Introduction
Tongue squamous cell carcinoma (TSCC) is the leading
type of cancer of the oral cavity and is characterized by a high
rate of tumor cell proliferation and metastasis (1). Approximately one-half of patients are
already in advanced stages of disease at the time of diagnosis
(2,3).
Elucidation of the molecular mechanism underlying the development
and progression of TSCC may aid improved understanding of the
pathogenesis of TSCC and improve the diagnosis, treatment and
prevention of the disease.
MicroRNAs (miRNAs) are a class of endogenously
expressed 22-nucleotide non-coding RNAs that are able to regulate
gene expression by binding the 3′-untranslated regions (3′-UTRs) of
their target messenger RNAs, resulting in translational repression
or mRNA degradation (4). It is
hypothesized that approximately one-third of human genes can be
regulated by miRNAs (4). Due to the
various reported roles of miRNAs in the regulation of gene
expression, miRNAs are considered to play key roles in numerous
biological processes, including cell growth, apoptosis, metabolism
and transformation (5–7). It has been reported that chromosome 7q32
is largely involved in the pathogenesis of malignancy (8). Previously, 8 miRNAs, consisting of
miR-593, miR-129-1, miR-335, miR-182, miR-96, miR-183, miR-29a and
miR-29b-1, were identified on this genomic locus, with certain
miRNAs being recognized either as oncogenes or tumor suppressor
genes (9–11). In the present study, 20 tumor and
adjacent non-cancerous control tissue samples were collected from
20 patients with histologically confirmed TSCC. Confirmatory
reverse transcription-quantitative polymerase chain reaction
(RT-qPCR) showed that miR-335 and miR-182 were significantly
downregulated in the tumor tissue samples compared with the
controls. Notably, survivin was identified as a potential shared
target of miR-335 and miR-182 by using online prediction tools
(mirdb.org/miRDB/) and in silico analysis.
Survivin is a member of the inhibitor of apoptosis protein (IAP)
family and is well-known for its exclusive expression in fetal
tissue and a wide spectrum of human cancers, but not in
non-proliferating adult tissue (12).
Survivin has also been reported to be a regulator of cellular
proliferation and metastasis in cancer (13), and an increasing number of studies
indicated that survivin was involved in various types of cancer
(14–16), particularly TSCC (2,3).
Based on the aforementioned evidence and the studies
reporting that significant underexpression of miR-335 or miR-182
was noted in various types of cancers, including glioma (17), breast cancer (18) and gastric cancer (19), the present study hypothesized that
miR-335 and miR-182 may function as regulators of the behavior of
TSCC cells by modulating survivin. To test this hypothesis, the
effect of miR-335 and miR-182 on the proliferation and invasion of
TSCC cells was evaluated. The expression patterns of miR-335,
miR-182 and survivin were also examined in the cancer tissues and
compared with adjacent non-cancerous tissues.
Materials and methods
Tissue samples
Paired primary TSCC samples and adjacent
histological normal tissues were collected from 20 TSCC patients,
consisting of 14 male and 6 female patients (age, 62.36±5.74
years), at the Department of Oral and Maxillofacial Surgery,
Guanghua School of Stomatology, Sun Yat-sen University (Guangzhou,
China), between September 2006 and December 2011. Tumor tissues and
adjacent normal tissues that were at least 1.5 cm distal to the
tumor margins were snap-frozen in liquid nitrogen and then stored
at −80°C until use. All samples were evaluated by two pathologists
according to the World Health Organization guidelines and
pathological tumor-node-metastasis Union for International Cancer
Control pathological staging criteria (20), and the samples were examined for the
expression level of survivin, miR-335 and miR-182. Patients that
received chemotherapy or radiotherapy prior to surgery were
excluded from the present study. Informed consent was obtained from
all patients, and the study was approved by the Human Research
Ethics Committee of Sun Yat-Sen University.
Cell culture
The human TSCC SCC-15 cell line was purchased from
American Type Culture Collection (ATCC, Manassas, VA, USA). The
SCC-15 cells were cultured in Dulbecco's modified Eagle's medium
(DMEM; Gibco; Thermo Fisher Scientific, Inc., Waltham, MA, USA)
supplemented with 10% heat-inactivated fetal bovine serum (FBS;
Gibco; Thermo Fisher Scientific, Inc.), 100 U/ml penicillin (Gibco;
Thermo Fisher Scientific, Inc.) and 100 µg/ml streptomycin (Gibco;
Thermo Fisher Scientific, Inc.) at 37°C in a 5% CO2
atmosphere.
Methyl thiazolyl tetrazolium (MTT)
assay
The SCC-15 cells were transfected with miR-335
mimics, miR-182 mimics or the negative control (scramble control
sequence) and incubated for 48 h at room temperature. Subsequently,
30 µl 0.5% MTT (Sigma-Aldrich, St. Louis, MO, USA) solution was
added to each well and the wells were incubated for another 4 h.
The medium was then replaced with 150 µl dimethyl sulfoxide (DMSO;
Sigma-Aldrich) in the microplate, and then agitated on a rotary
platform for 10 min. The survival rate was determined by measuring
the optical density (OD) values of the mimic-transfected cells
relative to the OD of the control at a 492-nm wavelength using a
Jenway spectrophotometer 7300 (67 series VU Visible Scanning
spectrophotometer; Bibby Scientific Ltd., Stone, Staffordshire,
UK).
Total RNA isolation and RT-qPCR
Total RNA was obtained using TRIzol One-Step RNA
Isolation kit (Invitrogen; Thermo Fisher Scientific, Inc.).
Complementary DNA specific for miR-335, miR-182, survivin and
glyceraldehyde 3-phosphate dehydrogenase (GAPDH), which acted as an
internal control, was synthesized from total RNA using
gene-specific primers with the TaqMan MicroRNA assay, according to
the manufacturer's protocol (Applied Biosystems; Thermo Fisher
Scientific, Inc.). Relative quantification of target miRNA
expression was measured using the 2−ΔΔCq method
(21). Each sample was examined in
triplicate and the raw data were expressed as the relative quantity
of target miRNA, normalized with respect to GAPDH.
Luciferase reporter assay
The SCC-15 cells were cultured in DMEM supplemented
with 10% FBS at 37°C in an atmosphere of 5% CO2 to
70–80% confluency. Using Lipofectamine 2000 (Invitrogen; Thermo
Fisher Scientific, Inc.), the cells were then transfected with the
WT-Sur-UTR DNA plasmid, which consisted of a firefly luciferase
reporter vector containing the wild-type survivin 3′-UTR, or the
mutant Sur-UTR DNA plasmids (Mut1 and Mut2), which consisted of a
firefly luciferase reporter vector containing the mutant survivin
3′-UTR, (Fig. 1) and the control
pRL-TK vector, which contained Renilla luciferase (Promega
Corporation, Madison, WI, USA). The firefly and Renilla
luciferase activities were measured consecutively using Dual Glo
Luciferase assays (Promega Corporation) 48 h subsequent to
transfection.
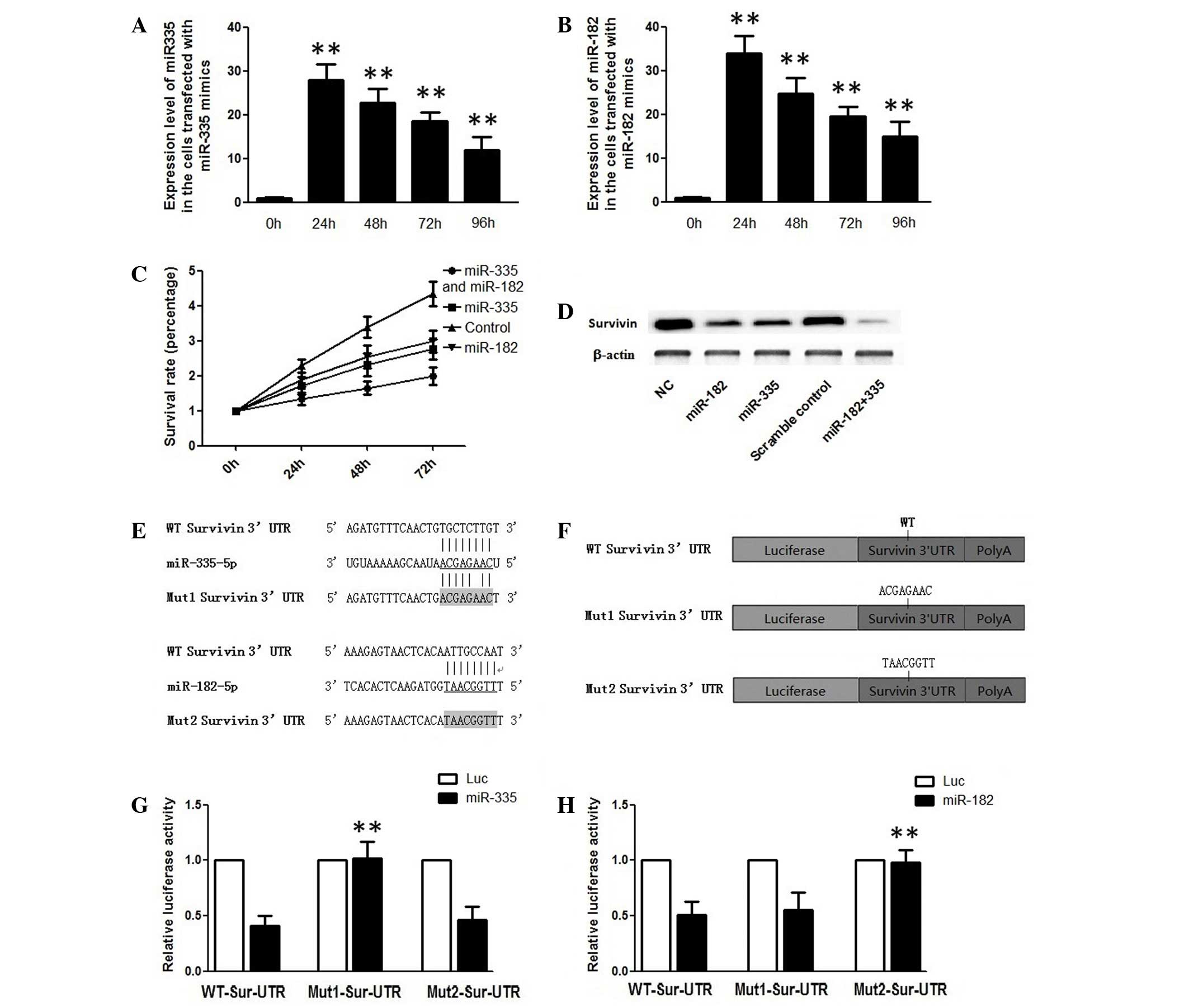 | Figure 1.(A) Expression levels of miR-335 24,
48, 72 and 96 h subsequent to transfection with miR-335 mimics. (B)
Expression levels of miR-182 24, 48, 72 and 96 subsequent to
transfection with miR-182 mimics. (C) Survival rate analysis of
TSCC cells transfected with miR-335 and/or miR-182 mimics, as
determined using a methyl thiazolyl tetrazolium assay 24, 48 and 72
h subsequent to transfection. (D) Suppression of the protein
expression levels of Sur by miR-335 and/or miR182 mimics in TSCC
cells. (E) Schematic representation of miR-335 or miR-182 and the
binding sequences of these miRs in the WT or Mut 3′-UTR of Sur
mRNA. (F) Schematic representation of the Mut 3′-UTR of Sur mRNA.
(G) Analysis of Luc activity in TSCC cells overexpressing miR-335
48 h subsequent to cotransfection with the control Renilla
Luc expression construct pRL-TK and firefly Luc reporter plasmids
containing either WT or Mut 3′-UTR of Sur. (H) Analysis of Luc
activity in miR-182-overexpressing TSCC cells 48 h subsequent to
cotransfection with the control Renilla Luc expression
construct pRL-TK and firefly Luc reporter plasmids containing
either WT or Mut 3′-UTR of Sur. **P<0.01 vs. control. miR,
microRNA; TSCC, tongue squamous cell carcinoma; UTR, untranslated
region; mRNA, messenger RNA; Mut, mutant; WT, wild-type; NC,
negative control; Luc, luciferase; Sur, survivin. |
Western blotting
Lysates were resolved on 12% sodium dodecyl
sulfate-polyacrylamide gels and the bands were transferred onto
polyvinylidene fluoride membranes (EMD Millipore, Billerica, MA,
USA). The membranes were blocked with 5% non-fat milk at room
temperature for 1 h followed by incubation with a rabbit anti-human
survivin antibody (catalog no., ab2050; Abcam, Cambridge, UK) under
room temperature for 2 h and subsequently incubated for 2 h with
horseradish peroxidase-conjugated anti-rabbit secondary antibody at
room temperature for 1 h. Chemical fluorescence was detected using
an Enhanced Chemiluminescence Western Blotting Detection Reagent
kit (GE Healthcare, Little Chalfont, Buckinghamshire, UK),
according to the manufacturer's protocol. The target bands
underwent densitometric analysis with Quantity One® 1-D
analysis software version 170-9600 (Bio-Rad Laboratories, Inc.,
Hercules, CA, USA) and were normalized against β-actin. A total of
3 independent experiments were performed.
Cell cycle analysis and apoptosis
analysis
The SCC-15 cells were digested with
trypsin-ethylenediaminetetraacetic acid solution (Beyotime
Institute of Biotechnology, Haimen, China), washed with
phosphate-buffered saline (PBS; Invitrogen; Thermo Fisher
Scientific, Inc.), and fixed in 70% ethanol 48 h subsequent to
transfection. Cell cycle analyses were conducted using propidium
iodide staining. FACSCalibur flow cytometer (BD Biosciences, San
Jose, CA, USA) was used to determine G0/G1, S and G2/M fractions.
For the apoptosis analysis, an Annexin V-FLUOS Staining kit
purchased from Roche (Mannheim, Germany) was used 48 h subsequent
to transfection, according to the manufacturer's protocol. The
results were analyzed using CellQuest Pro software (BD
Biosciences).
Cell invasion assay
For the cell invasion assay, 8-µm pore polycarbonate
membrane filters (BD Biosciences) pre-coated with 50 µl Matrigel
(1.25 mg/ml; BD Biosciences) were used. The cells transfected with
each miR were collected and seeded at a density of 1×105
cells/well with 1% FBS-supplemented DMEM in the cell culture
chambers, with the bottom chambers filled with 0.6 ml medium. The
chamber was incubated at 37°C for 48 h prior to the removal of the
Matrigel. The invaded cells were fixed, stained and calculated. The
invasiveness of cells was evaluated by the following equation:
Invasion index (%) = invaded cell number / total cell number ×
100.
Statistical analysis
The data were expressed as the mean ± standard
deviation. The differences were assessed for significance using
Student's t-test, χ2 test or one-way analysis of
variance followed by Fisher's least significant difference test, as
appropriate. The statistical analysis was conducted by SPSS 17.0
software (SPSS, Inc., Chicago, IL, USA). P<0.05 was considered
to indicate a statistically significant difference.
Results
Overexpression of miR-335 and miR-182
suppresses the growth of TSCC cells
To evaluate the effect of miR-335 and miR-182 on the
proliferation and invasion of TSCC cells, the SCC-15 cells were
transfected with miR-335 and miR-182 mimics. As shown in Fig. 1A and B, the levels of miR-335 and
miR-182 were increased >30-fold at 24 h subsequent to
transfection compared with the control. The MTT assay results
showed that exogenous overexpression of miR-335 and miR-182
individually and synergistically suppressed the proliferation of
SCC-15 cells compared with the control (Fig. 1C). In addition, the migratory ability
of the cells with or without overexpression of miR-335 and miR-182
was examined, and the introduction of either miR-335 or miR-182 was
not found to affect the migratory ability of the cells (data not
shown). The results indicated that miR-335 and miR-182 each played
inhibitory roles in the regulation of cell proliferation.
Exogenous overexpression of miR-335
and miR-182 led to G1 phase cell cycle arrest in TSCC cells
To investigate the mechanism underlying the effect
of miR-335 and miR-182 on cell proliferation, cell cycle
distribution analysis was performed in TSCC cells using flow
cytometry. The data revealed that overexpression of miR-335 and/or
miR-182 arrested the growth of TSCC cells in the G1 phase, with a
decrease in the proportion of cells in the G2/M and S phases
(Fig. 2). The result showed that
compared with the control group, the percentage of TSCC cells in
the G1 phase was increased between 61.35 and 75.14% in cells
transfected with miR-335 alone (P<0.01), 61.35 and 72.11% in
cells transfected with miR-182 alone (P<0.01) and 61.35 and
88.32% in cells transfected with miR-335 combined with miR-182
(P<0.01). The percentage of cells in the S phase decreased
between 29.26 and 16.24% in cells transfected with miR-335 alone
(P<0.01), 29.26 and 19.50% in cells transfected with miR-182
alone (P<0.01) and 29.26 and 4.23% in cells transfected with
miR-335 combined with miR-182 (P<0.01). The population of cells
in the G2 phase decreased between 9.39 and 8.63% in cells
transfected with miR-335 alone (P<0.01), between 9.39 and 8.39%
in cells transfected with miR-182 alone (P<0.01) and between
9.39 and 7.45% in cells transfected with miR-335 and miR-182
(P<0.01)(Fig. 2A-D). These data
demonstrated that overexpression of miR-335 and miR-182 suppressed
the proliferation of TSCC cells by arresting cell growth in the G1
phase.
Survivin is a shared direct target of
miR-335 and miR-182 in TSCC cells
The miRNA databases miRanda (22) and TargetScan (23) were searched for potential target genes
of miR-335 and miR-182, in order to identify the mediators of the
inhibitory effect of miR-335 and miR-182 on cell proliferation. By
screening the candidate genes that were identified based on their
functions combined with the information about the miRNA obtained
from the DIANA lab-based microRNA pathway analysis tool
(diana.imis.athena-innovation.gr/DianaTools/index.php), all
predicted genes were categorized functionally to identify candidate
genes that are functionally associated with the regulation of the
cell cycle. Finally, the present study focused on survivin, a key
cell cycle regulator and a putative shared target of miR-335 and
miR-182 (Fig. 1E). Therefore, it was
hypothesized that miR-335 and miR-182 may participate in the
regulation of the cell cycle by targeting survivin. To test the
hypothesis, the present study measured and compared the expression
level of survivin in the TSCC cells transfected with miR-335 and
miR-182 mimics using western blot analysis. The effects of these
mimics on survivin knockdown were comparable with the survivin
level in cells co-transfected with the two miRs, which demonstrated
an additional decrease (Fig. 1D).
Furthermore, a luciferase assay was conducted to confirm that
survivin is a direct shared target of miR-335 and miR-182.
Wild-type or mutant survivin 3′-UTR was cloned downstream of a
firefly luciferase gene (Fig. 1F),
and the constructs were transfected into miR-335 or
miR-182-overexpressing cells or the negative control. In addition,
overexpression of miR-335 significantly decreased the relative
luciferase activity of the wild-type and Mut1 survivin 3′-UTR, but
had minimal effects on the relative luciferase activity of the Mut2
survivin 3′-UTR (Fig. 1G).
Overexpression of miR-182 suppressed the luciferase activity of
wild-type and Mut2 (Fig. 1H), but the
luciferase activity of Mut1 was almost unaltered, suggesting that
miR-335 and miR-182 suppress the expression of survivin by binding
directly to the corresponding ‘seed sequence’ in the 3′-UTR of
survivin.
Examination and comparison of
expression patterns of survivin and miR-335/miR-182 in cancerous
tissue and adjacent normal tissue
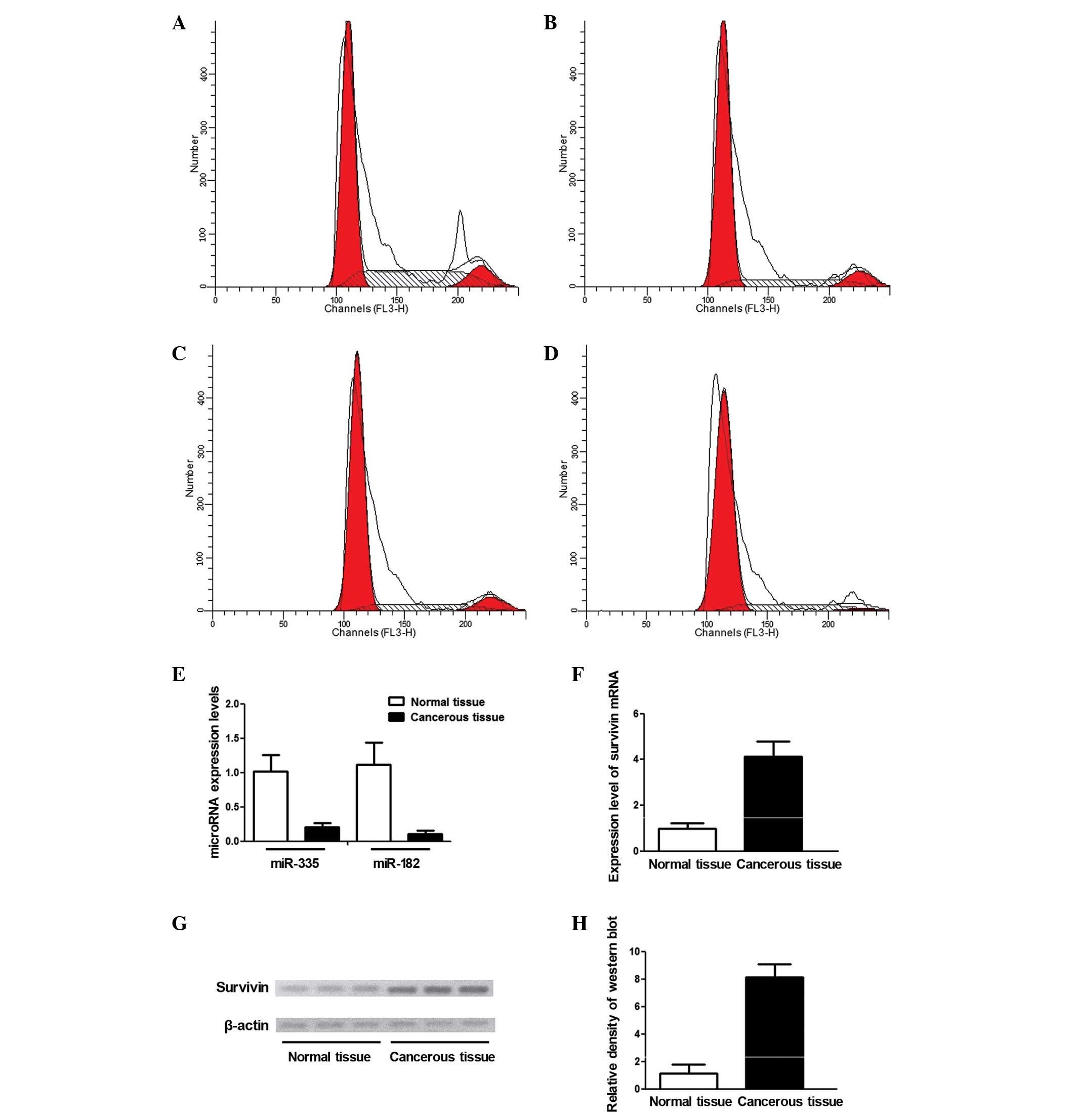
The levels of miR-335, miR-182 and survivin were
measured and compared in cancerous tissues (n=20) and adjacent
normal tissues (n=20). As shown in Fig.
2E, it was found that the relative expression level of miR-335
and miR-182 in cancerous tissue was significantly downregulated to
~20 and 10%, respectively, compared with normal tissues. The
expression level of survivin mRNA was measured by RT-qPCR, and it
was found that the mRNA level of survivin in the cancerous tissue
was significantly increased compared with normal tissues (Fig. 2F). The survivin protein level was also
measured using western blotting, and the relative density of
survivin in the cancerous tissue was almost 5-fold higher compared
with the normal control (Fig. 2G and
H).
Discussion
RNA-based therapeutic strategies, such as siRNA and
miRNA, depict a promising future for cancer treatment by regulating
target genes that are involved in cellular activities, such as cell
growth, apoptosis, metabolism and transformation (5–7).
Therefore, investigation of miRNAs and the targets and roles of
miRNAs in the regulation of cell behavior is crucial to improve the
understanding and facilitate the clinical application of RNA-based
therapeutic strategies.
At present, a panel of miRNAs have been reported as
either tumor suppressors, as they are downregulated, or oncogenes,
as they are upregulated, in multiple human cancers (24). miR-335 and miR-182 have been shown to
be involved in the regulation of cancer cell proliferation and
progression, but the roles of miR-335 and miR-182 in the
development of cancer were inconsistent among various cancer types
(25). miR-335 functions as a tumor
suppressor in osteosarcoma by regulating survivin expression
(25). However, Lu et al
(26) reported that overexpression of
miR-335 promotes cell proliferation and tumor growth of colorectal
carcinoma cells. Furthermore, miR-182 was reported to exhibit
anticancer activity in cervical cancer by inducing cell apoptosis
via the inhibition of DNMT3a expression (27). By contrast, miR-182 promotes cell
growth, migration and invasion of prostate cancer via the
suppression of FOXO1 expression (28). As described, miR-335 is down-regulated
in several types of tumors (29–32),
indicating complicated physiopathological roles of this miRNA
during tumorigenesis. In particular, it has been shown that miR-335
targets retinoblastoma 1 and regulates cell proliferation in a
p53-dependent manner by inducing apoptosis or cell cycle arrest
(33). miR-335 has previously been
reported to suppress metastasis of human breast cancer cells
through targeting of SRY-box 4, without affecting its proliferative
ability (34). Shu et al
reported that miR-335 functioned as a tumor promoter in conferring
tumorigenic features, such as growth and invasion on malignant
astrocytoma, and additional investigation revealed that miR-335
targets disheveled-associated activator of morphogenesis 1 at the
post-transcriptional level (17).
Human miR-182 is mapped to chromosome 7q32 and is
transcribed from the cluster of the miR-183 family that has been
extensively researched in a variety of human cancers (35,36). In
certain studies, miR-182 was reported to act as an oncogene in
several types of human cancers; by contrast, miR-182 demonstrated
tumor suppressive activity in human gastric adenocarcinoma, lung
adenocarcinomas and posterior uveal melanoma (37–39).
miRNAs have been shown to frequently regulate numerous target genes
with possible counteracting functions (40). Thus, the documented conflicting roles
of miR-335 and miR-182 in the development and progression of
malignancies may be attributed to the cell context-dependent
balance among the network of directly regulated genes and the
cancer type-specific activated or inactivated signaling pathways.
Even though the function of miR-335 and miR-182 has been
extensively studied in numerous cancer types, the roles of these
miRNAs in the development of TSCC remain mostly unknown.
In the present study increased miR-335 or miR-182
expression was identified to significantly suppress TSCC cell
proliferation in a TSCC cell line. The results of flow cytometry
analysis demonstrated that the growth of TSCC cells transfected
with the miR-335 or miR-182 mimic was arrested at the G1 phase with
a corresponding reduction in the percentage of cells in the S phase
(Fig. 1). By searching the miRNA
databases miRanda and TargetScan for the candidate genes of the two
miRs and functionally analyzing and screening the results, the
present study identified survivin as a mediator of the inhibitory
effect of the two miRs on the cell behavior.
Three basic roles have been identified for survivin
in the regulation of cell behavior, consisting of control of cell
division, promotion of cell cycle and inhibition of apoptosis
(41). Survivin, a member of the IAP
family, is well-known for its inhibitory role in the regulation of
apoptosis and the cell cycle (7–9). Studies
have demonstrated that elevated expression of survivin promoted the
development and progression of malignancy (42). It is generally agreed that the cell
cycle is under the control of a number of dynamically expressed
genes. Survivin is expressed at high levels during the G2/M phase
and has been shown to play an important role in cell cycle
progression (43). Survivin can
accelerate the S phase by interacting with cyclin-dependent kinase
4, a cell cycle regulator (44).
Accordingly, downregulation of wild-type survivin using RNA
interference evidently caused cell cycle arrested in the G1/S phase
(45). It has been shown that the
expression of survivin is an early event during malignant
transformation and conversion of the oral mucosa in rats exposed to
carcinogens (45). A well-studied G
to C polymorphism (rs9904341) in the promoter region of the
survivin gene may increase the expression of the mRNA and protein
of survivin by disrupting the binding site of the cell
cycle-dependent element/cell cycle gene homology region repressor,
and this polymorphism is associated with an increased risk of
various malignancies (46,47).
The expression patterns of miR-335, miR-182 and
survivin were also examined and compared between the cancerous
tissues and adjacent non-cancerous tissues, showing that miR-335
and miR-182 expression were significantly underexpressed and
survivin expression was significantly upregulated in the TSCC cell
line. The mechanism of underexpression of the two miRs in TSCC was
investigated and the present findings revealed that genetic copy
number loss at the locus of the two miRs was one mechanism by which
miR-335 expression is silenced in metastatic cells. In addition,
epigenetic modification also plays a role in the silencing of the
two miRs.
In conclusion, the present study revealed a negative
regulatory role of miR-335 and miR-182 in the proliferation of TSCC
cells via targeting of survivin. Therefore, miR-335 and miR-182 may
be novel therapeutic tools that may assist in the treatment of
TSCC.
Acknowledgements
The present study was fully sponsored by the Natural
Science Foundation of China (grant no. 81372844).
References
|
1
|
Jemal A, Siegel R, Ward E, Murray T, Xu J
and Thun MJ: Cancer statistics, 2007. CA Cancer J Clin. 57:43–66.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Yuen PW, Lam KY, Chan AC, Wei WI and Lam
LK: Clinicopathological analysis of local spread of carcinoma of
the tongue. Am J Surg. 175:242–244. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Po Wing Yuen A, Lam KY, Lam LK, Ho CM,
Wong A, Chow TL, Yuen WF and Wei WI: Prognostic factors of
clinically stage I and II oral tongue carcinoma-A comparative study
of stage, thickness, shape, growth pattern, invasive front
malignancy grading, Martinez-Gimeno score and pathologic features.
Head Neck. 24:513–520. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Bartel DP: MicroRNAs: Genomics,
biogenesis, mechanism and function. Cell. 116:281–297. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Thai TH, Calado DP, Casola S, Ansel KM,
Xiao C, Xue Y, Murphy A, Frendewey D, Valenzuela D, Kutok JL, et
al: Regulation of the germinal center response by microRNA-155.
Science. 316:604–608. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Brennecke J, Hipfner DR, Stark A, Russell
RB and Cohen SM: Bantam encodes a developmentally regulated
microRNA that controls cell proliferation and regulates the
proapoptotic gene hid in Drosophila. Cell. 113:25–36. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Poy MN, Eliasson L, Krutzfeldt J, Kuwajima
S, Ma X, Macdonald PE, Pfeffer S, Tuschl T, Rajewsky N, Rorsman P
and Stoffel M: A pancreatic islet-specific microRNA regulates
insulin secretion. Nature. 432:226–230. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Holland H, Koschny T, Ahnert P,
Meixensberger J and Koschny R: WHO grade-specific comparative
genomic hybridization pattern of astrocytoma-a meta-analysis.
Pathol Res Pract. 206:663–668. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Segura MF, Hanniford D, Menendez S, Reavie
L, Zou X, AlvarezDiaz S, Zakrzewski J, Blochin E, Rose A, Bogunovic
D, et al: Aberrant miR-182 expression promotes melanoma metastasis
by repressing FOXO3 and microphthalmia-associated transcription
factor. Proc Natl Acad Sci USA. 106:1814–1819. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Lowery AJ, Miller N, Dwyer RM and Kerin
MJ: Dysregulated miR-183 inhibits migration in breast cancer cells.
BMC Cancer. 10:5022010. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Yu S, Lu Z, Liu C, Meng Y, Ma Y, Zhao W,
Liu J, Yu J and Chen J: miRNA-96 suppresses KRAS and functions as a
tumor suppressor gene in pancreatic cancer. Cancer Res.
70:6015–6025. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Lu B, Mu Y, Cao C, Zeng F, Schneider S,
Tan J, Price J, Chen J, Freeman M and Hallahan DE: Survivin as a
therapeutic target for radiation sensitization in lung cancer.
Cancer Res. 64:2840–2845. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Kawasaki H, Toyoda M, Shinohara H, Okuda
J, Watanabe I, Yamamoto T, Tanaka K, Tenjo T and Tanigawa N:
Expression of survivin correlates with apoptosis, proliferation and
angiogenesis during human colorectal tumorigenesis. Cancer.
91:2026–2032. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Fan LF, Dong WG, Jiang CQ, Qian Q and Yu
QF: Role of Hypoxia-inducible factor-1 alpha and Survivin in
colorectal carcinoma progression. Int J Colorectal Dis.
23:1057–1064. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Nassar A, Sexton D, Cotsonis G and Cohen
C: Survivin expression in breast carcinoma: Correlation with
apoptosis and prognosis. Appl Immunohistochem Mol Morphol.
16:221–226. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Liu L, Zhang M and Zou P: Expression of
PLK1 and survivin in diffuse large B-cell lymphoma. Leuk Lymphoma.
48:2179–2183. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Shu M, Zheng X, Wu S, Lu H, Leng T, Zhu W,
Zhou Y, Ou Y, Lin X, Lin Y, et al: Targeting oncogenic miR-335
inhibits growth and invasion of malignant astrocytoma cells. Mol
Cancer. 10:592011. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Zhang S, Kim K, Jin UH, Pfent C, Cao H,
Amendt B, Liu X, WilsonRobles H and Safe S: Aryl hydrocarbon
receptor agonists induce microRNA-335 expression and inhibit lung
metastasis of estrogen receptor negative breast cancer cells. Mol
Cancer Ther. 11:108–118. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Xu Y, Zhao F, Wang Z, Song Y, Luo Y, Zhang
X, Jiang L, Sun Z, Miao Z and Xu H: MicroRNA-335 acts as a
metastasis suppressor in gastric cancer by targeting Bcl-w and
specificity protein 1. Oncogene. 31:1398–1407. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Shen Z, Zhan G, Ye D, Ren Y, Cheng L, Wu Z
and Guo J: MicroRNA-34a affects the occurrence of laryngeal
squamous cell carcinoma by targeting the antiapoptotic gene
survivin. Med Oncol. 29:2473–2480. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2−ΔΔCT method. Methods. 25:402–408. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
John B, Enright AJ, Aravin A, Tuschl T,
Sander C and Marks DS: Human MicroRNA targets. PLoS Biol.
2:e3632004. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Lewis BP, Burge CB and Bartel DP:
Conserved seed pairing, often flanked by adenosines, indicates that
thousands of human genes are microRNA targets. Cell. 120:15–20.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Ventura A and Jacks T: MicroRNAs and
cancer: Short RNAs go a long way. Cell. 136:586–591. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Liu ZF, Liang ZQ, Li L, Zhou YB, Wang ZB,
Gu WF, Tu LY and Zhao J: MiR-335 functions as a tumor suppressor
and regulates survivin expression in osteosarcoma. Eur Rev Med
Pharmacol Sci. 20:1251–1257. 2016.PubMed/NCBI
|
|
26
|
Lu Y, Yang H, Yuan L, Liu G, Zhang C, Hong
M, Liu Y, Zhou M, Chen F and Li X: Overexpression of miR-335
confers cell proliferation and tumour growth to colorectal
carcinoma cells. Mol Cell Biochem. 412:235–245. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Sun J, Ji J, Huo G, Song Q and Zhang X:
miR-182 induces cervical cancer cell apoptosis through inhibiting
the expression of DNMT3a. Int J Clin Exp Pathol. 8:4755–4763.
2015.PubMed/NCBI
|
|
28
|
Wallis CJ, Gordanpour A, Bendavid JS,
Sugar L, Nam RK and Seth A: MiR-182 Is associated with growth,
migration and invasion in prostate cancer via suppression of FOXO1.
J Cancer. 6:1295–1305. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Marsh EE, Lin Z, Yin P, Milad M,
Chakravarti D and Bulun SE: Differential expression of microRNA
species in human uterine leiomyoma versus normal myometrium. Fertil
Steril. 89:1771–1776. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Ronchetti D, Lionetti M, Mosca L, Agnelli
L, Andronache A, Fabris S, Deliliers GL and Neri A: An integrative
genomic approach reveals coordinated expression of intronic
miR-335, miR-342 and miR-561 with deregulated host genes in
multiple myeloma. BMC Med Genomics. 1:372008. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Uziel T, Karginov FV, Xie S, Parker JS,
Wang YD, Gajjar A, He L, Ellison D, Gilbertson RJ, Hannon G and
Roussel MF: The miR-17~92 cluster collaborates with the Sonic
Hedgehog pathway in medulloblastoma. Proc Natl Acad Sci USA.
106:2812–2817. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Wang YX, Zhang XY, Zhang BF, Yang CQ, Chen
XM and Gao HJ: Initial study of microRNA expression profiles of
colonic cancer without lymph node metastasis. J Dig Dis. 11:50–54.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Scarola M, Schoeftner S, Schneider C and
Benetti R: miR-335 directly targets Rb1 (pRb/p105) in a proximal
connection to p53-dependent stress response. Cancer Res.
70:6925–6933. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Tavazoie SF, Alarcón C, Oskarsson T, Padua
D, Wang Q, Bos PD, Gerald WL and Massagué J: Endogenous human
microRNAs that suppress breast cancer metastasis. Nature.
451:147–152. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Zhu W, Zhou K, Zha Y, Chen D, He J, Ma H,
Liu X, Le H and Zhang Y: Diagnostic Value of Serum miR-182,
miR-183, miR-210, and miR-126 Levels in Patients with Early-Stage
Non-Small Cell Lung Cancer. PLoS One. 11:e01530462016. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Jin ZB, Hirokawa G, Gui L, Takahashi R,
Osakada F, Hiura Y, Takahashi M, Yasuhara O and Iwai N: Targeted
deletion of miR-182, an abundant retinal microRNA. Mol Vis.
15:523–533. 2009.PubMed/NCBI
|
|
37
|
Yan D, Dong XD, Chen X, Yao S, Wang L,
Wang J, Wang C, Hu DN, Qu J and Tu L: Role of microRNA-182 in
posterior uveal melanoma: Regulation of tumor development through
MITF, BCL2 and cyclin D2. PLoS One. 7:e409672012. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Sun Y, Fang R, Li C, Li L, Li F, Ye X and
Chen H: Hsa-mir-182 suppresses lung tumorigenesis through down
regulation of RGS17 expression in vitro. Biochem Biophys Res
Commun. 396:501–507. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Kong WQ, Bai R, Liu T, Cai CL, Liu M, Li X
and Tang H: MicroRNA-182 targets cAMP-responsive element-binding
protein 1 and suppresses cell growth in human gastric
adenocarcinoma. FEBS J. 279:1252–1260. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Yi R, Li Y, Wang FL, Miao G, Qi RM and
Zhao YY: MicroRNAs as diagnostic and prognostic biomarkers in
colorectal cancer. World J Gastrointest Oncol. 8:330–340.
2016.PubMed/NCBI
|
|
41
|
Li F and Brattain MG: Role of the survivin
gene in pathophysiology. Am J Pathol. 169:1–11. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Altieri DC: Survivin, cancer networks and
pathway-directed drug discovery. Nat Rev Cancer. 8:61–70. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Li F and Altieri DC: The cancer
antiapoptosis mouse survivin gene: Characterization of locus and
transcriptional requirements of basal and cell cycle-dependent
expression. Cancer Res. 59:3143–3151. 1999.PubMed/NCBI
|
|
44
|
Ito T, Shiraki K, Sugimoto K, Yamanaka T,
Fujikawa K, Ito M, Takase K, Moriyama M, Kawano H, Hayashida M, et
al: Survivin promotes cell proliferation in human hepatocellular
carcinoma. Hepatology. 31:1080–1085. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Zhang R, Wang T, Li KN, Qin WW, Chen R,
Wang K, Jia LT, Zhao J, Wen WH, Meng YL, et al: A survivin double
point mutant has potent inhibitory effect on the growth of
hepatocellular cancer cells. Cancer Biol Ther. 7:547–554. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Xu Y, Fang F, Ludewig G, Jones G and Jones
D: A mutation found in the promoter region of the human survivin
gene is correlated to overexpression of survivin in cancer cells.
DNA Cell Biol. 23:527–537. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Jang JS, Kim KM, Kang KH, Choi JE, Lee WK,
Kim CH, Kang YM, Kam S, Kim IS, Jun JE, et al: Polymorphisms in the
survivin gene and the risk of lung cancer. Lung Cancer. 60:31–39.
2008. View Article : Google Scholar : PubMed/NCBI
|