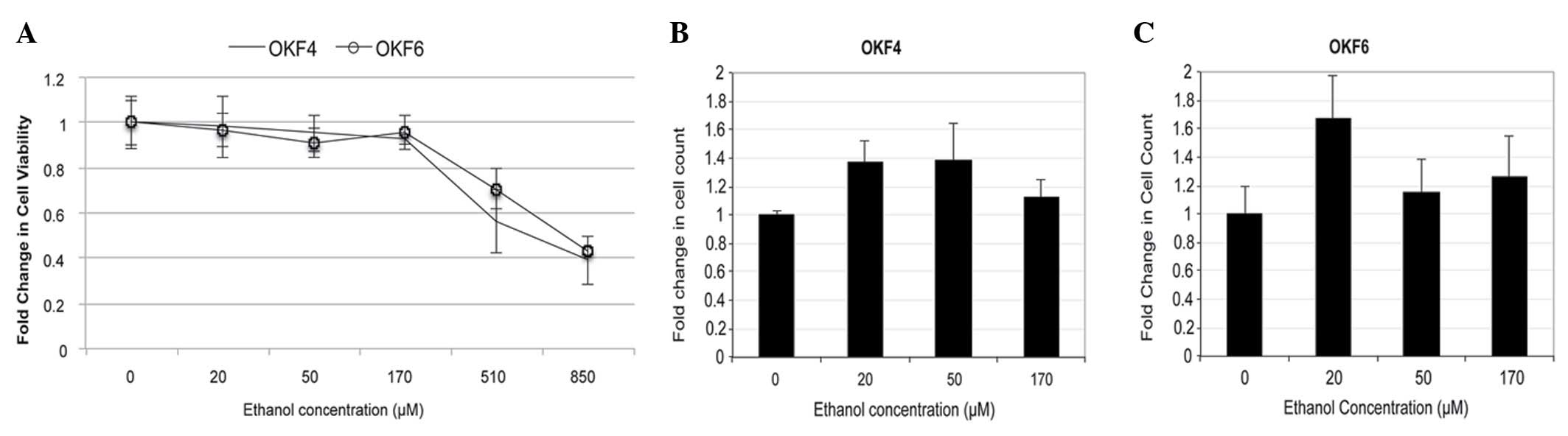
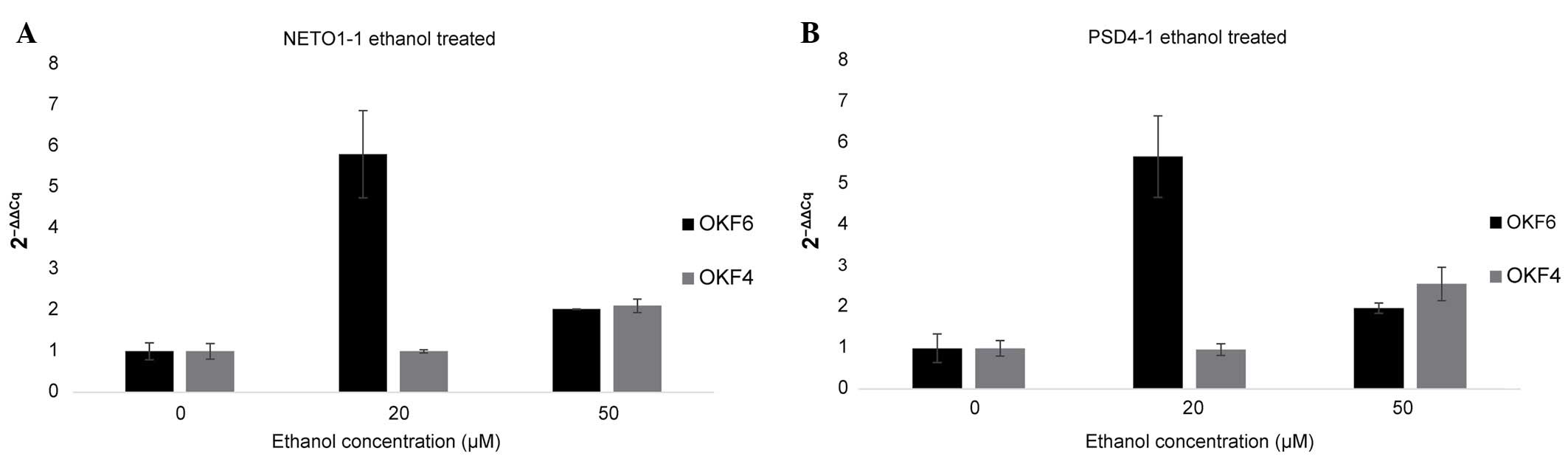
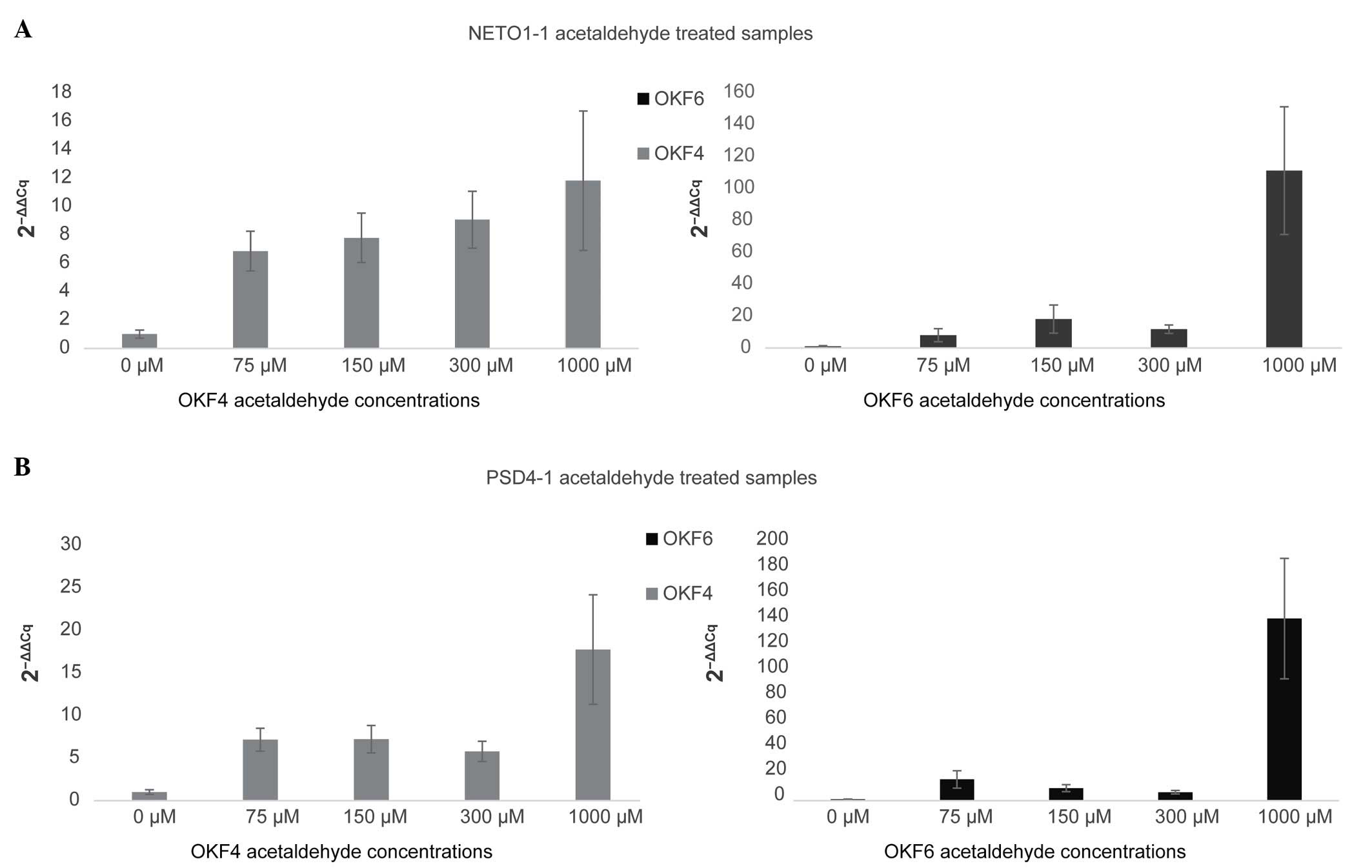
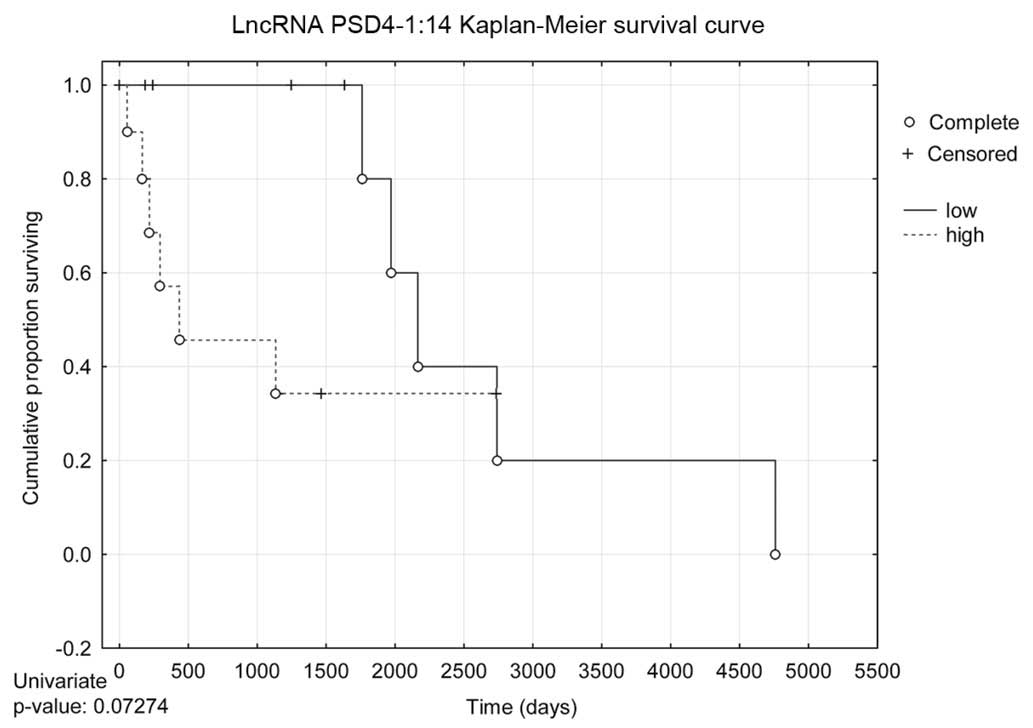
|
1
|
Hashibe M, Brennan P, Benhamou S,
Castellsague X, Chen C, Curado MP, Dal Maso L, Daudt AW, Fabianova
E, Fernandez L, et al: Alcohol drinking in never users of tobacco,
cigarette smoking in never drinkers and the risk of head and neck
cancer: Pooled analysis in the International Head And Neck Cancer
Epidemiology Consortium. J Natl Cancer Inst. 99:777–789. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Boffetta P and Hashibe M: Alcohol and
cancer. Lancet Oncol. 7:149–156. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Homann N, Jousimies-Somer H, Jokelainen K,
Heine R and Salaspuro M: High acetaldehyde levels in saliva after
ethanol consumption: Methodological aspects and pathogenetic
implications. Carcinogenesis. 18:1739–1743. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Rahimy E, Kuo SZ and Ongkeko WM:
Evaluation of non-coding RNAs as potential targets in head and neck
squamous cell carcinoma cancer stem cells. Curr Drug Targets.
15:1247–1260. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Deng G and Sui G: Noncoding RNA in
oncogenesis: A new era of identifying key players. Int J Mol Sci.
14:18319–18349. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Reilly M: Role of non-coding RNAs in the
neuroadaptation to alcoholism and fetal alcohol exposure. Front
Genet. 3:702012. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Laufer BI, Mantha K, Kleiber ML, Diehl EJ,
Addison SM and Singh SM: Long-lasting alterations to DNA
methylation and ncRNAs could underlie the effects of fetal alcohol
exposure in mice. Dis Model Mech. 6:977–992. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Spizzo R, Almeida MI, Colombatti A and
Calin GA: Long non-coding RNAs and cancer: A new frontier of
translational research? Oncogene. 31:4577–4587. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Dinger ME, Amaral PP, Mercer TR, Pang KC,
Bruce SJ, Gardiner BB, Askarian-Amiri ME, Ru K, Soldà G, Simons C,
et al: Long noncoding RNAs in mouse embryonic stem cell
pluripotency and differentiation. Genome Res. 18:1433–1445. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Guttman M, Amit I, Garber M, French C, Lin
MF, Feldser D, Huarte M, Zuk O, Carey BW, Cassady JP, et al:
Chromatin signature reveals over a thousand highly conserved large
non-coding RNAs in mammals. Nature. 458:223–227. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Saxena A and Carninci P: Long non-coding
RNA modifies chromatin: Epigenetic silencing by long non-coding
RNAs. BioEssays. 33:830–839. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Rinn JL and Chang HY: Genome regulation by
long noncoding RNAs. Annu Rev Biochem. 81:145–166. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Volders PJ, Helsens K, Wang X, Menten B,
Martens L, Gevaert K, Vandesompele J and Mestdagh P: LNCipedia: A
database for annotated human lncRNA transcript sequences and
structures. Nucleic Acids Res. 41:(Database issue). D246–D251.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Quinlan AR and Hall IM: BEDTools: A
flexible suite of utilities for comparing genomic features.
Bioinformatics. 26:841–842. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Robinson MD, McCarthy DJ and Smyth GK:
edgeR: A Bioconductor package for differential expression analysis
of digital gene expression data. Bioinformatics. 26:139–140. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Homann N, Tillonen J, Meurman JH,
Rintamäki H, Lindqvist C, Rautio M, Jousimies-Somer H and Salaspuro
M: Increased salivary acetaldehyde levels in heavy drinkers and
smokers: A microbiological approach to oral cavity cancer.
Carcinogenesis. 21:663–668. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2-δδCT method. Methods 25.4. 402–408. 2001.
|
|
18
|
Prensner JR, Iyer MK, Balbin OA,
Dhanasekaran SM, Cao Q, Brenner JC, Laxman B, Asangani IA, Grasso
CS, Kominsky HD, et al: Transcriptome sequencing across a prostate
cancer cohort identifies PCAT-1, an unannotated lincRNA implicated
in disease progression. Nat Biotechnol. 29:742–749. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Gupta RA, Shah N, Wang KC, Kim J, Horlings
HM, Wong DJ, Tsai MC, Hung T, Argani P, Rinn JL, et al: Long
non-coding RNA HOTAIR reprograms chromatin state to promote cancer
metastasis. Nature. 464:1071–1076. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Yang QQ and Deng YF: Long non-coding RNAs
as novel biomarkers and therapeutic targets in head and neck
cancers. Int J Clin Exp Pathol. 7:1286–1292. 2014.PubMed/NCBI
|
|
21
|
Morris KV: Long antisense non-coding RNAs
function to direct epigenetic complexes that regulate transcription
in human cells. Epigenetics. 4:296–301. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Carrieri C, Cimatti L, Biagioli M, Beugnet
A, Zucchelli S, Fedele S, Pesce E, Ferrer I, Collavin L, Santoro C,
et al: Long non-coding antisense RNA controls Uchl1 translation
through an embedded SINEB2 repeat. Nature. 491:454–457. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Geisler S and Coller J: RNA in unexpected
places: Long non-coding RNA functions in diverse cellular contexts.
Nat Rev Mol Cell Biol. 14:699–712. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Kent WJ, Sugnet CW, Furey TS, Roskin KM,
Pringle TH, Zahler AM and Haussler D: The human genome browser at
UCSC. Genome Res. 12:996–1006. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
di Magliano M Pasca, Di Lauro R and
Zannini M: Pax8 has a key role in thyroid cell differentiation.
Proc Natl Acad Sci USA. 97:13144–13149. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Narlis M, Grote D, Gaitan Y, Boualia SK
and Bouchard M: Pax2 and pax8 regulate branching morphogenesis and
nephron differentiation in the developing kidney. J Am Soc Nephrol.
18:1121–1129. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Muratovska A, Zhou C, He S, Goodyer P and
Eccles MR: Paired-Box genes are frequently expressed in cancer and
often required for cancer cell survival. Oncogene. 22:7989–7997.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Laury AR, Perets R, Piao H, Krane JF,
Barletta JA, French C, Chirieac LR, Lis R, Loda M, Hornick JL, et
al: A comprehensive analysis of PAX8 expression in human epithelial
tumors. Am J Surg Pathol. 35:816–826. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Liliac L, Carcangiu ML, Canevari S,
Căruntu ID, Apostol DG Ciobanu, Danciu M, Onofriescu M and Amălinei
C: The value of PAX8 and WT1 molecules in ovarian cancer diagnosis.
Rom J Morphol Embryol. 54:17–27. 2013.PubMed/NCBI
|
|
30
|
Brunner AH, Riss P, Heinze G, Meltzow E
and Brustmann H: Immunoexpression of PAX 8 in endometrial cancer:
Relation to high-grade carcinoma and p53. Int J Gynecol Pathol.
30:569–575. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Xiang L and Kong B: PAX8 is a novel marker
for differentiating between various types of tumor, particularly
ovarian epithelial carcinomas. Oncol Lett. 5:735–738.
2013.PubMed/NCBI
|