|
1
|
Xia H, Ng SS, Jiang S, Cheung WK, Sze J,
Bian XW, Kung HF and Lin MC: miR-200a-mediated downregulation of
ZEB2 and CTNNB1 differentially inhibits nasopharyngeal carcinoma
cell growth, migration and invasion. Biochem Biophys Res Commun.
391:535–541. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Zhang F and Zhang J: Clinical hereditary
characteristics in nasopharyngeal carcinoma through Ye-Liang's
family cluster. Chin Med J (Engl). 112:185–187. 1999.PubMed/NCBI
|
|
3
|
Lo KW, Chung GT and To KF: Deciphering the
molecular genetic basis of NPC through molecular, cytogenetic, and
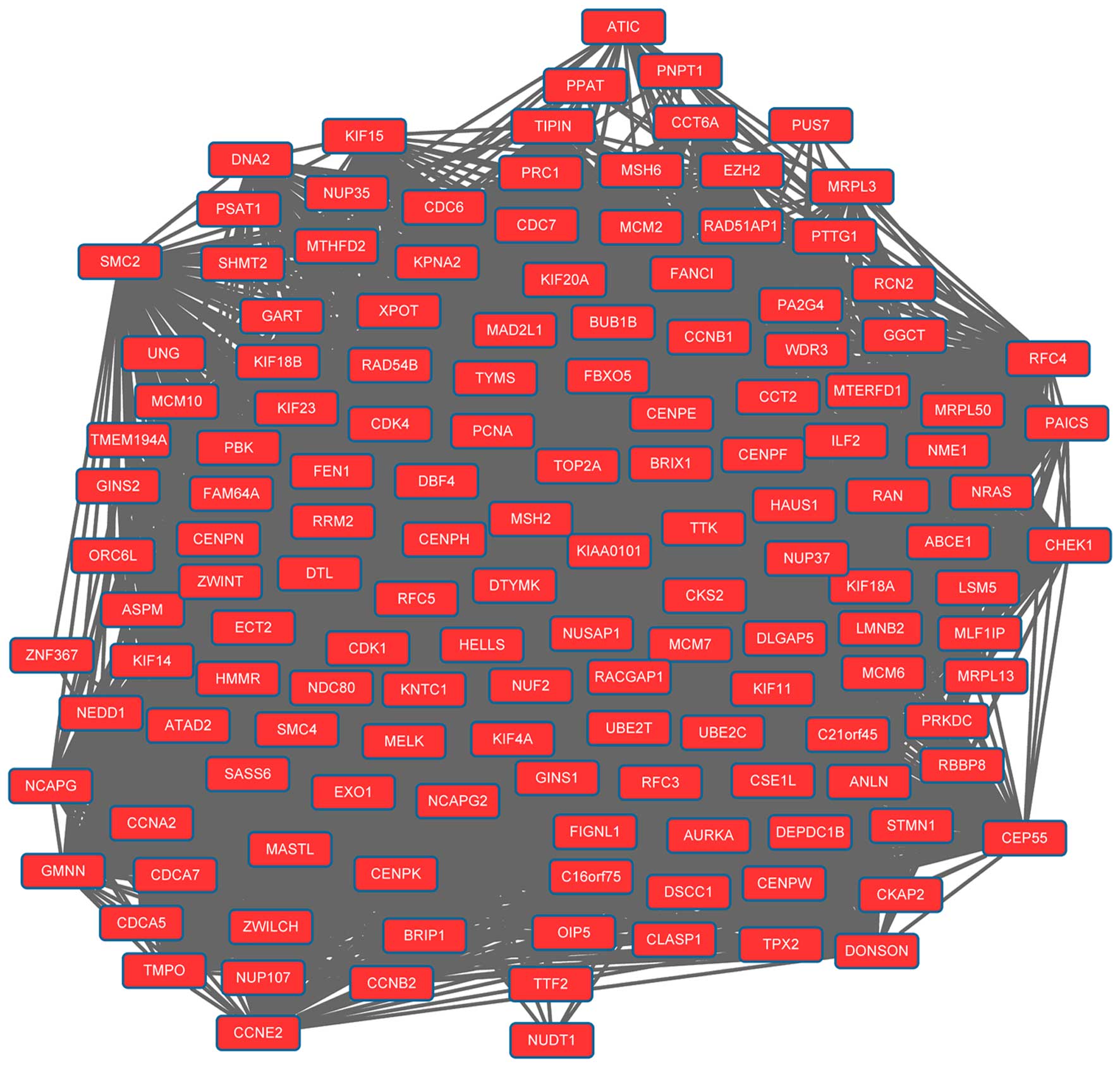
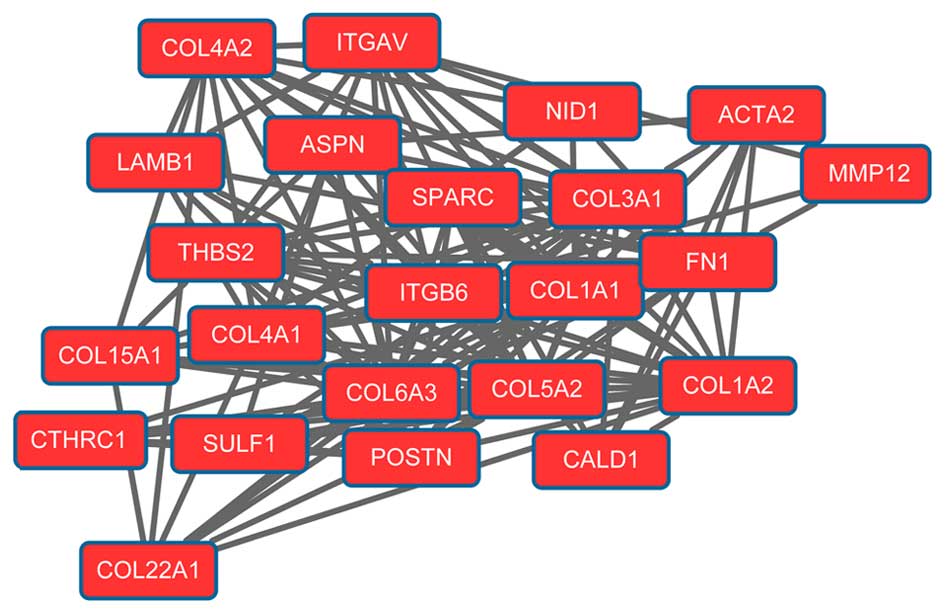
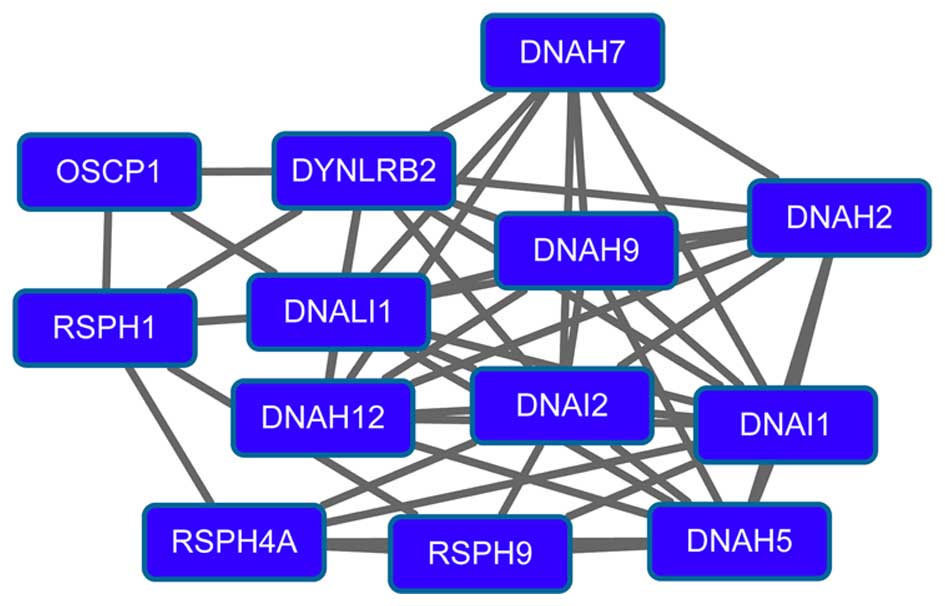
epigenetic approaches. Semin Cancer Biol. 22:79–86. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Yu MC, Ho JH, Lai SH and Henderson BE:
Cantonese-style salted fish as a cause of nasopharyngeal carcinoma:
Report of a case-control study in Hong Kong. Cancer Res.
46:956–961. 1986.PubMed/NCBI
|
|
5
|
Lozano R, Naghavi M, Foreman K, Lim S,
Shibuya K, Aboyans V, Abraham J, Adair T, Aggarwal R, Ahn SY, et
al: Global and regional mortality from 235 causes of death for 20
age groups in 1990 and 2010: A systematic analysis for the global
burden of disease study 2010. Lancet. 380:2095–2128. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Wei MX, Moulin JC, Decaussin G, Berger F
and Ooka T: Expression and tumorigenicity of the Epstein-Barr virus
BARF1 gene in human Louckes B-lymphocyte cell line. Cancer Res.
54:1843–1848. 1994.PubMed/NCBI
|
|
7
|
Decaussin G, SbihLammali F, de
Turenne-Tessier M, Bouguermouh A and Ooka T: Expression of BARF1
gene encoded by Epstein-Barr virus in nasopharyngeal carcinoma
biopsies. Cancer Res. 60:5584–5588. 2000.PubMed/NCBI
|
|
8
|
Seto E, Yang L, Middeldorp J, Sheen TS,
Chen JY, Fukayama M, Eizuru Y, Ooka T and Takada K: Epstein-Barr
virus (EBV)-encoded BARF1 gene is expressed in nasopharyngeal
carcinoma and EBV-associated gastric carcinoma tissues in the
absence of lytic gene expression. J Med Virol. 76:82–88. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Zhou L, Jiang W, Ren C, Yin Z, Feng X, Liu
W, Tao Q and Yao K: Frequent hypermethylation of RASSF1A and TSLC1,
and high viral load of Epstein-Barr Virus DNA in nasopharyngeal
carcinoma and matched tumor-adjacent tissues. Neoplasia. 7:809–815.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Lo KW, Kwong J, Hui AB, Chan SY, To KF,
Chan AS, Chow LS, Teo PM, Johnson PJ and Huang DP: High frequency
of promoter hypermethylation of RASSF1A in nasopharyngeal
carcinoma. Cancer Res. 61:3877–3881. 2001.PubMed/NCBI
|
|
11
|
Feng P, Ren EC, Liu D, Chan SH and Hu H:
Expression of Epstein-Barr virus lytic gene BRLF1 in nasopharyngeal
carcinoma: Potential use in diagnosis. J Gen Virol. 81:2417–2423.
2000. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Horikawa T, Sheen TS, Takeshita H, Sato H,
Furukawa M and Yoshizaki T: Induction of c-Met proto-oncogene by
Epstein-Barr virus latent membrane protein-1 and the correlation
with cervical lymph node metastasis of nasopharyngeal carcinoma. Am
J Pathol. 159:27–33. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Heussinger N, Büttner M, Ott G, Brachtel
E, Pilch BZ, Kremmer E and Niedobitek G: Expression of the
Epstein-Barr virus (EBV)-encoded latent membrane protein 2A (LMP2A)
in EBV-associated nasopharyngeal carcinoma. J Pathol. 203:696–699.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Sengupta S, den Boon JA, Chen IH, Newton
MA, Dahl DB, Chen M, Cheng YJ, Westra WH, Chen CJ, Hildesheim A, et
al: Genome-wide expression profiling reveals EBV-associated
inhibition of MHC class I expression in nasopharyngeal carcinoma.
Cancer Res. 66:7999–8006. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Gautier L, Cope L, Bolstad BM and Irizarry
RA: Affy-analysis of Affymetrix GeneChip data at the probe level.
Bioinformatics. 20:307–315. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Bolstad BM, Irizarry RA, Åstrand M and
Speed TP: A comparison of normalization methods for high density
oligonucleotide array data based on variance and bias.
Bioinformatics. 19:185–193. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Smyth GK: Limma: Linear models for
microarray dataBioinformatics and Computational Biology Solutions
using R and Bioconductor. Gentleman R, Carey V, Dudoit S, Irizarry
R and Huber W: Springer; New York: pp. 397–420. 2005, View Article : Google Scholar
|
|
18
|
Tweedie S, Ashburner M, Falls K, Leyland
P, McQuilton P, Marygold S, Millburn G, OsumiSutherland D,
Schroeder A, Seal R, et al: FlyBase: Enhancing Drosophila gene
ontology annotations. Nucleic Acids Res. 37:D555–D559. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Kanehisa M and Goto S: KEGG: kyoto
encyclopedia of genes and genomes. Nucleic Acids Res. 28:27–30.
2000. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Dennis G Jr, Sherman BT, Hosack DA, Yang
J, Gao W, Lane HC and Lempicki RA: DAVID: Database for annotation,
visualization, and integrated discovery. Genome Biol. 4:P32003.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Franceschini A, Szklarczyk D, Frankild S,
Kuhn M, Simonovic M, Roth A, Lin J, Minguez P, Bork P, von Mering C
and Jensen LJ: STRING v9. 1: Protein-protein interaction networks,
with increased coverage and integration. Nucleic Acids Res.
41:D808–D815. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Smoot ME, Ono K, Ruscheinski J, Wang PL
and Ideker T: Cytoscape 2.8: New features for data integration and
network visualization. Bioinformatics. 27:431–432. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Nepusz T, Yu H and Paccanaro A: Detecting
overlapping protein complexes in protein-protein interaction
networks. Nat Methods. 9:471–472. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Cheung HW, Jin DY, Ling MT, Wong YC, Wang
Q, Tsao SW and Wang X: Mitotic arrest deficient 2 expression
induces chemosensitization to a DNA-damaging agent, cisplatin, in
nasopharyngeal carcinoma cells. Cancer Res. 65:1450–1458. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Wang X, Jin DY, Wong YC, Cheung AL, Chun
AC, Lo AK, Liu Y and Tsao SW: Correlation of defective mitotic
checkpoint with aberrantly reduced expression of MAD2 protein in
nasopharyngeal carcinoma cells. Carcinogenesis. 21:2293–2297. 2000.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Wang X, Jin DY, Wong HL, Feng H, Wong YC
and Tsao SW: MAD2-induced sensitization to vincristine is
associated with mitotic arrest and Raf/Bcl-2 phosphorylation in
nasopharyngeal carcinoma cells. Oncogene. 22:109–116. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Huang TS, Shu CH, Chao Y, Chen SN and Chen
LL: Activation of MAD 2 checkprotein and persistence of cyclin
B1/CDC 2 activity associate with paclitaxel-induced apoptosis in
human nasopharyngeal carcinoma cells. Apoptosis. 5:235–241. 2000.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Tsai ST, Jin YT, Leung HW, Wang ST, Tsao
CJ and Su IJ: Bcl-2 and proliferating cell nuclear antigen (PCNA)
expression correlates with subsequent local recurrence in
nasopharyngeal carcinomas. Anticancer Res. 18:2849–2854.
1998.PubMed/NCBI
|
|
29
|
Lian B, Wang JQ and Jin L: Effects of
siRNA targeting PCNA gene on nasopharyngeal carcinoma CNE2 cell
cycle. Chin J Pathophysiol. 25:1533–1537. 2009.(In Chinese).
|
|
30
|
Luo WR and Yao KT: Cancer stem cell
characteristics, ALDH1 expression in the invasive front of
nasopharyngeal carcinoma. Virchows Arch. 464:35–43. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Wu A, Luo W, Zhang Q, Yang Z, Zhang G, Li
S and Yao K: Aldehyde dehydrogenase 1, a functional marker for
identifying cancer stem cells in human nasopharyngeal carcinoma.
Cancer Lett. 330:181–189. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Luo WR, Gao F, Li SY and Yao KT: Tumour
budding and the expression of cancer stem cell marker aldehyde
dehydrogenase 1 in nasopharyngeal carcinoma. Histopathology.
61:1072–1081. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Hou W, He W, Li Y, Ma R, Wang Z, Zhu X, Fu
Q, Wen Y, Li H and Wen W: Increased expression of aldehyde
dehydrogenase 1 A1 in nasopharyngeal carcinoma is associated with
enhanced invasiveness. Eur Arch Otorhinolaryngol. 271:171–179.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
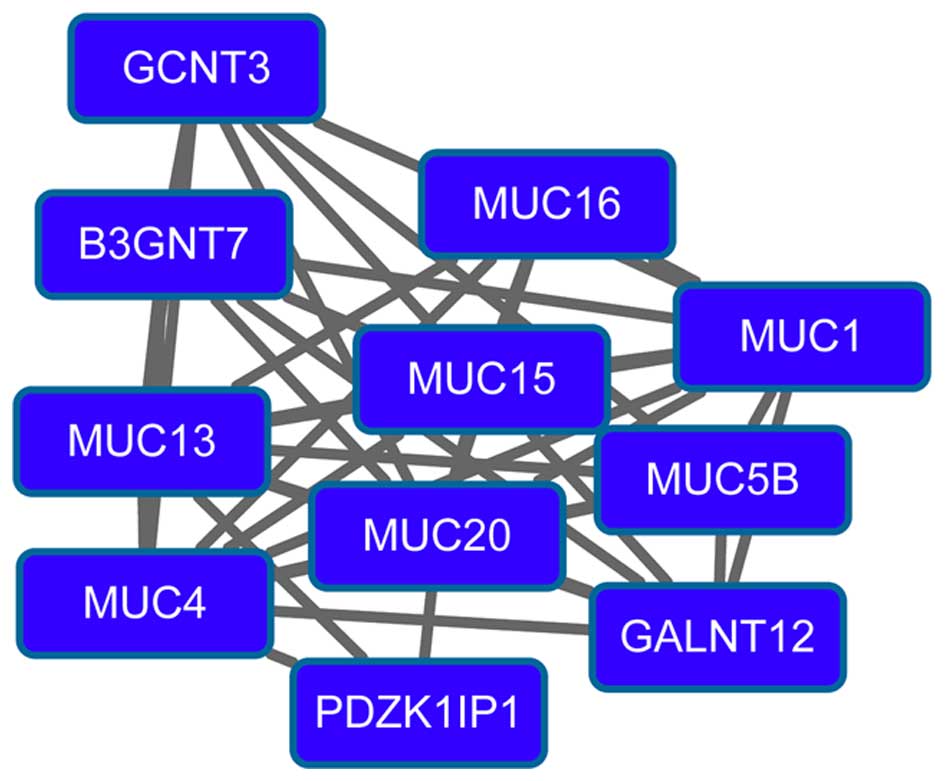
Kondo S, Yoshizaki T, Wakisaka N, Horikawa
T, Murono S, Jang KL, Joab I, Furukawa M and Pagano JS: MUC1
induced by Epstein-Barr virus latent membrane protein 1 causes
dissociation of the cell-matrix interaction and cellular
invasiveness via STAT signaling. J Virol. 81:1554–1562. 2007.
View Article : Google Scholar : PubMed/NCBI
|