Introduction
Glioma is the most common primary tumor of the
central nervous system with an annual incidence of 3–5 cases per
100,000 individuals, accounting for ~46% of all intracranial tumors
in the United States (1,2). Glioma is the second leading cause of
mortality in young cancer patients (<34 years of age) (2). High morbidity and mortality rates make
glioma the fourth most fatal malignant cancer (3). Currently, the standard treatment for
glioma is surgical resection followed by combined administration of
radiation and adjuvant chemotherapy (4,5). Despite
current therapeutic efforts, the median survival time (~14 months)
has not changed significantly in the past decades (6). The poor prognosis and low 5-year
survival rate (<10%) highlights the urgent requirement for the
development of novel therapeutic methods against glioma (2,7). At
present, cancer treatment studies predominantly focus on molecular
therapies, thus the identification of efficient targets is of great
importance.
Cyclooxygenase-2 (COX-2) is an important enzyme that
regulates the conversion of arachidonic acid to prostaglandin E2
(PGE2) (8). Previous studies have
demonstrated that PGE2, which is produced normally by epithelial
cells and at extremely high levels in a variety of malignant
tumors, upregulates the activity of T-regulatory cells, which
suppress immune function, resulting in tumor progression and poor
disease outcome (8–11). Martín Sanz et al (12) revealed that as single molecule
modification of COX-2 may induce tumorigenesis in COX-2 transgenic
mice without other genetic modifications. Krzystyniak (13) reported that COX-2 upregulation affects
both angiogenesis and the production of specific proteases that are
critical for glioma cancer growth and metastasis (13). Based on these previous studies, the
downregulation of COX-2/PGE2 signaling may present a promising and
efficient method for the prevention and treatment of malignant
tumors.
MicroRNAs (miRNAs) are small, non-coding RNAs (~22
nucleotides in length) that mediate the post-transcriptional
silencing of specific target mRNAs (14,15).
miRNAs are currently recognized as important regulators of various
biological functions in a number of cellular processes, including
cellular proliferation, differentiation, metabolism and apoptosis
(14,15). To date, >400 human miRNAs have been
identified and >1,000 have been postulated to exist (16). It is hypothesized that ≤30% of human
genes are regulated by miRNAs (17).
Each miRNA regulates a variety of targeting genes, and an
individual mRNA may be regulated by distinct miRNAs, giving rise to
an extensive regulatory network (15,18). A
number of miRNAs, including miR-21, miR-221/222, miR-124 and
miR-128, are aberrantly expressed in malignant gliomas and are
considered to have important functions in determining the degree of
malignancy (19–22).
The present study determined the relative
expressions and functions of miR-101 and COX-2 in glioma cells, as
well as the regulatory roles between them, thus, providing novel
possibilities for the prevention and treatment of glioma.
Materials and methods
Patients and specimens
Fresh-frozen human glioma tissues (n=40) and matched
distant normal tissues were obtained from patients who underwent
radical surgery between January 1 and October 1, 2013, at the
Affiliated Hospital of Hainan Medical College (Haikou, China). The
patients included 24 men and 16 women, with a mean age of 51 years.
None of these patients received radiotherapy or chemotherapy prior
to surgical resection. All 40 cases were reviewed for histological
subtype, differentiation and tumor stage (23). Written informed consent was obtained
from all patients.
Cell culture
The human glioma cell lines U373-MG and U87-MG, and
the normal human glial cell line C6, were obtained from the
American Type Culture Collection (Manassas, VA, USA). Cells were
cultured in Dulbecco's modified Eagle's medium (DMEM) (Gibco;
Thermo Fisher Scientific, Inc., Waltham, MA, USA) supplemented with
10% bovine calf serum (Gibco; Thermo Fisher Scientific, Inc.) and
maintained at 37°C in a humidified atmosphere of 5%
CO2.
Reverse transcription
(RT)-quantitative polymerase chain reaction (qPCR)
Total RNA was extracted from malignant and normal
tissues (which were soaked in formalin solution for 1 h and stored
at −80°C prior to use) and cells using Isogen reagent (Nippon Gene
Co., Ltd., Toyama, Japan). Total RNA (3 µg) was reverse-transcribed
using MultiScribe Reverse Transcriptase (Applied Biosystems; Thermo
Fisher Scientific, Inc.), according to the manufacturer's
instructions. miR-101 and COX-2 mRNA expression levels were
analyzed using TaqMan® Small RNA assays (Applied
Biosystems; Thermo Fisher Scientific, Inc.) and a MiniOpticon
Real-Time PCR System (Bio-Rad Laboratories, Inc., Hercules, CA,
USA) according to the manufacturer's instructions. The PCR cycling
conditions included initial denaturation at 95°C for 3 min followed
by 40 cycles of 95°C denaturation for 12 sec and 62°C annealing for
40 sec. Expression values were normalized against glyceraldehyde
3-phosphate dehydrogenase (GAPDH) mRNA expression levels by the
2−ΔΔCq methods. The oligonucleotide primer sequences of
miR-101 and COX-2 are shown in Table
I.
 | Table I.Oligonucleotide primer sequences used
for reverse transcription-polymerase chain reaction in the present
study. |
Table I.
Oligonucleotide primer sequences used
for reverse transcription-polymerase chain reaction in the present
study.
|
Oligonucleotide | Primer sequence
(5′-3′) |
|---|
|
Cyclooxygenase-2 |
|
|
Forward |
GCACCCCGACATAGAGAGC |
|
Reverse |
CTGCGGAGTGCAGTGTTCT |
| miR-101 |
|
|
Forward |
ACGGGCGAGCTACAGTACTGTG |
|
Reverse |
CCAGTGCAGGGTCCGAGGTA |
Western blot analysis
Cell or tissue lysates were separated by 10% sodium
dodecyl sulfate-polyacrylamide gel electrophoresis and transferred
to a polyvinylidene difluoride membrane (EMD Millipore, Darmstadt,
Germany). The membrane was blocked with 5% non-fat milk and
incubated with rabbit primary antibodies against COX-2 (rabbit,
anti-human, monoclonal; catalog no. 12282; 1:2,000 dilution) and
GAPDH (rabbit, anti-human, monoclonal; catalog no. 5174; 1:3,000
dilution) (Cell Signaling Technology, Danvers, MA, USA) at 4°C
overnight. After three washes with Tris-buffered saline containing
0.1% Tween-20, the membrane was incubated with corresponding
horseradish peroxidase-conjugated secondary antibody (goat
anti-rabbit IgG-horseradish peroxidase; catalog no. sc-2030;
1:3,000 dilution; Santa Cruz Biotechnology, Inc., Dallas, TX, USA)
for 1 h at room temperature. Immunoreactive bands were visualized
with a Pierce enhanced chemiluminescence system (Thermo Fisher
Scientific, Inc.).
Vector construction
miRNA expression plasmids were constructed using the
pRNAT-U6.1/Neo vector (GenScript, Piscataway, NJ, USA). The
pre-miRNAs were synthesized (Table
II) and cloned into the pRNAT-U6.1/Neo vector. The luciferase
reporter vector was constructed using the pGL3-basic vector
(Promega Corporation, Madison, WI, USA) and the 3′-untranslated
region of COX-2 was amplified by RT-PCR. The COX-2 small
interfering RNA (siRNA) was synthesized by Invitrogen (Thermo
Fisher Scientific, Inc.).
 | Table II.miRNA sequence used for vector
transfection. |
Table II.
miRNA sequence used for vector
transfection.
| miRNA | Sequence
(5′-3′) |
|---|
| miR-101 |
TGCCCTGGCTCAGTTATCACAGTGC |
|
|
TGATGCTGTCTATTCTAAAGGTACA |
|
|
GTACTGTGATAACTGAAGGATGGCA |
Cell transfection
Cells were grown to 70–80% confluence and
respectively transfected with miR-101 or mock vehicles using
Lipofectamine 2000 (Invitrogen; Thermo Fisher Scientific, Inc.)
according to the manufacturer's instructions. Cells transfected
with miR-101 and mock vehicles were termed U373(87)/miR-101 and
U373(87)/NC, respectively. After 48 h, cells were harvested, and
underwent limited dilution in a 96-well plate for the generation of
individual clones. Three weeks later, the level of COX-2 mRNA in
the cell clones transfected with miR-101 was analyzed by
RT-qPCR.
Dual-luciferase reporter assay
A dual-luciferase assay was performed as described
previously (24). Briefly, cells were
seeded in a 24-well plate at ~80% confluence and transfected with
miRNA expression vector, reporter vector and the pRL-TK control
vector encoding Renilla luciferase (Biovector Science Lab,
Inc., Beijing, China), using Invitrogen Lipofectamine 2000
according to the manufacturer's instructions. Following 24 h of
transfection, cells were harvested, lysed by CyQuant®
cell Lysis Buffer (Thermo Fisher Scientific, Inc.) and analyzed
using the Dual-Luciferase Reporter Assay System kit (Promega
Corporation). All experiments were performed in triplicate.
Cell proliferation assays
Cell proliferation was analyzed using Cell Counting
Kit-8 (CCK-8; Dojindo Molecular Technologies, Inc., Rockville MD,
USA) as previously described (25).
Briefly, 3,000 cells/well were seeded in a 96-well plate. At daily
time intervals (days 1–7), 10 µl CCK-8 was added to each well,
followed by incubation for 2 h in the dark. The absorbance (Ab) was
recorded at a wavelength of 450 nm by a microplate reader (Thermo
Multiskan MK3; Thermo Fisher Scientific, Inc.) and cell viability
was calculated using the following equation: Cell viability (%) =
[(Absample - Abblank) / (Abcontrol
- Abblank)] × 100%, (where Absample,
Abcontrol and Abblank are the Ab values of
each sample, the cells cultured in culture medium without any
additional substances and the culture medium without cells in
wells, respectively).
Transwell assays
A cell migration assay was performed using 24-well
Transwell plates as previously described (26). Briefly, the under surface of the
membrane was coated with human fibronectin (10 mg/ml; Gene
Operation, Ann Arbor, MI, USA) in phosphate-buffered saline at 37°C
for 2 h. The lower chamber was filled with 0.6 ml DMEM supplemented
with 10% fetal bovine serum (Gibco; Thermo Fisher Scientific,
Inc.). Prior to the experiment, cells were serum-starved overnight
[DMEM plus 0.5% bovine serum albumin (BSA; Beyotime Biotechnology,
Shanghai, China], then 1×106 cells in 0.1 ml migration
medium (DMEM plus 0.5% BSA) were added to the upper chamber. After
incubation at 37°C for 12 h, cells on the upper surface of the
membrane were removed. The migrated cells that had attached to the
lower surface were fixed in 10% formalin and stained with a
solution containing 1% crystal violet and 2% ethanol in 100 mmol/l
borate buffer (pH, 9.0; Beyotime Biotechnology). The number of
migrated cells on the lower surface of the membrane was counted
under a light microscope.
In vivo tumor growth assays
Four-week-old female BALB/c nude mice (mean weight,
14.3 g) were purchased from Dashuo Biotechnology Co., Ltd.
(Chengdu, China) and raised in specific pathogen-free conditions at
26°C. A total of 5×106 control and miR-101
overexpressing cells were injected subcutaneously into BALB/c nude
mice (age, 6–8 weeks), respectively. The tumor size was measured in
two perpendicular diameters with precision calipers and calculated
using the following equation (25):
Tumor volume (mm3) = (length × width2) / 2.
At the end of the experiments, all the mice were sacrificed by
cervical vertebra dislocation.
Statistical analysis
The Student's t-test was performed to compare
differences between two groups, whereas analysis of variance
followed by Dunnett's multiple comparison test was used to compare
more than two groups. Data are presented as mean ± standard
deviation. All data analyses were performed using SPSS version 10.0
(SPSS, Inc., Chicago, IL, USA). P<0.05 was considered to
indicate a statistically significant difference.
Results
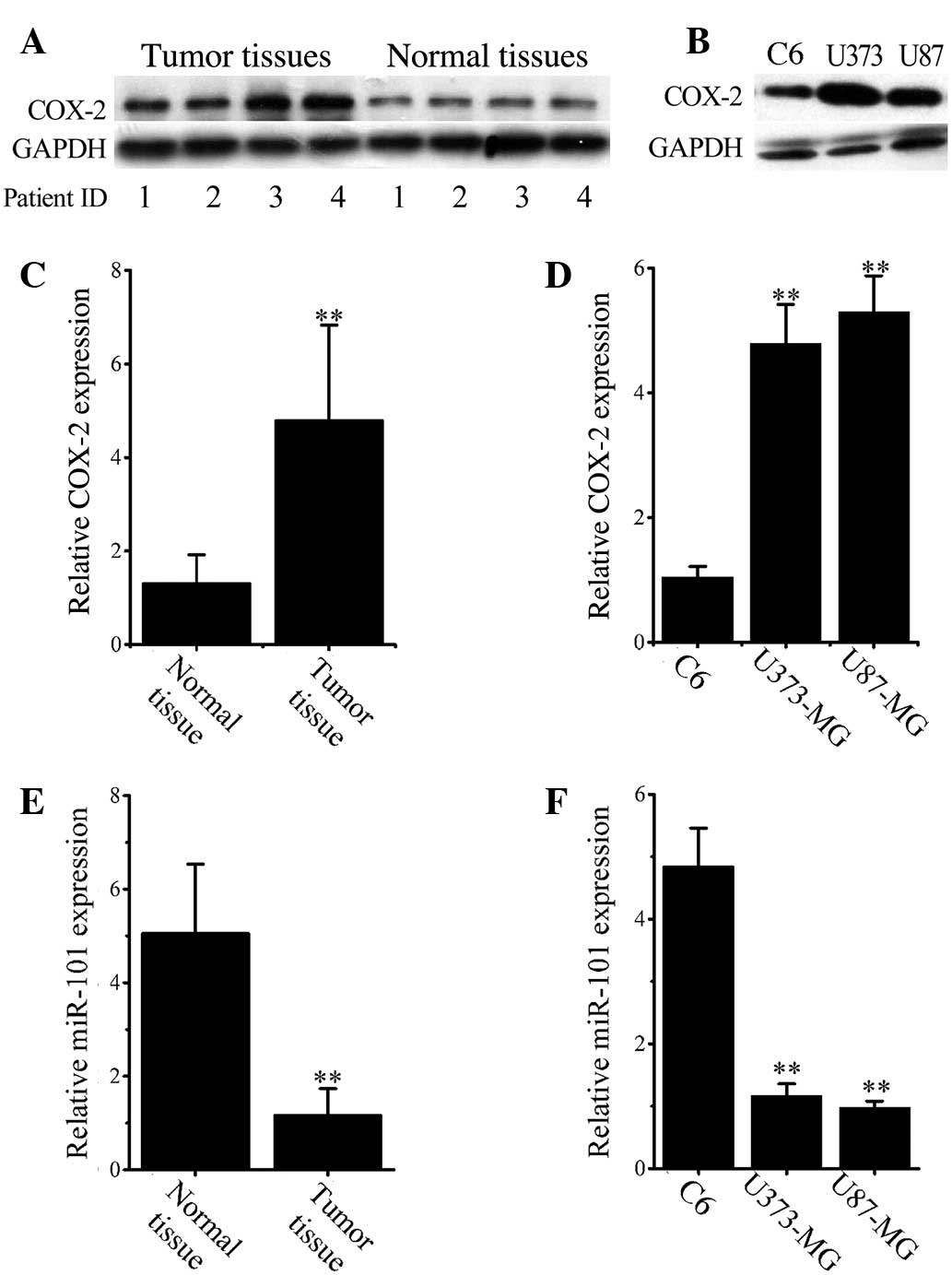
Elevated COX-2 expression in human
glioma tissues and cells
To identify aberrant COX-2 expression in glioma, the
COX-2 protein levels in normal and malignant tissues in 33 glioma
and adjacent normal tissues were analyzed by western blotting.
Representative images (Fig. 1A)
demonstrate that COX-2 is significantly overexpressed in glioma
tissues when compared with that in normal tissues. Consistent
results were obtained in cultured cell lines as shown in Fig. 1B, which revealed that COX-2 was
significantly overexpressed in both U373 and U87 glioma cell lines
compared with C6 cells. COX-2 overexpression in malignant tissues
and cells was confirmed at the mRNA level by RT-qPCR analysis
(Fig. 1C and D).
miR-101 downregulation in glioma
tissues and cell lines
RT-qPCR results (Fig.
1E) demonstrated that the mean expression of miR-101 was
significantly decreased in all glioma cancer tissues when compared
with normal tissues (P<0.001). Similarly, the levels of miR-101
in both U373 and U87 cells were significantly lower than that in C6
cells (P<0.001; Fig. 1F).
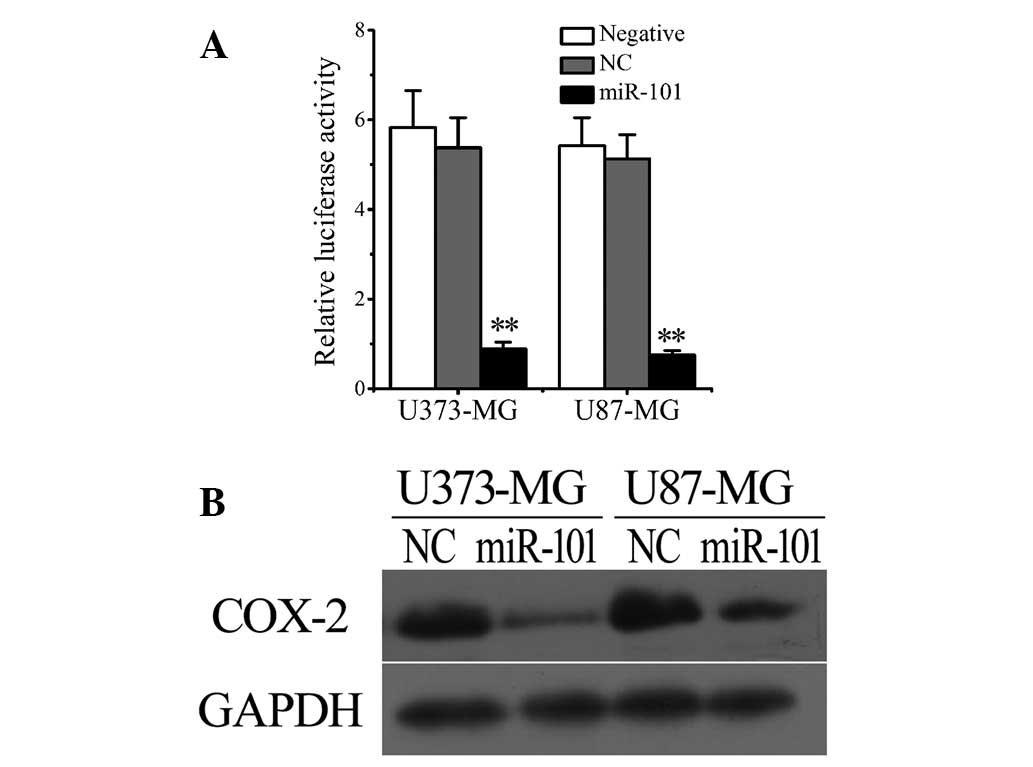
Overexpression of miR-101
significantly reduces COX-2 levels in glioma cells
Considering the downregulation of miR-101 levels
(Fig. 1E and F) and upregulation of
COX-2 expression in human glioma cells and tissues (Fig. 1A-D), we hypothesized that a negative
correlation may exist between the intracellular levels of COX-2 and
miR-101. To verify this hypothesis, the luciferase reporter system
was used to investigate the regulatory function of miR-101 on the
expression of COX-2. The results revealed that transfection with
miR-101, but not empty vectors [negative control (NC)],
significantly reduced the luciferase levels in U373 and U87 cells
(Fig. 2A). The results were confirmed
by western blot analysis. As shown in Figure 2B, miR-101 transfection markedly
downregulated COX-2 expression in both U373 and U87 glioma
cells.
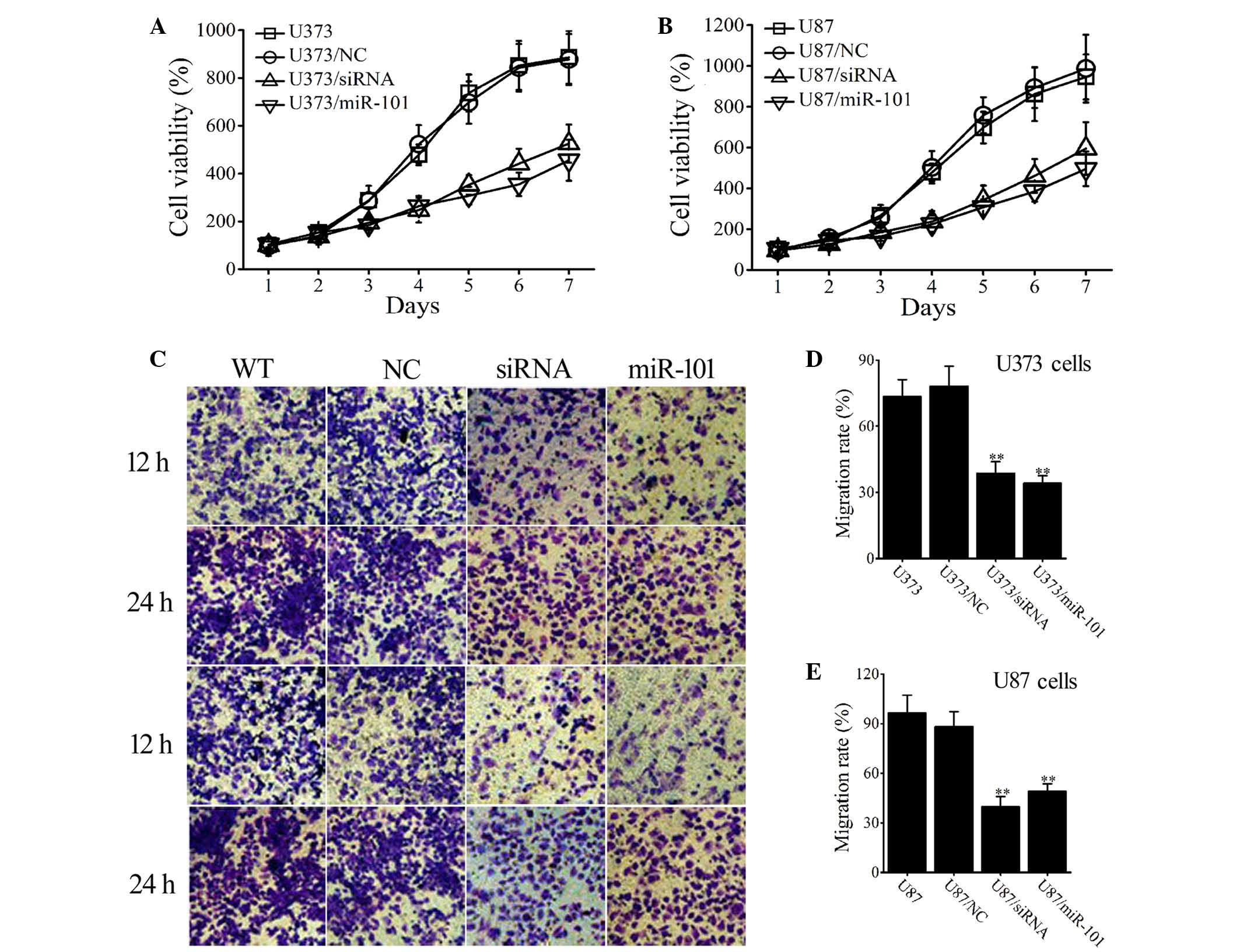
Overexpression of miR-101 inhibits
cell proliferation and migration of glioma cells
To investigate the effect of miR-101, cell
proliferation was assessed in wild type (WT) and
miR-101-overexpressing glioma cells. Cells transfected with empty
vectors were used as an NC. Figure 3A
illustrates that after a 7-day period of proliferation, cell
proliferation was lower in the miR-101-transfected group
(U373/miR-101) than in the non-transfected (U373) and mock-vehicle
groups (U373/NC; P<0.001). Similar results were obtained in U87
cells (Fig. 3B).
Transwell assays were used to measure the effect of
miR-101 on tumor cell migration. After 12 h, the number of migrated
cells was markedly decreased in miR-101-overexpressing U373 and U87
cells (Fig. 3C) and this effect was
also observed after 24 h. After 24 h, the migration rate of U373
(U373/miR-101) and U87 (U87/miR-101) cells was significantly
reduced when compared with that of WT and NC-transfected cells
(P<0.001; Fig. 3D and E). The
results indicate that miR-101 overexpression inhibits cellular
proliferation and migration of U373 and U87 glioma cells.
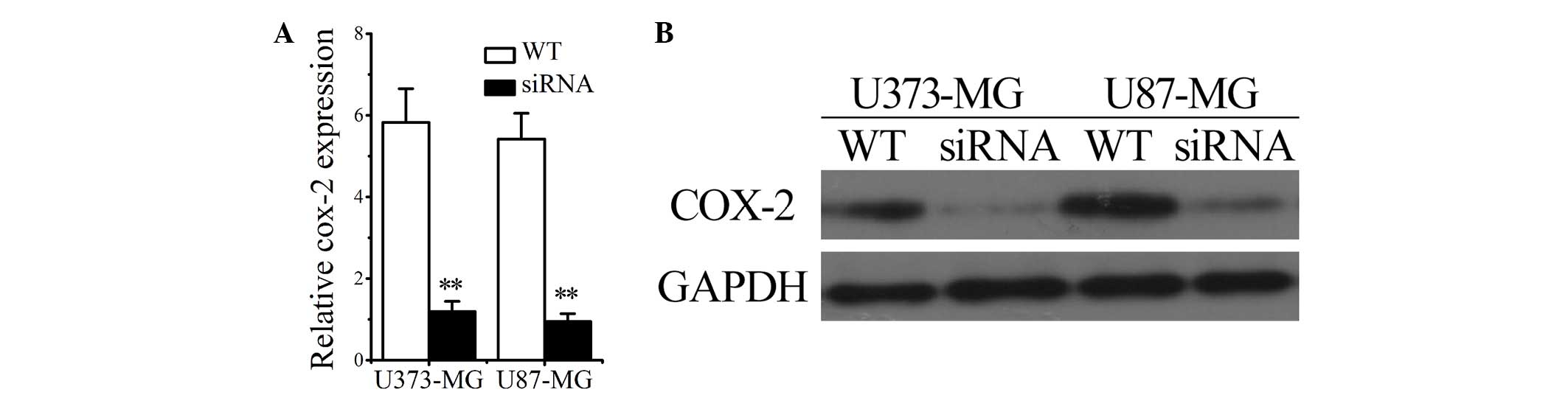
COX-2 interference inhibits cell
proliferation and migration of glioma cells
To determine the impact of COX-2 on the
proliferation and invasion of glioma cancer cells, glioma cells
were transfected with COX-2 siRNAs and the proliferation and
invasion abilities were compared between WT and siRNA-transfected
glioma cells (U373/siRNA and U87/siRNA groups). The successful
transfection of siRNAs into U373 and U87 cells was confirmed by
RT-qPCR and western blotting, the results of which revealed that
COX-2 mRNA and protein levels were significantly decreased
(Fig. 4A and B). The results shown in
Fig. 3 suggest that transfection with
COX-2 siRNA negatively influences the proliferation (Fig. 3A and B) and invasion (Fig. 3D-F) of human U373 and U87 glioma
cells.
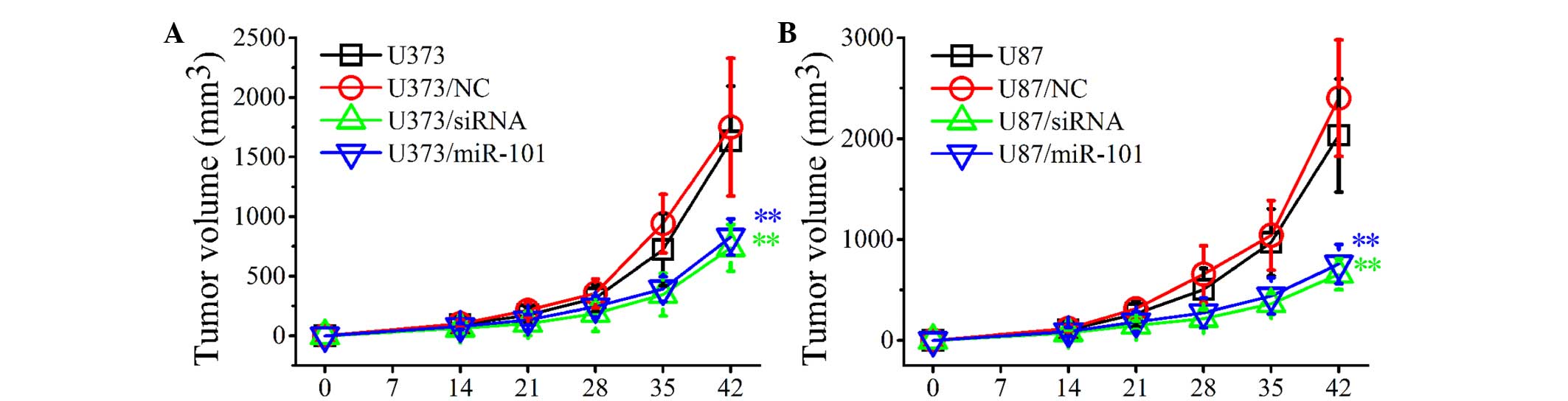
Overexpression of miR-101 inhibits the
development of established tumors in a localized human glioma
xenotransplant model
To investigate the inhibitory effects of miR-101
overexpression on the growth of established tumors, control and
miR-101-transfected U373 and U87 cells were injected subcutaneously
into BALB/c nude mice. The development of established tumors was
monitored every day for 6 weeks. As shown in Fig. 5, the development of solid tumors was
first visible at ~14 days post-inoculation and tumors grew rapidly
in WT (U373 and U87) and NC-transfected (U373/NC and U87/NC)
groups. By contrast, in the U373/miR-101 and U87/miR-101 groups,
the tumor growth was markedly slower and the tumor volume was
significantly decreased compared with control groups (Fig. 5A and B). Similar results were obtained
when glioma cells were transfected with COX-2 siRNAs. The in
vivo experiments indicated that miR-101 overexpression may
significantly inhibit tumor growth in vivo and thus may
present a useful treatment for glioma.
Discussion
Gliomas are the most malignant tumors of the central
nervous system. Although conventional therapies have significantly
advanced in recent years, the prognosis of glioma remains poor
(1–3).
Understanding of the molecular mechanisms of glioma development has
improved and, subsequently, molecular targeted therapy has been
proposed for the treatment of gliomas (1,15,27,28).
Molecular targeted therapy requires the identification of novel and
efficient therapeutic targets for the diagnosis and treatment of
the disease.
Increasing evidence has demonstrated that
up/downregulation of miRNAs is involved in the initiation and
progression of various types of malignancy via dysregulation of
targeting oncogenes and/or tumor suppressor genes (29,30).
Investigation of differentially expressed miRNAs in various cancer
types has yielded significant information with regard to
carcinogenesis, thus providing potential tumor suppressors for
treating malignancies (14,31,32).
miR-101 has been reported to be frequently
downregulated in various tumor types, including breast, bladder,
gastric, prostate, cervical and liver cancers (33–38). To
the best of our knowledge, studies regarding the effect of miR-101
in glioma are limited. In the present study, it was demonstrated
that miR-101 functions as an effective tumor suppressor via the
repression of COX-2 in gliomas.
COX-2, a key enzyme in the production of
prostaglandins, is involved in numerous biological processes that
are associated with tumorigenesis, including cell proliferation,
differentiation, migration and the regulation of antitumor immunity
(8,11–13). The
regulation of COX-2 expression in cancer has been the focus of
numerous studies (11–14). The results of the present study
demonstrated that COX-2 levels were significantly increased in
glioma tissues and cell lines. Furthermore, miR-101 overexpression
caused downregulation of COX-2 in glioma cells and tissues.
Notably, COX-2 interference in glioma cells following siRNA or
miR-101 transfection resulted in markedly decreased proliferation
and migration rates in vitro. The results also suggest that
miR-101 overexpression in both U373 and U87 cells results in
significant inhibition of tumor growth in localized glioma
xenotransplant models.
To the best of our knowledge, this is the first
study to demonstrate that miR-101 downregulation contributes to
cell proliferation, migration and invasion in gliomas via
upregulation of COX-2 expression. The identification of candidate
target genes of miR-101 may aid in elucidating potential
carcinogenic mechanisms in human gliomas. In summary, these
findings indicate that miR-101 may present a novel potential target
that may potentially be exploited for the treatment of glioma.
Acknowledgements
This study was supported by funds for the
modernization of traditional Chinese medicine in Hainan Province
(grant no. 2015zy23).
References
|
1
|
Gu X, Wang C, Wang X, Ma G, Li Y, Cui L,
Chen Y, Zhao B and Li K: Efficient inhibition of human glioma
development by RNA interference-mediated silencing of PAK5. Int J
Biol Sci. 11:230–237. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Meyer MA: Malignant gliomas in adults. N
Engl J Med. 359:1850author reply 1850. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Ostrom QT, Gittleman H, Liao P, Rouse C,
Chen Y, Dowling J, Wolinsky Y, Kruchko C and Barnholtz-Sloan J:
CBTRUS statistical report: Primary brain and central nervous system
tumors diagnosed in the United States in 2007-2011. Neuro Oncol.
16(Suppl 4): iv1–iv63. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Huang FY, Lee TW, Chang CH, Chen LC, Hsu
WH, Chang CW and Lo JM: Evaluation of (188) Re-labeled PEGylated
nanoliposome as a radionuclide therapeutic agent in an orthotopic
glioma-bearing rat model. Int J Nanomedicine. 10:463–473. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Vredenburgh JJ, Desjardins A, Reardon DA,
Peters KB, Herndon JE II, Marcello J, Kirkpatrick JP, Sampson JH,
Bailey L, Threatt S, et al: The addition of bevacizumab to standard
radiation therapy and temozolomide followed by bevacizumab,
temozolomide and irinotecan for newly diagnosed glioblastoma. Clin
Cancer Res. 17:4119–4124. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Stupp R, Mason WP, van den Bent MJ, Weller
M, Fisher B, Taphoorn MJ, Belanger K, Brandes AA, Marosi C, Bogdahn
U, et al: Radiotherapy plus concomitant and adjuvant temozolomide
for glioblastoma. N Engl J Med. 352:987–996. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Quick A, Patel D, Hadziahmetovic M,
Chakravarti A and Mehta M: Current therapeutic paradigms in
glioblastoma. Rev Recent Clin Trials. 5:14–27. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Karavitis J and Zhang M: COX2 regulation
of breast cancer bone metastasis. Oncoimmunology. 2:e231292013.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Tsujii M and DuBois RN: Alterations in
cellular adhesion and apoptosis in epithelial cells overexpressing
prostaglandin endoperoxide synthase 2. Cell. 83:493–501. 1995.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Howe LR: Inflammation and breast cancer.
Cyclooxygenase/prostaglandin signaling and breast cancer. Breast
Cancer Res. 9:2102007. View
Article : Google Scholar : PubMed/NCBI
|
|
11
|
Tyagi A, Agarwal C, DwyerNield LD, Singh
RP, Malkinson AM and Agarwal R: Silibinin modulates TNF-α and IFN-γ
mediated signaling to regulate COX2 and iNOS expression in
tumorigenic mouse lung epithelial LM2 cells. Mol Carcinog.
51:832–842. 2012. View
Article : Google Scholar : PubMed/NCBI
|
|
12
|
Martín Sanz P, Hortelano S, Bosca L and
Casado M: Cyclooxygenase 2: Understanding the pathophysiological
role through genetically altered mouse models. Front Biosci.
11:2876–2888. 2006. View
Article : Google Scholar : PubMed/NCBI
|
|
13
|
Krzystyniak KL: Current strategies for
anticancer chemoprevention and chemoprotection. Acta Pol Pharm.
59:473–478. 2002.PubMed/NCBI
|
|
14
|
Farh KK, Grimson A, Jan C, Lewis BP,
Johnston WK, Lim LP, Burge CB and Bartel DP: The widespread impact
of mammalian MicroRNAs on mRNA repression and evolution. Science.
310:1817–1821. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Yamashita D, Kondo T, Ohue S, Takahashi H,
Ishikawa M, Matoba R, Suehiro S, Kohno S, Harada H, Tanaka J and
Ohnishi T: MiR-340 suppresses the stem-like cell function of
glioma-initiating cells by targeting tissue plasminogen activator.
Cancer Res. 75:1123–1133. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Wu D, Zhou Y, Pan H, Zhou J, Fan Y and Qu
P: MicroRNA-99a inhibiting cell proliferation, migration and
invasion by targeting fibroblast growth factor receptor 3 in
bladder cancer. Oncol Lett. 7:1219–1224. 2014.PubMed/NCBI
|
|
17
|
Slaby O, Svoboda M, Fabian P, Smerdova T,
Knoflickova D, Bednarikova M, Nenutil R and Vyzula R: Altered
expression of miR-21, miR-31, miR-143 and miR-145 is related to
clinicopathologic features of colorectal cancer. Oncology.
72:397–402. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Calin GA and Croce CM: MicroRNA signatures
in human cancers. Nat Rev Cancer. 6:857–866. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Silber J, Lim DA, Petritsch C, Persson AI,
Maunakea AK, Yu M, Vandenberg SR, Ginzinger DG, James CD, Costello
JF, et al: MiR-124 and miR-137 inhibit proliferation of
glioblastoma multiforme cells and induce differentiation of brain
tumor stem cells. BMC Med. 6:142008. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Chan JA, Krichevsky AM and Kosik KS:
MicroRNA-21 is an antiapoptotic factor in human glioblastoma cells.
Cancer Res. 65:6029–6033. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Zhang Y, Chao T, Li R, Liu W, Chen Y, Yan
X, Gong Y, Yin B, Liu W, Qiang B, et al: MicroRNA-128 inhibits
glioma cells proliferation by targeting transcription factor E2F3a.
J Mol Med (Berl). 87:43–51. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Lorimer IA: Regulation of p27Kip1 by miRNA
221/222 in glioblastoma. Cell Cycle. 8:26852009.PubMed/NCBI
|
|
23
|
Lou M and Zhao Y: Satisfactory therapy
results of combining nimustine with nicardipine against glioma at
advanced stage. J Cancer Res Ther. 11:10302015. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Dews M, Homayouni A, Yu D, Murphy D,
Sevignani C, Wentzel E, Furth EE, Lee WM, Enders GH, Mendell JT and
Thomas-Tikhonenko A: Augmentation of tumor angiogenesis by a
Myc-activated microRNA cluster. Nat Genet. 38:1060–1065. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Wu C, Li H, Zhao H, Zhang W, Chen Y, Yue
Z, Lu Q, Wan Y, Tian X and Deng A: Potentiating antilymphoma
efficacy of chemotherapy using a liposome for integration of CD20
targeting, ultra-violet irradiation polymerizing and controlled
drug delivery. Nanoscale Res Lett. 9:4472014. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Bandrés E, Cubedo E, Agirre X, Malumbres
R, Zárate R, Ramirez N, Abajo A, Navarro A, Moreno I, Monzó M and
García-Foncillas J: Identification by Real-time PCR of 13 mature
microRNAs differentially expressed in colorectal cancer and
non-tumoral tissues. Mol Cancer. 5:292006. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Li J, Yen C, Liaw D, Podsypanina K, Bose
S, Wang SI, Puc J, Miliaresis C, Rodgers L, McCombie R, et al:
PTEN, a putative protein tyrosine phosphatase gene mutated in human
brain, breast and prostate cancer. Science. 275:1943–1947. 1997.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Hermanson M, Funa K, Hartman M,
ClaessonWelsh L, Heldin CH, Westermark B and Nistér M:
Platelet-derived growth factor and its receptors in human glioma
tissue: Expression of messenger RNA and protein suggests the
presence of autocrine and paracrine loops. Cancer Res.
52:3213–3219. 1992.PubMed/NCBI
|
|
29
|
Bartel DP: MicroRNAs: Genomics,
biogenesis, mechanism and function. Cell. 116:281–297. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
He L and Hannon GJ: MicroRNAs: Small RNAs
with a big role in gene regulation. Nat Rev Genet. 5:522–531. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Gao J, Li L, Wu M, Liu M and Xie X, Guo J,
Tang H and Xie X: MiR-26a inhibits proliferation and migration of
breast cancer through repression of MCL-1. PLoS One. 8:e651382013.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Lin Y, Chen H, Hu Z, Mao Y, Xu X, Zhu Y,
Xu X, Wu J, Li S, Mao Q, et al: MiR-26a inhibits proliferation and
motility in bladder cancer by targeting HMGA1. FEBS Lett.
587:2467–2473. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Deng M, Tang HL, Lu XH, Liu MY, Lu XM, Gu
YX, Liu JF and He ZM: MiR-26a suppresses tumor growth and
metastasis by targeting FGF9 in gastric cancer. PLoS One.
8:e726622013. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Zhao S, Ye X, Xiao L, Lian X, Feng Y, Li F
and Li L: MiR-26a inhibits prostate cancer progression by
repression of Wnt5a. Tumour Biol. 35:9725–9733. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Verghese ET, Drury R, Green CA, Holliday
DL, Lu X, Nash C, Speirs V, Thorne JL, Thygesen HH, Zougman A, et
al: MiR-26b is down-regulated in carcinoma-associated fibroblasts
from ER-positive breast cancers leading to enhanced cell migration
and invasion. J Pathol. 231:388–399. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Liang X, Liu Y, Zeng L, Yu C, Hu Z, Zhou Q
and Yang Z: MiR-101 inhibits the G1-to-S phase transition of
cervical cancer cells by targeting Fos. Int J Gynecol Cancer.
24:1165–1172. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Sheng Y, Li J, Zou C, Wang S, Cao Y, Zhang
J, Huang A and Tang H: Downregulation of miR-101-3p by hepatitis B
virus promotes proliferation and migration of hepatocellular
carcinoma cells by targeting Rab5a. Arch Virol. 159:2397–2410.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Edge SB, Sobin LH, Page DL, Gospodarowicz
MK, Greene FL and Winchester DP: Re: Colon cancer survival rates
with the new American Joint Committee on Cancer sixth edition
staging. J Natl Cancer Inst. 97:463–464. 2005. View Article : Google Scholar : PubMed/NCBI
|