Introduction
Breast cancer is one of the most common types of
cancer in women, with >200,000 new cases annually in the USA
(1). Women over the age of 60 years
have a greater likelihood of developing cancer; however, for
younger females, the development of cancer may be due to inherited
genetic variants, also referred to as polymorphisms (2). These polymorphisms have been identified
in high-penetrance genes, including ATM serine/threonine kinase,
tumor protein P53, and breast cancer 1 and 2, which only account
for a small percentage of the breast cancer cases that cause the
familial/early onset tumors. Other risks for breast cancer in young
females may be due to polymorphisms in genes of moderate/low
penetrance.
The faciogenital dysplasia 1 (FGD1) gene encodes for
a guanine nucleotide exchange factor (GEF) protein, which is a
member of a family of proteins that activate the Rho GTPases
(3). Fgd1 protein specifically
activates the cell division cycle 42 (Cdc42) GTPase. Cdc42 signals
for cellular migration by regulating cytoskeleton restructuring,
gene transcription and cell morphology, extension and adhesion. In
cancer cells, Cdc42 modulates tumor cell migration and invasiveness
(4,5).
Researchers have identified that several GEF proteins, such as
leukemia-associated Rho-GEF and Rho/Rac GEF 2, have a similar
sequence to Fgd1. These GEF proteins are linked to the upregulation
of GTPases in tumor cells and have been labeled as potential
oncogenes in advanced cancers (6,7).
Recently, researchers have detected overexpression
of the Fgd1 protein in infiltrating and poorly differentiated
breast and invasive prostate tumors (8). Missense mutations in the FGD1 gene were
identified in late-stage tumors of numerous types of cancer tissue,
including ovarian cancer, prostate cancer, melanoma, uterine
cancer, head and neck cancer (9,10).
Amplification at Xp22.2, the FGD1 gene locus, has also been
reported in several types of cancer, including breast, uterine,
hepatocellular and lung cancers (9,10). Along
with somatic alterations, polymorphisms in the FGD1 gene have been
linked to an inherited disease and thyroid cancer. Polymorphisms in
the FGD1 gene have been associated with a rare X-linked disorder
known as faciogenital dysplasia or Aarskog-Scott syndrome (3). This disorder is characterized by short
stature and congenital anomalies of the face, skeleton and genitals
(11–13). Malformations are consistent with a
loss of cellular migration during embryonic development (14). Many of the germline variants are
present as either an insertion or a deletion in the FGD1 gene,
which results in a frameshift causing inactivation of Fgd1 protein.
Several missense changes have also been linked to the disorder. In
a recent study, researchers have identified two polymorphisms,
rs1126744 and rs12011120, in thyroid cancer (15). However, the status of genetic
variants, somatic and germline, in the FGD1 gene has not been
studied in breast cancer samples. This purpose of the present study
was to examine the association of genetic variants in the FGD1 gene
with early-onset status using primary breast tumors.
Materials and methods
Tissue sections and DNA isolation
Frozen tissue sections from breast tumors and
corresponding normal breast tissue were obtained through the South
Carolina Biorepository System at the Lexington Regional Medical
Center (Lexington, SC, USA). The 46 matched-pair samples were
de-identified with clinical information for pathological stage and
estrogen receptor (ER)/progesterone receptor (PR)/human epidermal
growth factor receptor 2 status of the cancer, which is provided in
Table I. The frozen tissue sections
were prepared for sectioning with Optimal Cutting Temperature
Medium (Sakura Finetek, Torrence, CA, USA). The tissue samples were
cut into 15-µm slices and fixed onto microscope slides using
ethanol. The fixed slides were stained with Mayer's hematoxylin
(Sigma-Aldrich, St. Louis, MO, USA) and eosin (Harleco; EMD
Millipore, Lawrence, KS, USA) to distinguish between tumor and
normal cells. Subsequently, tumor and normal cells were extracted
from the slides using micro-dissection with an optical microscope.
DNA of the micro-dissected cells was isolated using the
phenol-chloroform protocol (16). DNA
was measured using a Nanodrop spectrophotometer (Thermo Fisher
Scientific, Inc., Grand Island, NY, USA). DNA from tumor and normal
samples was diluted to a final standard concentration of 10 ng/µl.
The present study was approved by an Institutional Review
Board.
 | Table I.Summary of the breast cancer
patients. |
Table I.
Summary of the breast cancer
patients.
| Patient ID | Age at diagnosis
(years) | Race | Stage | Estrogen receptor
status | Progesterone receptor
status | HER-2/Neu receptor
status | Vital status | Date and type of
first recurrence | PIK3CA mutations | FGD1
polymorphisms |
|---|
| 585 | 82 | Caucasian | 2A | + | + | Borderline | Deceased; cancer | No recurrence | − | − |
| 594 | 54 | Caucasian | 2B | + | − | − | Alive | Never
disease-free | − | 51451 T->C
(697Thr) Hetero |
|
|
|
|
|
|
|
|
|
|
| 51496 A->G
(712Pro) Hetero |
| 597 | 72 | Caucasian | 2A | + | − | − | Alive | 06/30/2010 (local
recurrence) | − | − |
| 5106 | 85 | Caucasian | 2A | + | + | Borderline | Deceased; cancer | Never
disease-free | − | − |
| 5130 | 45 | African-American | N/A | − | + | − | Alive | Never
disease-free | − | 51451 T->C
(697Thr) Hetero |
| 5134 | 85 | Caucasian | 2A | − | − | + | Deceased; cancer | Never
disease-free | 90775 A->G | − |
|
|
|
|
|
|
|
|
|
|
(1047His->Arg) |
|
| 5139 | 67 | Caucasian | 1 | + | + | − | Alive | Never
disease-free | − | − |
| 5151 | 52 | Caucasian | 1 | + | + | − | Alive | Never
disease-free | 74781 G->A | − |
|
|
|
|
|
|
|
|
|
| (545Glu->Lys) |
|
| 5153 | 51 | Caucasian | 1 | + | + | − | Alive | Never
disease-free | − | − |
| 5165 | 69 | Caucasian | 1 | + | + | − | Alive | Never
disease-free | − | − |
| 5167 | 40 | Caucasian | 1 | + | + | − | Alive | 05/18/2010 & | 90775 A->G | 51451 T->C
(697Thr) Homo |
|
|
|
|
|
|
|
|
| Distant
recurrence |
(1047His->Arg) | 30726 G->A
(226Ala->Thr) Hetero |
| 5169 | 80 | Caucasian | 1 | + | + | − | Alive | No recurrence | − | − |
| 5170 | 61 |
African-American | 1 | + | + | − | Alive | Never
disease-free | − | 51451 T->C
(697Thr) Hetero |
|
|
|
|
|
|
|
|
|
|
| 51496 A->G
(712Pro) Homo |
| 5172 | 78 | Caucasian | 2A | + | + | − | Alive | Never
disease-free | 90775 A->G | − |
|
|
|
|
|
|
|
|
|
|
(1047His->Arg) |
|
| 5177 | 44 |
African-American | 2A | + | + | + | Alive | Never
disease-free | − | 51451 T->C
(697Thr) Hetero |
|
|
|
|
|
|
|
|
|
|
| 51496 A->G
(712Pro) Hetero |
| 5180 | 61 | Caucasian | 2A | − | − | − | Alive | No recurrence | − | − |
| 5181 | 49 | Caucasian | 2B | − | + | + | Deceased;
cancer | 07/13/2009 | − | − |
|
|
|
|
|
|
|
|
| (recurrence, site
unknown) |
|
| 5183 | 71 | Caucasian | 1 | + | + | − | Deceased | Never
disease-free | 90775 A->G | − |
|
|
|
|
|
|
|
|
|
|
(1047His->Arg) |
|
| 5187 | 65 | Caucasian | 1 | − | − | − | Alive | Never
disease-free | 90775 A->G | − |
|
|
|
|
|
|
|
|
|
|
(1047His->Arg) |
|
| 5192 | 49 | Caucasian | 2A | + | + | Borderline | Alive | Never
disease-free | 90775 A->G | − |
|
|
|
|
|
|
|
|
|
|
(1047His->Arg) |
|
| 5195 | 60 | Caucasian | 2A | − | − | − | Alive | No recurrence | − | − |
| 5226 | 41 |
African-American | 2B | − | − | − | Deceased;
cancer | No recurrence | − | 51451 T->C
(697Thr) Hetero |
| 5231 | 84 |
African-American | N/A | Test not ordered or
performed | Test not ordered or
performed | Test not
performed | Deceased;
cancer | No recurrence | 74781 G->A | 51451 T->C
(697Thr) Homo |
|
|
|
|
|
|
|
|
|
|
(545Glu->Lys) | 51496 A->G
(712Pro) Hetero |
| 5237 | 46 | Caucasian | 1 | + | + | − | Alive | Never
disease-free | − | − |
| 5239 | 41 | Caucasian | 1 | + | + | − | Alive | No recurrence | 74781 G->A | 51451 T->C
(697Thr) Hetero |
|
|
|
|
|
|
|
|
|
|
(545Glu->Lys) | 51496 A->G
(712Pro) Hetero |
| 5260 | 68 | Caucasian | 2B | + | + | + | Alive | No recurrence | − | − |
| 5262 | 46 |
African-American | 2A | + | + | Borderline | Alive | Never
disease-free | 90775 A->G | 51451 T->C
(697Thr) Homo |
|
|
|
|
|
|
|
|
|
|
(1047His->Arg) | 51496 A->G
(712Pro) Homo |
| 5263 | 71 | Caucasian | 1 | + | − | − | Alive | No recurrence | − | 51451 T->C
(697Thr) Homo |
|
|
|
|
|
|
|
|
|
|
| 51496 A->G
(712Pro) Homo |
| 5284 | 59 |
African-American | 1 | − | − | − | Deceased;
cancer | Never
disease-free | − | 51451 T->C
(697Thr) Homo |
|
|
|
|
|
|
|
|
|
|
| 51496 A->G
(712Pro) Homo |
| 5288 | 68 | Caucasian | 1A | + | + | − | Alive | Never
disease-free | − | 51451 T->C
(697Thr) Hetero |
|
|
|
|
|
|
|
|
|
|
| 51496 A->G
(712Pro) Hetero |
| 5300 | 50 |
African-American | 1A | + | + | − | Alive | Never
disease-free | − | − |
| 5303 | 90 | Caucasian | 2A | + | + | − | Deceased | Never
disease-free | − | − |
| 5312 | 57 | Pacific Islander,
NOS | 2A | + | + | − | Alive | Never
disease-free | − | 51451 T->C
(697Thr) Hetero |
|
|
|
|
|
|
|
|
|
|
| 51496 A->G
(712Pro) Hetero |
| 5317 | 70 | Caucasian | 1A | + | + | − | Alive | Never
disease-free | − | 51451 T->C
(697Thr) Hetero |
| 5320 | 74 |
African-American | 1A | + | + | − | Alive | Never
disease-free | 90775 A->G | 51451 T->C
(697Thr) Hetero |
|
|
|
|
|
|
|
|
|
|
(1047His->Leu) | 51496 A->G
(712Pro) Hetero |
| 5331 | 62 | Caucasian | 2A | + | + | Borderline | Alive | Never
disease-free | − | − |
| 5333 | 67 | Caucasian | 0 | + | + | − | Alive | Never
disease-free | 90775 A->G | − |
|
|
|
|
|
|
|
|
|
|
(1047His->Arg) |
|
| 5357 | 50 | Caucasian | 1A | + | + | − | Alive | No recurrence | 90775 A->G | 51451 T->C
(697Thr) Hetero |
|
|
|
|
|
|
|
|
|
|
(1047His->Arg) | 51496 A->G
(712Pro) Hetero |
| 5380 | 65 | Caucasian | 2A | + | + | − | Alive | Never
disease-free | − | − |
| 5385 | 46 | Caucasian | 2A | − | − | + | Alive | Never
disease-free | − | − |
| 5387 | 71 | Caucasian | 1A | + | − | − | Alive | Never
disease-free | − | 54929 G->A
(919Ala) Hetero |
| 5391 | 67 | Caucasian | 4 | + | − | − | Alive | Never
disease-free | − | 51496 A->G
(712Pro) Hetero |
| 5394 | 42 |
African-American | 1A | + | + | Borderline | Alive | Never
disease-free | − | 51451 T->C
(697Thr) Hetero |
|
|
|
|
|
|
|
|
|
|
| 51496 A->G
(712Pro) Hetero |
| 5395 | 44 |
African-American | 2B | − | − | − | Alive | Never
disease-free | − | 51451 T->C
(697Thr) Hetero |
|
|
|
|
|
|
|
|
|
|
| 51496 A->G
(712Pro) Hetero |
| 5402 | 77 |
African-American | 1 | + | + | − | Alive | Never
disease-free | − | − |
| 5443 | 50 |
African-American | 3A | − | − | − | Alive | Never
disease-free | − | − |
Preparation of polymerase chain
reaction (PCR) amplicons for sequencing
The exons of the FGD1 gene and the ‘hotspots’ for
phosphatidylinositol-4,5-bisphosphate 3-kinase, catalytic subunit α
(PIK3CA) and AKT1 were identified using the Ensembl website
(17). The PCR primers for the exons
of the FGD1 gene and hotspots of PIK3CA and v-Akt murine thymoma
viral oncogene homolog 1 (AKT1) genes were designed with the
PRIMER3 website with a restriction of 300 bps in size (18). P1 and A tag sequences were added to
the forward and reverse primers (fusion primer tags) for these
genes as described in the Ion Torrent Library Preparation (Fusion
Method; Thermo Fisher Scientific, Inc.) (19). PCR of the exons was performed using
KAPA HiFi PCR kits (Kapa Biosystems, Wilmington, MA, USA), with a
reaction system consisting of 2.5 µl of 2 µM forward primer, 2.5 µl
of 2 µM reverse primer, 2.4 µl of PCR water (Integrated DNA
Technologies, Inc., Coralville IA, USA), and 1.5 µl of DNA template
(>10 ng). Thermal cycling was performed using a 96CFX Thermal
Cycler (Bio-Rad Laboratories, Inc., Hercules CA, USA) with the
following touchdown protocol: 1 cycle of 95°C for 2 min; 3 cycles
of 94°C for 10 sec, 64°C for 10 sec and 70°C for 30 sec; 3 cycles
of 94°C for 10 sec, 61°C for 10 sec and 70°C for 30 sec; 3 cycles
of 94°C for 10 sec, 58°C for 10 sec and 70°C for 30 sec; and 50
cycles of 94°C for 10 sec, 57°C for 10 sec and 70°C for 30 sec. The
PCR products or amplicons were purified using SPRI Ampure beads
(Beckman Coulter Inc., Beverly, MA, USA) following the
manufacturers protocol. The purified amplicons were analyzed by
electrophoresis on a 3% agarose gel and photographed using a UVP
and BioDoc-it Imaging System (UVP LLC, Upland, CA, USA).
Sequencing with Ion Torrent Personal
Genome Machine (PGM) system
The purified amplicons (FGD1, PIK3CA and AKT exons),
which consisted of 24 amplicons from each sample, were pooled into
two categories. These two amplicon pools were identified as tumor
or normal amplicons. The pools of amplicons were sent to the
Medical University of South Carolina Proteogenomics Core
(Charleston, SC, USA) for evaluation. The amplicons were measured
for size using the Agilent Bioanalyzer (Agilent Technologies, Santa
Clara, CA, USA). Emulsion PCR of amplicons was performed on the Ion
OneTouch 2 instrument, and the amplicons were cleaned and enriched
on the OneTouch Enrichment System instrument (Thermo Fisher
Scientific, Inc.). The Qubit Fluorometer (Thermo Fisher Scientific,
Inc.) was used to determine the amount of amplicons recovered from
the enrichment. Sequencing of the tumor and normal samples was
performed using two 318 chips on the Ion Torrent PGM instrument
(Thermo Fisher Scientific, Inc.).
Variant detection analysis
Data from the Ion Torrent PGM system was analyzed
with the CLC Genomics Workbench 6 software (Qiagen CLC Bio, Aarhus,
Denmark). The sequencing reads were aligned to the FGD1, PIK3CA and
AKT1 reference templates from the National Center for Biotechnology
Information (NCBI) (hg19 Build 37) with the Next-Generation
Sequencing Tool on the CLC software. Polymorphisms and somatic
mutations on the three genes were detected through Probabilistic
Variant Detection on the CLC software. Variants that were observed
in the tumor and normal reads were confirmed with the NCBI or the
1000 Genomes SNP database websites (20,21).
Mutations detected in the tumor sequencing reads were compared to
somatic mutations on the cBioPortal and Sanger UK-COSMIC somatic
mutation database websites (9,10).
Sanger sequencing of variants
The variants detected in the tumor and normal
sequencing reads were identified in the exons of FGD1 and PIK3CA
genes. The exons containing variants were amplified using 6.25 µl
of BioRad EvaGreen Supermix (Bio-Rad Laboratories, Inc.) 2.5 µl of
2 µM of forward primer, 2.5 µl of 2 µM reverse primer, 2.4 µl of
PCR water and 1.5 µl of DNA template (>5 ng). Thermal cycling
was performed using the 96CFX Thermal Cycler with the following
touchdown protocol: 1 cycle of 98°C for 2 min; 3 cycles of 98°C for
10 sec, 64°C for 10 sec and 70°C for 30 sec; 3 cycles of 98°C for
10 sec, 61°C for 10 sec and 70°C for 30 sec; 3 cycles of 94°C for
10 sec, 58°C for 10 sec and 70°C for 30 sec; and 50 cycles of 94°C
for 10 sec, 57°C for 10 sec and 70°C for 30 sec. The amplicons were
sent to the Beckman Coulter Sequencing Facility (Beverly, MA, USA)
for Sanger sequencing, and were sequenced using the P1 and A tags
from the fusion primers. Sequences were analyzed with the CLC
Genomics Workbench.
Statistical analysis
Statistical analysis of genotype frequency,
ethnicity and age at diagnosis were performed using the Student's
t-test in the Social Science Statistics calculator
(http://www.socscistatistics.com/tests/studentttest/Default2.aspx),
and Fisher's exact test in GraphPad Prism software (http://www.graphpad.com/).
Results
Somatic variants
The purpose of this sequencing study was to identify
somatic mutations and novel polymorphisms of the FGD1 gene in the
tissues samples of 46 breast cancer patients. The exons or coding
region sequences of the FGD1 gene were sequenced in the tumor and
matched normal tissue samples. The 21 primer pairs with Ion Torrent
Tags (A and P1) targeted the coding regions of the FGD1 gene, which
consists of 18 exons. As positive controls for somatic mutations
for next-generation sequencing, exons 10 and 21 of the PIK3CA gene
and exon 4 of the AKT1 gene were sequenced in the tumor DNA of the
tissue samples. These three exons were selected for the study since
positions E542K, E545K, H1047R and H1047L in PIK3CA, and E17K in
AKT1 are frequently mutated in tumor DNA sequences of breast cancer
patents (9,10). These 24 PCR amplicons were quantitated
and equal amounts were pooled together into two groups, tumor and
normal, for the next-generation sequencing. The tumor and normal
groups of amplicons were bi-directionally sequenced with two runs
on the Ion Torrent Sequencer, which produced 3×106
individual sequencing reads with an average read length of 250
bps.
For somatic mutations in PIK3CA and AKT1 for the 46
tumor samples, there were 3 samples with an E545K mutation, 9
samples with a H1047R mutation, and 1 sample with a H1047L
mutation. None of the samples had somatic mutations in E542K of the
PIK3CA gene or E17K in the AKT1 gene (Table I). These frequencies of PIK3CA
mutations were similar to the frequencies that were observed in the
cBioPortal and Sanger UK-COSMIC somatic mutation databases
(9,10).
Germline variants
The Ion Torrent sequencing of the FGD1 gene resulted
with an average depth 170,000 sequencing reads for each of the
exons and the reads covered of all 18 exons in the gene. The
sequencing of the tumor DNA detected no somatic mutations in any of
the 46 tumor DNA samples; however, 3 synonymous variants and 1
missense variant were observed in the tumor and corresponding
normal sequencing pools compared with the Ensembl reference
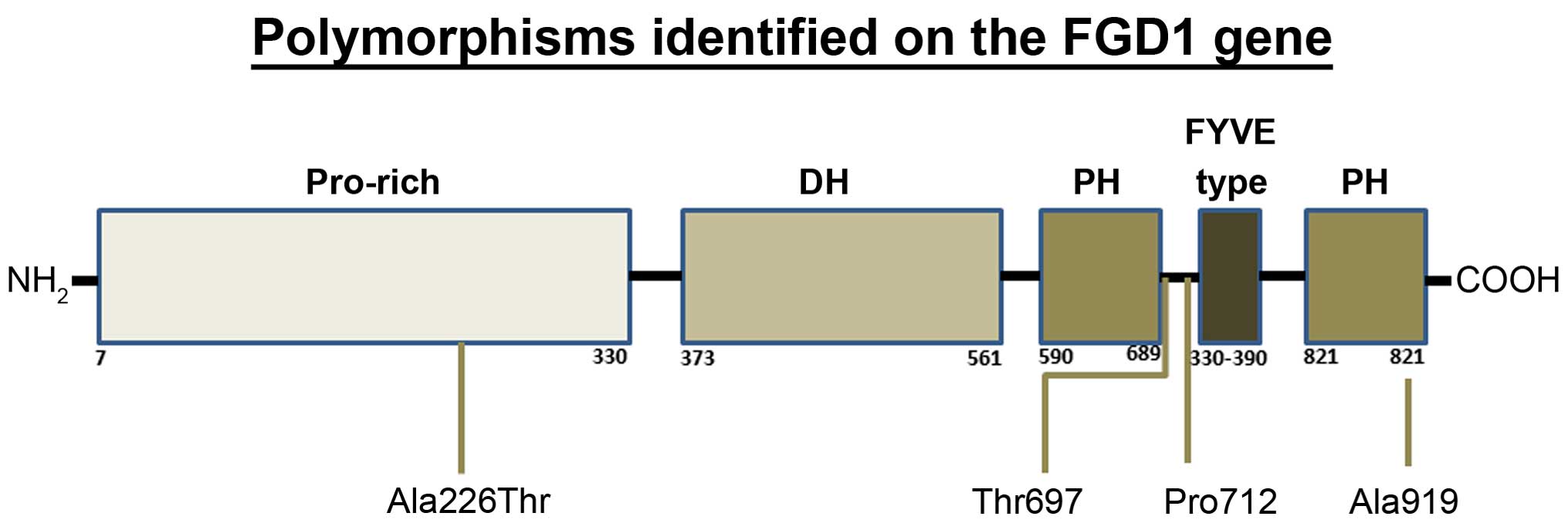
sequence of the FGD1 gene (ENSG00000102302; Fig. 1) (17).
Two of the variants were detected in the tumor and normal
sequencing pools at a frequency of <1%. The other two variants
were identified in exon 14 at frequency of >20% in both pools.
All of the polymorphisms were confirmed and identified in each of
the patient samples using Sanger sequencing (Table I). One of the low-frequency variants
from the sequencing study was a G to A change at position 54446238
in exon 18. This synonymous change (Ala919; rs61734180) was
identified in 1 patient who was heterozygous for the polymorphism.
The second low-frequency variant was observed in 1 patient who was
also heterozygous for the change. The variant was a missense change
(Ala226Thr; rs138723423) of an A to a G at position 54470441 in
exon 4. This missense polymorphism was identified in a 40-year-old
woman whose tumor was ER/PR positive and it was later discovered
that she had recurrence.
The silent polymorphism, Thr697 (rs12011120), is
found in exon 14 with the change of an A to G at position 54449671.
In this sequencing project, 18 breast cancer patients possessed the
Thr697 variant: 10 African-American patients, 7 Caucasian patients
and 1 Pacific Islander patient, with an average age at diagnosis of
55 years. Notably, the frequency of this polymorphism was higher
among African-American patients (0.76) than in the Caucasian
patients (0.22). This difference was statistically significant
(two-tailed P=0.0018). Although the frequencies between the two
ethnic groups differed, the average ages at diagnosis of the
African-American and Caucasian patients with the polymorphism were
observed to be similar (54 vs. 56 years, respectively). However,
the average age at diagnosis of patients without the polymorphism
was 10 years older (65 years). A Student's t-test was
performed to compare the age at diagnosis of patients with the
Thr697 polymorphism vs. patients without the polymorphism, and was
found to be statistically significant (P=0.0175).
The second polymorphism, Pro712 (rs1126744), is also
found in exon 14 with a T to C change at position 54449716 in the
FGD1 gene. Pro712 was detected in 15 breast cancer patients (8
African-American patients, 6 Caucasian patients and 1 Pacific
Islander patient) with an average age of 58 years. Although the
polymorphism was found in a large number of patients, the average
age at diagnosis was 58 years, and the age of the Pro712 group was
not significantly different when compared with the patients without
the polymorphisms (P>0.05).
Results of the 1000 Genome Project have determined
that these two synonymous polymorphisms, Pro712 and Thr697, have a
strong linkage of disequilibrium in the general population, with an
r-value of 0.89 (21). In the
current study, these two polymorphisms were observed together in 13
out of 46 breast cancer patients (28%); this percentage of the two
variant haplotypes agrees with the general population percentage of
~31% (22). The average age at
diagnosis of the two polymorphisms together in the breast cancer
patients was not significantly different when compared with the
patients without the two polymorphisms, based on a Student's
t-test (P>0.05).
Discussion
For several years, FGD1 has been postulated to play
a role in diseases involving protein-damaging polymorphisms, as in
Aarskog-Scott syndrome, and in the somatic alterations observed in
several late-stage/invasive cancer projects (8–10,12). The present study sequenced the FGD1
gene for possible somatic and germline variants that may be
associated with tumor development in breast cancer patients.
Although somatic alterations have been observed in other studies in
late-stage breast cancer (9,10), no somatic FGD1 mutations were
identified in the present study. A possible explanation for this
difference may be that a majority (90%) of the cancer patients in
this project were diagnosed with an early stage of breast cancer,
with no distant tumor growth or node involvement (stage 1–2). As
Fgd1 is associated with late-stage tumor development (8–10), the
somatic alterations in the FGD1 gene would not have occurred until
the tumor was ready to detach from the primary tumor and migrate
through the tissue.
The sequencing of the tumor and normal DNA in 46
breast cancer patients revealed 4 polymorphisms, with 3 silent
(Ala919, Thr697 and Pro712) and 1 missense (Ala226Thr) variants.
The Ala919 variant was identified in a 70-year-old female patient
with an early-stage cancer. This polymorphism is located in the
Pleckstrin homology 2 domain and does not appear to play a
significant role in tumor development. The two silent variants,
Thr697 and Pro712, were observed together in >20% of the breast
cancer patients; however, they appeared to differ with regard to
the age at diagnosis of the patients, with Thr697 being associated
with breast cancer in patients who were at least 10 years younger
compared with the control (without the polymorphisms), while the
age at diagnosis of patients with Pro712 was closer (<10 years)
to the control patients. The Thr697 polymorphism also was detected
in a higher percentage of African-American patients. Taken
together, these results suggest that this polymorphism may play a
role in setting the background for subsequent changes that
influence early-stage tumor development in breast cancer, and, in
particular, may influence early onset in African-American breast
cancer patients.
The only missense polymorphism, Ala226Thr, was
identified in a 40-year-old female patient with tumor recurrence,
which suggests that the variant may be associated with an
aggressive form of breast cancer.
In conclusion, although no somatic mutations of FGD1
were observed in the studied population of breast cancer patients,
the present results support the hypothesis that the somatic
alterations observed in breast cancer may be a late-stage molecular
event in tumor development. Three of the FGD1 polymorphisms
(Ala226Thr, Thr697 and Pro712) that were identified in the breast
cancer patients may play role in early onset, or may ultimately
influence development of a more invasive form of breast cancer.
References
|
1
|
Siegel R, Naishadham D and Jemal A: Cancer
Statistics, 2012. CA Cancer J Clin. 62:10–29. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
American Cancer Society: Breast Cancer -
Facts & Figures 2011–2012. American Cancer Society, Inc.
Atlanta, GA: 2011.http://www.cancer.org/acs/groups/content/@epidemiologysurveilance/documents/document/acspc-030975.pdf
|
|
3
|
Pasteris NG, Cadle A, Logie LJ, Porteous
ME, Schwartz CE, Stevenson RE, Glover TW, Wilroy RS and Gorski JL:
Isolation and characterization of the faciogenital dysplasia
(Aarskog-Scott syndrome) gene: A putative Rho/Rac guanine
nucleotide exchange factor. Cell. 79:669–678. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Yamazaki D, Kurisu S and Takenawa T:
Regulation of cancer cell motility through actin reorganization.
Cancer Sci. 96:379–386. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Nobes CD and Hall A: Rho, rac, and cdc42
GTPases regulate the assembly of multimolecular focal complexes
associated with actin stress fibers, lamellipodia, and filopodia.
Cell. 81:53–62. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Mizuarai S, Kazunori Y and Hidehito K:
Mutant p53 induces the GEF-H1 oncogene, a guanine nucleotide
exchange factor-H1 for RhoA, resulting in accelerated cell
proliferation in tumor cells. Cancer Res. 66:6319–6326. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Kourlas PJ, Strout MP, Becknell B,
Veronese ML, Croce CM, Theil KS, Krahe R, Ruutu T, Knuutila S,
Bloomfield CD and Caligiuri MA: Identification of a gene at 11q23
encoding a guanine nucleotide exchange factor: Evidence for its
fusion with MLL in acute myeloid leukemia. Proc Natl Acad Sci USA.
97:2145–2150. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Ayala I, Giacchetti G, Caldieri G,
Attanasio F, Mariggiò S, Tetè S, Polishchuk R, Castronovo V and
Buccione R: Faciogenital dysplasia protein Fgd1 regulates
invadopodia biogenesis and extracellular matrix degradation and is
up-regulated in prostate and breast cancer. Cancer Res. 69:747–752.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Gao J, Aksoy BA, Dogrusoz U, Dresdner G,
Gross B, Sumer SO, Sun Y, Jacobsen A, Sinha R, Larsson E, et al:
Integrative analysis of complex cancer genomics and clinical
profiles using the cBioPortal. Sci Signal. 6:pl12013. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Forbes SA, Beare D, Gunasekaran P, Leung
K, Bindal N, Boutselakis H, Ding M, Bamford S, Cole C, Ward S, et
al: COSMIC: Exploring the world's knowledge of somatic mutations in
human cancer. Nucleic Acids Res. 43(Database issue): D805–D811.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Orrico A, Galli L, Cavaliere ML, Garavelli
L, Fryns JP, Crushell E, Rinaldi MM, Medeira A and Sorrentino V:
Phenotypic and molecular characterization of the Aarskog-Scott
syndrome: A survey of the clinical variability in light of FGD1
mutation analysis in 46 patients. Eur J Hum Genet. 12:16–23. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Orrico A, Galli L, Faivre L, Clayton-Smith
J, Azzarello-Burri SM, Hertz JM, Jacquemont S, Taurisano R, Carrera
Arroyo I, Tarantino E, et al: Aarskog-Scott syndrome: Clinical
update and report of nine novel mutations of the FGD1 gene. Am J
Med Genet A. 152A:313–318. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Aarskog D: A familial syndrome of short
stature associated with facial dysplasia and genital anomalies. J
Pediatr. 77:856–861. 1970. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Schwartz CE, Gillessen-Kaesbach G, May M,
Cappa M, Gorski J, Steindl K and Neri G: Two novel mutations
confirm FGD1 is responsible for the Aarskog syndrome. Eur J Hum
Genet. 8:869–874. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Murugan AK, Yang C and Xing M: Mutational
analysis of the GNA11, MMP27, FGD1, TRRAP and GRM3 genes in thyroid
cancer. Oncol Lett. 6:437–441. 2013.PubMed/NCBI
|
|
16
|
Sambrook J and Russell DW: Purification of
nucleic acids by extraction with phenol: Chloroform. CSH Protoc.
2006:pii2006.
|
|
17
|
Cunningham F, Amode MR, Barrell D, Beal K,
Billis K, Brent S, Carvalho-Silva D, Clapham P, Coates G,
Fitzgerald S, et al: Ensembl 2015. Nucleic Acids Res. 43(Database
issue): D662–D669. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Untergrasser A, Cutcutache I, Koressaar T,
Ye J, Faircloth BC, Remm M and Rozen SG: Primer3 - new capabilities
and interfaces. Nucleic Acids Res. 40:e1152012. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Beadling C, Neff TL, Heinrich MC, Rhodes
K, Thornton M, Leamon J, Andersen M and Corless CL: Combining
highly multiplexed PCR with semiconductor-based sequencing for
rapid cancer genotyping. J Mol Diagn. 15:171–176. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Sherry ST, Ward MH, Kholodov M, Baker J,
Phan L, Smigielski EM and Sirotkin K: dbSNP: The NCBI database of
genetic variation. Nucleic Acids Res. 29:308–311. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
1,000 Genomes Project Consortium. Abecasis
GR, Auton A, Brooks LD, DePristo MA, Durbin RM, Handsaker RE, Kang
HM, Marth GT and McVean GA: An integrated map of genetic variation
from 1,092 human genomes. Nature. 491:56–65. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Fu W, O'Connor TD, Jun G, Kang HM,
Abecasis G, Leal SM, Gabriel S, Rieder MJ, Altshuler D, Shendure J,
et al: Analysis of 6,515 exomes reveals the recent origin of most
human protein-coding variants. Nature. 493:216–220. 2013.
View Article : Google Scholar : PubMed/NCBI
|