study analyzed the mRNA expression of a broad panel of
MMPs, TIMPs and EMMPRIN following TSA and aza treatment.
For optimum treatment, the two inhibitors were
combined and this treatment combination exhibited the strongest
antiproliferative effects in all cell lines. This has already been
reported by Karam et al (19)
in urothelial carcinoma cell lines and Cecconi et al
(20) in an endocrine pancreatic
carcinoma cell line. The effect of the inhibitors may be associated
with the re-expression of cell cycle regulatory proteins and the
activation of key genes of the apoptotic cascade (21). In the present study, treatment with
aza and TSA combination on the low-proliferating RT-4 cell line
resulted in clear antiproliferative effects. This may be due to the
switch-off of additional cell-cycle-promoting proteins or the
switch-on of cell-cycle-controlling proteins. The requirement of a
combined epigenetic modification, leading to a decreased
proliferation in the other cell lines (RT-112, VMCUB-1 and T-24),
suggests that there are pre-existing structural gene alterations of
cell cycle promoting or controlling proteins, whose effects are
affected by combined TSA and aza treatment.
Invasiveness is a complex function of migration and
extracellular matrix remodelling. As summarized in Fig. 4 aza/TSA-treatment stimulated the
expression of the majority of MMPs and TIMPs. Sato et al
(22) observed increased invasiveness
in pancreatic cancer cells and an induction of MMP-1, −2, −3, −7,
−9 and −14 following aza treatment. In contrast to the other cell
lines, in RT112 cells, aza/TSA-treatment suppressed MMP-2 and
MMP-28 mRNA expression. Shukeir et al (23) identified a decreased MMP 2 expression
triggered by gene methylation in highly invasive prostate cancer
cell lines. Couillard et al (21) described an increased MMP-3 expression
in colon cancer cell lines following demethylating treatment with
aza. However, in that study MMP-10 expression was not affected by
aza treatment. Furthermore, in B-cell lymphoma cells a MMP-10
induction, but not a MMP-3 induction, was observed (21). Chen et al (24) described a clear inhibitory effect on
the invasiveness of bladder cancer cells treated with the histone
deacetylase inhibitor valproic acid; however, inhibition of
migration was not observed. Since, epigenetic modifiers induce
antiproliferative effects, it is necessary to dissect
antiproliferative from anti-migratory and anti-invasive effects.
Therefore, the present study corrected migration and invasion
assays against antiproliferative effects. With the exception of
VMCUB-1 cells, a significant inhibition in migration and invasion
was observed in all cell lines, and VMCUB-1 cells exhibited a trend
towards reduced invasion. These various, somewhat contradictory,
results suggest an epigenetic effect on the invasiveness and
migratory behavior in a cell- or tissue-specific manner.
In the present study, MMP-14 suppression by aza/TSA
treatment was a constant effect in all tested cell lines. MMP-14,
also known as MT1-MMP, and TIMP-2 form an activating complex with
proMMP-2 on the cell surface (25).
As shown by qPCR in the present study, MMP-14 mRNA expression was
suppressed, possibly resulting in a reduction of activating
complexes. Selective inhibition of MMP-14 is capable of blocking
invasion and tumour growth (26).
Itoh and Seiki (27) underlined the
essential role of MMP-14 in the regulation of invasion and
migration in tumour cells via homodimerization on the cell surface
for pericellular collagenolysis, a mechanism that has also been
identified for the collagenase MMP-13 (28). Notably, MMP-13 was significantly
induced in all cell lines in the present study following treatment
with aza and TSA combination. Therefore, the reduction in invasion
and migration in the present experiments may be explained by the
inhibition of MMP-14 expression. Kitagawa et al (29) found that the mRNA of MMP-14 and MMP-15
was significantly overexpressed in urothelial carcinoma in
comparison with normal mucosa. High mRNA expression of the two MMPs
was associated with multilocular tumours (29). In contrast, mRNA expression of MMP-16,
which is also a proMMP-2 activator, was detected at a much lower
level without any particular association (29). In the present study, MMP-16 was
significantly stimulated in the low grade cell lines RT-4 and
RT-112. In contrast, this MMP-16 mRNA expression was suppressed in
the high grade cell lines VMCUB-1 and T-24. In colorectal cancer
and cancer cell lines, MMP-16 promoter hypermethylation was
reported to be associated with a decreased mRNA expression
(30). Treatment with 5-aza
2′-deoxycytidine restored this mRNA expression in colorectal cancer
cell lines (30). Knockdown of mRNA
expression was associated with cell migration, re-expression and
overexpression of genes and proteins, with reduced migration
(30). These observations confirm the
findings from the present study regarding mRNA expression of MMP-16
in the low grade urothelial cancer cell lines, RT-4 and RT-112, but
are in contrast to the findings of the present study that regard
migration and the effects in the high grade cell lines, VMCUB-1 and
T-24. Therefore, it is likely that regulatory pathways are more
complex. Furthermore, Moon et al (30) did not consider the potential
anti-proliferative effects of aza treatment. TIMP proteins are
genuine inhibitors of MMPs, but they also perform additional
functions. TIMP-3 overexpression is associated with the induction
of apoptosis (31,32), while TIMP-1 and TIMP-2 protect B cells
and melanoma cells, respectively, from apoptosis (33). In the present study, TIMP mRNA
expression was increased in all cell lines. An alteration in the
MMP/TIMP ratio towards TIMPs and suppression of MMP-14, coupled
with inhibited migratory properties, may be essential for the
observed reduced invasiveness of RT-4, RT-112 and T-24 cell lines.
In VMCUB-1 cells, no effects on migration and invasion were
observed; therefore, other extracellular matrix remodelling
proteinases, including urokinase and plasminogen activator-1, may
be of importance in this cell line.
In conclusion, the present study revealed that the
epigenetic modifications of aza and TSA suppressed the motility and
invasiveness of three out of four urothelial cancer cell lines. The
inhibitory effect on cell motility appears to be crucial for
reduced invasive properties. However, even a broad spectrum mRNA
analysis of the MMP/TIMP axis does not sufficiently explain the
loss of invasiveness, since it leaves no scope for functional
conclusions, such as the homodimerization of MMP-14 or MMP-13.
Further complex urothelial tumour models should be applied to
investigate whether epigenetic therapeutic approaches may be used
in urothelial cancer.
References
|
1
|
Kanwal R and Gupta S: Epigenetic
modifications in cancer. Clin Genet. 81:303–311. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Ades L and Santini V: Hypomethylating
agents and chemotherapy in MDS. Best Pract Res Clin Haematol.
26:411–419. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Kim TK, Gore SD and Zeidan AM: Epigenetic
therapy in acute myeloid leukemia: Current and future directions.
Semin Hematol. 52:172–183. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Boumber Y and Issa JP: Epigenetics in
cancer: What's the future? Oncology (Williston Park). 25:220–226,
228. 2011.PubMed/NCBI
|
|
5
|
Montero AJ, Diaz-Montero CM, Mao L,
Youssef EM, Estecio M, Shen L and Issa JP: Epigenetic inactivation
of EGFR by CpG island hypermethylation in cancer. Cancer Biol Ther.
5:1494–1501. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Bauman J, Verschraegen C, Belinsky S,
Muller C, Rutledge T, Fekrazad M, Ravindranathan M, Lee SJ and
Jones D: A phase I study of 5-azacytidine and erlotinib in advanced
solid tumor malignancies. Cancer Chemother Pharmacol. 69:547–554.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Vigushin DM, Ali S, Pace PE, Mirsaidi N,
Ito K, Adcock I and Coombes RC: Trichostatin A is a histone
deacetylase inhibitor with potent antitumor activity against breast
cancer in vivo. Clin Cancer Res. 7:971–976. 2001.PubMed/NCBI
|
|
8
|
Ailenberg M and Silverman M: Trichostatin
A-histone deacetylase inhibitor with clinical therapeutic
potential-is also a selective and potent inhibitor of gelatinase A
expression. Biochem Biophys Res Commun. 298:110–115. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Christman JK: 5-Azacytidine and
5-aza-3′-deoxycytidine as inhibitors of DNA methylation:
mechanistic studies and their implications for cancer therapy.
Oncogene. 21:5483–5495. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Ferguson AT, Lapidus RG, Baylin SB and
Davidson NE: Demethylation of the estrogen receptor gene in
estrogen receptor-negative breast cancer cells can reactivate
estrogen receptor gene expression. Cancer Res. 55:2279–2283.
1995.PubMed/NCBI
|
|
11
|
Yuecheng Y, Hongmei L and Xiaoyan X:
Clinical evaluation of E-cadherin expression and its regulation
mechanism in epithelial ovarian cancer. Clin Exp Metastasis.
23:65–74. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Kang SH, Choi HH, Kim SG, Jong HS, Kim NK,
Kim SJ and Bang YJ: Transcriptional inactivation of the tissue
inhibitor of metalloproteinase-3 gene by dna hypermethylation of
the 3′-CpG island in human gastric cancer cell lines. Int J Cancer.
86:632–635. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Izawa JI, Slaton JW, Kedar D, Karashima T,
Perrotte P, Czerniak B, Grossman HB and Dinney CP: Differential
expression of progression-related genes in the evolution of
superficial to invasive transitional cell carcinoma of the bladder.
Oncol Rep. 8:9–15. 2001.PubMed/NCBI
|
|
14
|
Hara I, Miyake H, Hara S, Arakawa S and
Kamidono S: Significance of matrix metalloproteinases and tissue
inhibitors of metalloproteinase expression in the recurrence of
superficial transitional cell carcinoma of the bladder. J Urol.
165:1769–1772. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
National Institute for Health and Care
Excellence: Bladder cancer: Diagnosis and management. NICE
Guideline 2. National Collaborating Centre for Cancer (Cardiff).
2015.https://www.nice.org.uk/guidance/ng2/resources/bladder-cancer-diagnosis-and-management-of-bladder-cancer-51036766405Accessed.
June 10–2016
|
|
16
|
Elmamoun MH, Christmas TJ and Woodhouse
CR: Destruction of the bladder by single dose Mitomycin C for
low-stage transitional cell carcinoma (TCC) - avoidance,
recognition, management and consent. BJU Int. 113:E34–E38. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Pommier JD, Ben Lasfar N, Van Grunderbeeck
N, Burdet C, Laouénan C, Rioux C, Pierre-Audigier C, Meybeck A,
Choudat L, Benchikh A, et al: Complications following intravesical
bacillus Calmette-Guerin treatment for bladder cancer: A case
series of 22 patients. Infect Dis (Lond). 47:725–731. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) Method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Karam JA, Fan J, Stanfield J, Richer E,
Benaim EA, Frenkel E, Antich P, Sagalowsky AI, Mason RP and Hsieh
JT: The use of histone deacetylase inhibitor FK228 and DNA
hypomethylation agent 5-azacytidine in human bladder cancer
therapy. Int J Cancer. 120:1795–1802. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Cecconi D, Donadelli M, Dalla Pozza E,
Rinalducci S, Zolla L, Scupoli MT, Righetti PG, Scarpa A and
Palmieri M: Synergistic effect of trichostatin A and
5-aza-3′-deoxycytidine on growth inhibition of pancreatic endocrine
tumour cell lines: A proteomic study. Proteomics. 9:1952–1966.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Couillard J, Demers M, Lavoie G and
St-Pierre Y: The role of DNA hypomethylation in the control of
stromelysin gene expression. Biochem Biophys Res Commun.
342:1233–1239. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Sato N, Maehara N, Su GH and Goggins M:
Effects of 5-aza-3′-deoxycytidine on matrix metalloproteinase
expression and pancreatic cancer cell invasiveness. J Natl Cancer
Inst. 95:327–330. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Shukeir N, Pakneshan P, Chen G, Szyf M and
Rabbani SA: Alteration of the methylation status of tumor-promoting
genes decreases prostate cancer cell invasiveness and tumorigenesis
in vitro and in vivo. Cancer Res. 66:9202–9210. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Chen CL, Sung J, Cohen M, Chowdhury WH,
Sachs MD, Li Y, Lakshmanan Y, Yung BY, Lupold SE and Rodriguez R:
Valproic acid inhibits invasiveness in bladder cancer but not in
prostate cancer cells. J Pharmacol Exp Ther. 319:533–542. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Kinoshita T, Sato H, Okada A, Ohuchi E,
Imai K, Okada Y and Seiki M: TIMP-2 promotes activation of
progelatinase A by membrane-type 1 matrix metalloproteinase
immobilized on agarose beads. J Biol Chem. 273:16098–16103. 1998.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Devy L, Huang L, Naa L, Yanamandra N,
Pieters H, Frans N, Chang E, Tao Q, Vanhove M, Lejeune A, et al:
Selective inhibition of matrix metalloproteinase-14 blocks tumor
growth, invasion, and angiogenesis. Cancer Res. 69:1517–1526. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Itoh Y and Seiki M: MT1-MMP: A potent
modifier of pericellular microenvironment. J Cell Physiol. 206:1–8.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Leeman MF, Curran S and Murray GI: The
structure, regulation, and function of human matrix
metalloproteinase-13. Crit Rev Biochem Mol Biol. 37:149–66. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Kitagawa Y, Kunimi K, Ito H, Sato H,
Uchibayashi T, Okada Y, Seiki M and Namiki M: Expression and tissue
localization of membrane-types 1, 2, and 3 matrix
metalloproteinases in human urothelial carcinomas. J Urol.
160:1540–1545. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Moon JW, Choi JH, Lee SK, Lee YW, Lee JO,
Kim N, Lee HJ, Seo JS, Kim J, Kim HS, et al: Promoter
hypermethylation of membrane type 3 matrix metalloproteinase is
associated with cell migration in colorectal adenocarcinoma. Cancer
Genet. 208:261–270. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Baker AH, George SJ, Zaltsman AB, Murphy G
and Newby AC: Inhibition of invasion and induction of apoptotic
cell death of cancer cell lines by overexpression of TIMP-3. Br J
Cancer. 79:1347–1355. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Fata JE, Leco KJ, Voura EB, Yu HY,
Waterhouse P, Murphy G, Moorehead RA and Khokha R: Accelerated
apoptosis in the Timp-3-deficient mammary gland. J Clin Invest.
108:831–841. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Valente P, Fassina G, Melchiori A,
Masiello L, Cilli M, Vacca A, Onisto M, Santi L, Stetler-Stevenson
WG and Albini A: TIMP-2 over-expression reduces invasion and
angiogenesis and protects B16F10 melanoma cells from apoptosis. Int
J Cancer. 75:246–253. 1998. View Article : Google Scholar : PubMed/NCBI
|