Introduction
Cervical cancer is the fourth leading cause of
cancer-associated mortality in females worldwide (1). The development of invasive cervical
cancer is strongly associated with human papillomavirus (HPV)
infection (2,3). The presence of high-risk HPV DNA, viral
oncogene expression (E6 and E7) and interaction of viral
oncoproteins with growth-regulating host-cell proteins has been
established as the major risk factors for cervical cancer
development (3). A well-known
consequence of deregulated expression of E6 and E7 is chromosome
instability, which contributes to the accumulation of aberrations
in host cell genes over time (4,5). Following
HPV infection, cervical cancer develops through a series of
subsequent steps, including development of precancerous lesions,
cervical intraepithelial neoplasia (CIN) grades 1–3 (CIN 1–3) and
progression to cervical cancer (6).
To overcome the limitations of morphological diagnosis, molecular
diagnostic tests have been developed as a complementary form of
testing, and HPV tests have been introduced into different testing
algorithms, particularly in countries with underdeveloped health
systems (7). However, it is known
that only a small fraction of patients with high-risk HPV infection
develop clinically relevant cervical lesions, and usually these
viruses are eventually cleared from the tissue (7,8).
Therefore, broad application of cervical cytology screening has
been associated with a dramatic reduction in cervical cancer
incidence and mortality (9). The
Bethesda system, used to communicate accurately the risk of
cervical cancer, enables to classify cytological samples into six
categories (10). The second most
common abnormal cytology results are low-grade squamous
intraepithelial lesions (LSILs), and the risk of CIN 2–3+ at
initial colposcopy following an LSIL result is 15–30% in the
majority of studies (11,12). CIN 2 or 3 has been reported in ≥70% of
women with cytology results of high-grade SILs (HSILs) (13). Since the sensitivity range of
conventional cytology is very broad (30–70%), it has limited
efficacy as a single screening method, and all abnormal cytology
results must be evaluated by histopathology (11,12). The
combination of cytology and HPV testing has markedly increased the
sensitivity of early detection of cervical cancer (12).
Infection with high-risk HPV is not sufficient for
cancer development, and the clearance of HPV infection is mediated
by the hosts' immune system, particularly by migration of
Langerhans cells (LCs) within the infected epithelium (14). LCs interact with keratinocytes trough
E-cadherin-mediated contact (15),
which is important for maintaining the immune response during
chronic HPV infection (14). The
deficit of molecules responsible for adhesion may be important in
the development of cervical cancer (16). E-cadherin, encoded by the cadherin 1
(CDH1) gene, is a transmembrane glycoprotein localized at
the surface of epithelial cells, and plays a pivotal role in
cell-cell adhesion dependent on calcium ions (Ca2+)
(17). E-cadherin is important for
the maintenance of normal tissue architecture (18), and therefore, it is considered as a
suppressor of invasiveness and metastasis (19).
Intensive studies on multiple types of human cancer
detected reduced or lost expression of E-cadherin, thus,
disturbance in E-cadherin expression may be one of the main events
in the early and late steps of cancer development (20). It is known that the expression of
numerous genes is affected by the presence of hypermethylation of
cytosine residues within CpG islands of the promoter region,
resulting in loss of function or inactivation of tumor suppressor
genes (21,22). Aberrant methylation patterns have been
described for a diverse number of tumor suppressor genes in CIN
lesions and in cervical cancer (21–23). One
of the most frequently methylated genes in transforming CIN lesions
is CDH1 (24). The comparable
CDH1 methylation frequency in primary breast tumors and
paired sentinel lymph node metastases indicates its important role
in the development of metastasis, which may be clinically used for
patient prognosis and for predicting early regional metastases
(19). CDH1 gene
hypermethylation was detected in ~51.1% of cervical cancer tissue
samples (24).
The aim of the present study was to investigate the
methylation pattern of the CDH1 promoter in order to
identify potential novel factors involved in cervical
carcinogenesis using only cytological samples, which could
contribute to an improved sensitive detection of early cervical
cancer. However, the current study did not identify any association
between CDH1 promoter hypermethylation, CDH1 gene
expression or HPV infection in cytological cervical specimens
obtained from patients with different stages of SIL or
carcinoma.
Materials and methods
Patients and clinicopathological
findings
Cervical specimens were obtained from 93 patients
with cervical lesions who underwent colposcopy or surgical
treatment of the cervical lesion at the Department of Obstetrics
and Gynaecology of Jessenius Faculty of Medicine in Martin (Martin,
Slovakia) between January 2010 and August 2013. Clinical diagnosis
was verified by histological examination. Other cervical specimens
were collected from 47 patients with normal cervical cytology
(controls), who were HPV-negative and had no previous history of
cervical lesion treatment. All patients agreed to be included in
the study and signed the informed consent form, which was approved
by the Regional Ethical Committee at the Jessenius Faculty of
Medicine, Comenius University in Bratislava (Martin, Slovakia).
Cervical samples were collected by Dacron™-tipped swabs (BD
Biosciences, Franklin Lakes, NJ, USA), and transferred to the
medium used for transportation. Cytological samples were classified
according to the Bethesda classification of 2001 (10). The present study included 34 LSIL
samples, 46 HSIL samples and 13 invasive squamous cervical cancer
(SCC) samples. The presence of the HPV genotype was determined by a
method described previously (25).
Nucleic acid extraction and bisulfite
conversion
Nucleic acids were extracted from cervical cells
using MasterPure Complete DNA and RNA Purification kits (Epicentre,
Madison, WI, USA). RNA was treated with DNase to eliminate DNA
contamination, and all samples were stored at −80°C. DNA was
quantified by ultraviolet absorption, and 1–2 µg were used for
bisulfite conversion, where unmethylated cytosines were converted
to uracil using an EpiTect Bisulfite kit (Qiagen GmbH, Hilden,
Germany) according to the manufacturer's protocol.
Methylation-specific polymerase chain
reaction (PCR) (MSP)
The methylation status of the CDH1 promoter
was investigated by MSP using a nested PCR approach. In the
first step of the nested PCR, degenerated primers were used that
flank the CpG-rich promoter region; do not discriminate between
methylated and unmethylated nucleotides; and cover nucleotides −57
to +110 around the translational start region of CDH1. The
PCR products of the first step were diluted 1:1,000 and subjected
to the second step of MSP, which applied sets of specific primers
for unmethylated or methylated DNA. The sequences of the primers
used (which are shown in Table I)
were previously published in the literature (26). The first step of the nested PCR was
conducted in a 25-µl total reaction volume, containing Taq DNA
polymerase (Roche Diagnostics, Indianapolis, IN, USA), 10X PCR
buffer, 2.5 mmol/l MgCl2, 0.5 mmol/l of each of the four
deoxynucleotides (Applied Biosystems; Thermo Fisher Scientific,
Inc., Waltham, MA, USA) and 10 pmol/l of each forward and reverse
primer for methylated and unmethylated DNA. The PCR was performed
at 95°C for 10 min, followed by 25 cycles of 95°C for 30 sec, 62°C
for 30 sec and 72°C for 30 sec. PCR was performed in a thermocycler
with an annealing temperature of 60°C. The PCR products were
analyzed on a 1.75% agarose gel with ethidium bromide staining. The
first-step PCR products were diluted 500-fold, and 1 µl of this
dilution was added to the second PCR step in a 25-µl reaction
volume, containing specific primers for methylated or unmethylated
alleles. Amplification was performed in 25 cycles with an annealing
temperature of 62°C. Similarly, 5 µl of PCR products were loaded
onto 1.75% agarose gels with ethidium bromide staining for
analysis.
 | Table I.Specific primers used in MSP and
pyrosequencing, and sizes of the PCR products. |
Table I.
Specific primers used in MSP and
pyrosequencing, and sizes of the PCR products.
| Type of primer | Sequence | Size (bp) | Number of analyzed
CpGs |
|---|
| MSP primer
setsa |
|
|
|
|
External primer set |
|
|
|
|
Forward |
5′-GTGTTTTYGGGGTTTATTTGGTTGT-3′ | 186 |
|
|
Reverse |
5′-TACRACTCCAAAAACCCATAACTAACC-3′ |
|
|
|
Internal methylated primer
set |
|
|
|
|
Forward |
5′-TGTAGTTACGTATTTATTTTTAGTGGCGTC-3′ | 112 |
|
|
Reverse |
5′-CGAATACGATCGAATCGAACCG-3′ |
|
|
|
Internal unmethylated primer
set |
|
|
|
|
Forward |
5′-TGGTTGTAGTTATGTATTTATTTTTAGTGGTGTT-3′ | 120 |
|
|
Reverse |
5′-ACACCAAATACAATCAAATCAAACCAAA-3′ |
|
|
| Pyrosequencing
primer sets |
|
|
|
| PCR
primer set |
|
|
|
|
Forward |
5′-GATTGGTTGTGGTCGGTAGGTGAATTTT-3′ |
235 | 19 |
|
Reverse |
5′-btn-ACTCCAAAAACCCATAACTAACC-3′ |
|
|
|
Pyrosequencing primers |
|
|
|
|
Sequencing primer
1 |
5′-GTAGGTGAATTTTTAGTTAATTAG-3′ |
| 7 |
|
Sequencing primer
2 |
5′-GTTTGCGGAAGTTAGTTTAGATT-3′ |
| 11 |
|
Sequencing primer
3 |
5′-GTGTTTTCGGGGTTTATTTGGTTGT-3′ |
| 5 |
Quantitative pyrosequencing
Pyrosequencing was used to determine the percentage
of methylation of 19 CpG islands within the minimal promoter region
of the CDH1 gene (−68 to +124 bp relative to the
transcription start site). In total, 20 ng of bisulfite-converted
DNA was amplified using PyroMark PCR kit (Qiagen GmbH) and a primer
set with biotin-labelled reverse primer. The PCR primer set and the
sequencing primers (Table I) were
designed using PyroMark Assay Design software version 2.0.1.15
(Qiagen GmbH), covering nucleotides from −57 to +116 around the
CDH1 translational start site. Pyrosequencing was performed
according to the manufacturer's protocol, using three assays with 4
pmol of the respective sequencing primer on a PyroMark Q96 ID
System (Qiagen GmbH) with PyroMark Gold Q96 Reagents. Target CpGs
were evaluated with the instrument software (PyroMark Q96 software
version 2.5.8; Qiagen GmbH), which calculates the proportion of
methylation at each CpG site as a C/T ratio according to the peak
height. All replicates contained dilution series of control
methylated DNA (0, 25, 50, 75 and 100%) mixed with unmethylated DNA
following bisulfite conversion (Qiagen GmbH).
CDH1 gene expression analysis
The effect of CDH1 gene hypermethylation on
CDH1 gene expression was evaluated by relative
quantification. For CDH1 gene expression analysis,
complementary (c)DNA was synthesized from 1 µg of total RNA using
the High-Capacity cDNA Reverse Transcription kit (Applied
Biosystems; Thermo Fisher Scientific, Inc.) in a total volume of 20
µl. Multiplex quantitative PCR reactions were performed in
triplicate in a final volume of 20 µl using 50 ng of cDNA,
TaqMan® Gene Expression Master Mix (Thermo Fisher
Scientific, Inc.) and TaqMan assays Hs01013959_m1 (CDH1) and
Hs99999903_m1 (actin beta; ACTB; Thermo Fisher Scientific,
Inc.). Relative gene expression of CDH1 was normalized to
that of the endogenous control ACTB. Both assays
(Hs01013959_m1 and Hs99999903_m1) were verified using the standard
curve method and normalized to ACTB.
Statistical analysis
All statistical analyses were conducted in R version
3.2.1 (www.r-project.org), with the aid of
libraries MASS (cran.r-project.org/web/packages/MASS/index.html),
RVAideMemoire (cran.r-project.org/web/packages/RVAideMemoire/index.html;
version 0.9–57), ridge (cran.r-project.org/src/contrib/Archive/ridge/;
version 2.1–3), earth (cran.r-project.org/web/packages/earth/index.html;
version 4.4.4), robustbase (cran.r-project.org/web/packages/robustbase/index.html;
version 0.92–5), mgcv (cran.r-project.org/web/packages/mgcv/index.html),
randomForest (cran.r-project.org/web/packages/randomForest/index.html)
and Deducer (www.jstatsoft.org/article/view/v049i08). Plots were
produced by R version 3.2.1 libraries ggplot2 (ggplot2.org/) and plotmo (cran.r-project.org/web/packages/plotmo/index.html;
verions 3.1.4), and multiple comparisons with the binomial exact
test with false discovery rate (FDR) correction were conducted.
P<0.05 was considered to indicate a statistically significant
difference.
Results
Patients and clinicopathological
findings
The promoter hypermethylation status of the
CDH1 gene was examined in 47 specimens of normal cervical
squamous epithelium and 93 specimens of SIL or carcinoma. The
average age of patients with normal cervical epithelium and
cervical lesion was 52.4 years (range, 25–77 years) and 41 years
(range, 18–75 years), respectively. HPV infection was detected in
67.6% (23/34) of LSIL, 84.8% (39/46) of HSIL and 84.6% (11/13) of
SCC. Control specimens with negative cytology were also negative
for HPV infection.
Detection of CDH1 promoter
hypermethylation by MSP
In order to determine the methylation status of the
CDH1 promoter, 93 samples from patients with cervical
dysplasia and 47 control samples were analyzed by MSP. Methylation
was detected in 0% (0/47) of normal cervical epithelium samples,
and, of 93 cervical lesions, 20.6% (7/34) of LSIL, 21.7% (10/46) of
HSIL and 46.2% (6/13) of SCC exhibited methylation of CpG islands
in the CDH1 promoter. Using the non-parametric χ2
test, it was demonstrated that the methylation in LSIL, HSIL and
SCC was significantly different from that in controls [P=0.01774
and 95% confidence interval (CI), 0.0172–0.3520 for LSIL; P=0.00914
and 95% CI, 0.0485–0.03438 for HSIL; and P=0.00011 and 95% CI,
0.1100–0.7635 for SSC]. However, multiple comparison by the
binomial exact test with FDR correction supported the claim that
the presence of CDH1 methylation in SCC is equally probable
as its absence (presence of methylation, <50% of SCC; P=1.0000),
which supports the evidence that methylation occurs rather randomly
in these cells and is not a marker of SCC. The observed relative
frequencies of methylation in LSIL and HSIL were half of the
expected value, and the unequal probability was statistically
significant (P=0.0012 and P=0.0005, respectively), which suggests
that methylation in this lesions is not randomly, and may represent
an early event.
The correlation of CDH1 methylation with age
(threshold for age, 50 years) demonstrated a statistically
significant difference in the control group and in the patients
groups when χ2 test with Yates' continuity correction
was used. CDH1 promoter hypermethylation in the control
group was almost absent (P=0.5000; one-sided 95% CI, −1.0000 to
0.055573), thus demonstrating no correlation with patient's age.
However, CDH1 methylation was more frequent in patients older than
50 years (P=0.01085; one-sided 95% CI, −1.0000 to −0.0514).
When the frequency of HPV infection was compared in
cervical lesions with methylated and unmethylated CDH1
promoters, no significant difference between both groups was
observed (χ2 test with Yates' continuity correction;
P=0.6270 and one-sided 95% CI, for the probabilities of HPV
infection −1.0000 to 0.2596). No change was observed when the
cervical specimens were divided according to the severity of SIL
(data not shown).
Detection of CDH1 promoter
hypermethylation by pyrosequencing
To verify the presence of CDH1 promoter
hypermethylation and to specify the affected nucleotides,
quantitative pyrosequencing was used to assess the methylation
level in 19 CpG islands of the CDH1 promoter region, which
are covered by a previous MSP assay in 38 (all MSP-negative)
control samples and in 43 (17 MSP-positive and 26 MSP-negative)
samples from cervical lesions. Other samples were excluded from the
analysis due to the low quality of the pyrosequencing data. The
average of percentage methylation for each of the 19 CpG islands
was calculated for cervical lesions and control samples. Certain
level of basal methylation was detected in control samples that
were unmethylated by MSP. However, the average methylation levels
of CpG islands in cervical lesions were significantly higher
(P<0.05; Table II) for all
nucleotides, with the exception of nucleotides −52 and −36,
compared with the control group. When cervical lesions were divided
according to severity (LSIL, HSIL and SCC) and compared with
control samples, the mean methylation status for each CpG island
was significantly higher in cervical lesions for the nucleotides
described in Table II with
significant P-values (as marked by asterisks).
 | Table II.Significance of the mean methylation
level of investigated CpG islands according to the severity of the
cervical lesion, compared with the mean methylation level of the
control samples. |
Table II.
Significance of the mean methylation
level of investigated CpG islands according to the severity of the
cervical lesion, compared with the mean methylation level of the
control samples.
| Nucleotide | LSILa | HSILa | SCCa |
|---|
| −57 | 0.0133*;
0.0047 | 0.0226*;
0.0023 | 0.0961;
−0.0125 |
| −52 | 0.4386;
−0.0139 | 0.0384*;
0.0010 | 0.1943;
−0.0299 |
| −45 | 0.0310*;
0.0019 | 0.0562;
−0.0004 | 0.0259*;
0.0087 |
| −36 | 0.1371;
−0.0096 | 0.2003;
−0.0105 | 0.2236;
−0.0150 |
| −13 | 0.1587;
−0.8431 | 0.0177*;
0.0093 | 0.1098;
−0.0125 |
| +6 | 0.0760;
−0.0023 | 0.0483*;
0.0013 | 0.1890;
−0.0137 |
| +9 | 0.0485*;
0.0002 | 0.0097*;
0.0053 | 0.0695;
−0.0039 |
| +36 | 0.0303*;
0.0036 | 0.1038;
−0.0053 | 0.1442;
−0.0178 |
| +60 | 0.0371*;
0.0022 | 0.0587;
−0.0013 | 0.1230;
−0.0133 |
| +70 | 0.0300*;
0.0040 | 0.0302*;
0.0039 | 0.1380;
−0.0179 |
| +75 | 0.0027*;
0.0121 | 0.0096*;
0.0073 | 0.1298;
−0.0150 |
| +80 | 0.0033*;
0.0159 | 0.0317*;
0.0032 | 0.1056;
−0.0118 |
| +84 | 0.0461*;
0.0004 | 0.0998;
−0.0039 | 0.1767;
−0.0262 |
| +90 | 0.0063*;
0.0129 | 0.0575;
−0.0010 | 0.1123;
−0.0142 |
| +93 | 0.0245*;
0.0027 | 0.0129*;
0.0038 | 0.2644;
−0.0281 |
| +103 | 0.1904;
−0.0161 | 0.0593;
−0.0015 | 0.0867;
−0.0098 |
| +107 | 0.0872;
−0.0034 | 0.0900;
−0.0036 | 0.1218;
−0.0167 |
| +110 | 0.0050*;
0.0057 | 0.0079*;
0.0035 | 0.1123;
−0.0087 |
| +116 | 0.0563;
−0.0009 | 0.0716;
−0.0027 | 0.1199;
−0.0189 |
The methylation level of MSP-methylated cervical
lesions was also significantly higher in nucleotides −57, −45, −13,
+9, +70, +75, +80, +93 and +110 compared with MSP-unmethylated
control samples (Table III).
 | Table III.Significance of the mean methylation
of MSP-methylated cervical lesions compared with that of
MSP-unmethylated control samples for the investigated
nucleotides. |
Table III.
Significance of the mean methylation
of MSP-methylated cervical lesions compared with that of
MSP-unmethylated control samples for the investigated
nucleotides.
| Nucleotide | P-value and lower
bound of 95% CI |
|---|
| −57 | 0.0409*;
0.0017 |
| −52 | 0.0648;
−0.0025 |
| −45 | 0.0093*;
0.0107 |
| −36 | 0.1375;
−0.0080 |
| −13 | 0.0196*;
0.0090 |
| +6 | 0.0654;
−0.0017 |
| +9 | 0.0462*;
0.0005 |
| +36 | 0.1032;
−0.0069 |
| +60 | 0.1086;
−0.0078 |
| +70 | 0.0358*;
0.0033 |
| +75 | 0.0381*;
0.0021 |
| +80 | 0.0278*;
0.0053 |
| +84 | 0.0683;
−0.0031 |
| +90 | 0.0671;
−0.0029 |
| +93 | 0.0438*;
0.0010 |
| +103 | 0.1383;
−0.0127 |
| +107 | 0.1963;
−0.0153 |
| +110 | 0.0243*;
0.0034 |
| +116 | 0.1256;
−0.0114 |
Association between MSP and
pyrosequencing results
To validate the results obtained in MSP and
pyrosequencing analyses, the results of both methods were processed
in a random forest analysis with MSP as response. This analysis
identified the most important nucleotides, and the realistic
(out-of-bag) estimate of the misclassification error was observed
to be 37.5%. Subsequent multivariate logistic regression and Akaike
information criterion (AIC) model selection narrowed down the set
of important nucleotides to nucleotides −45, +70, +90 and +107
(P=0.0122, 0.0819, 0.1749 and 0.1240, respectively). The selected
model was used for the receiver operating characteristic curve
construction (Fig. 1), with an area
under the curve of 79.6%.
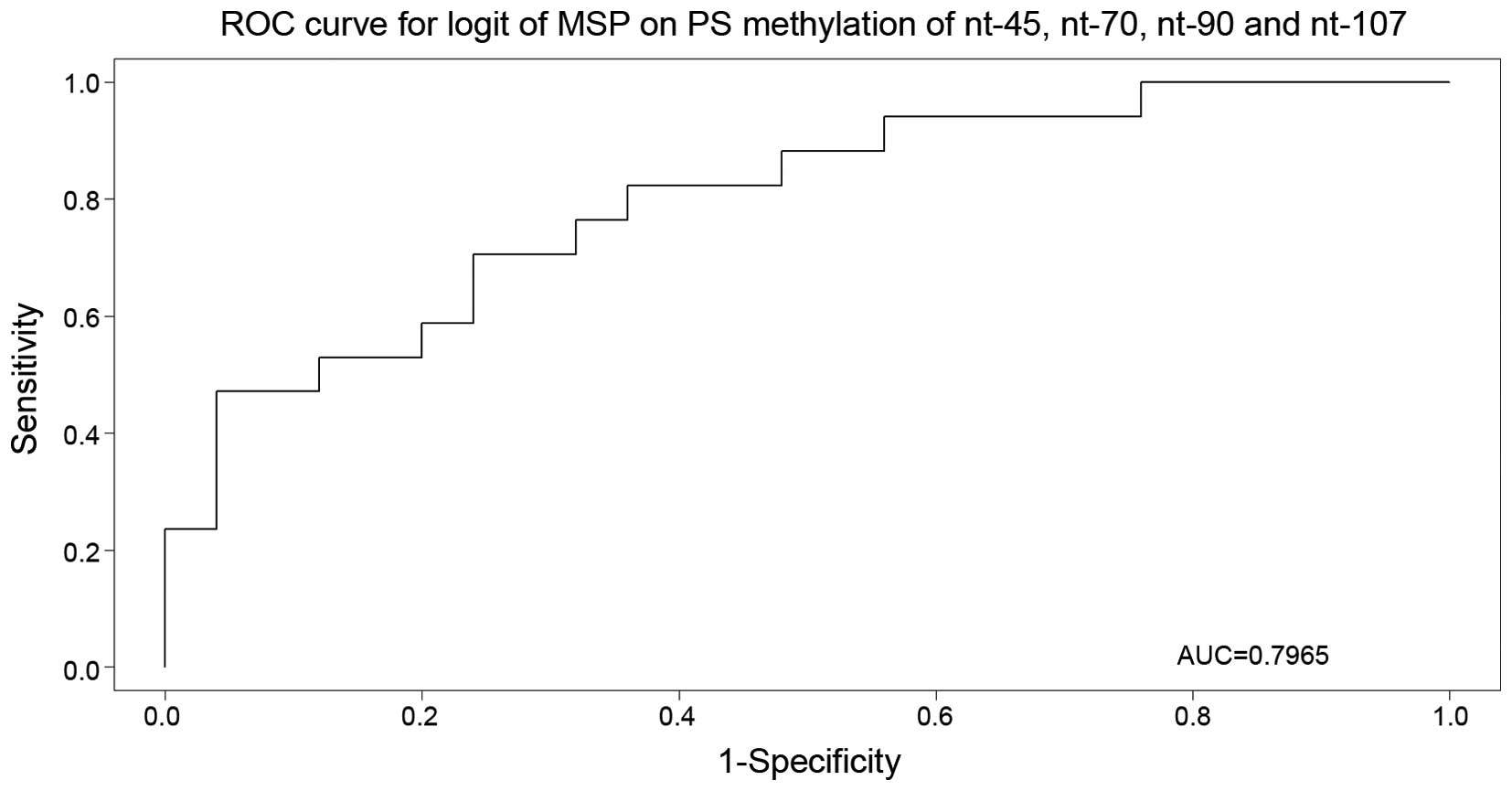 | Figure 1.Receiver operating characteristic
curve for nucleotides −45, +70, +90 and +107 (P=0.0122, 0.0819,
0.1749 and 0.1240, respectively), with area under the curve equal
to 79.6%. ROC, receiver operating characteristic; AUC, area under
the curve; MSP, methylation-specific polymerase chain reaction; nt,
nucleotide; logit, logistic regression; PS, pyrosequencing. |
Effect of CDH1 hypermethylation on
CDH1 gene expression
The observed methylation pattern of the CDH1
promoter region was compared with the relative CDH1 gene
expression. The mean level of CDH1 gene expression was
2.4153 in the control group (negative for intraepithelial lesion or
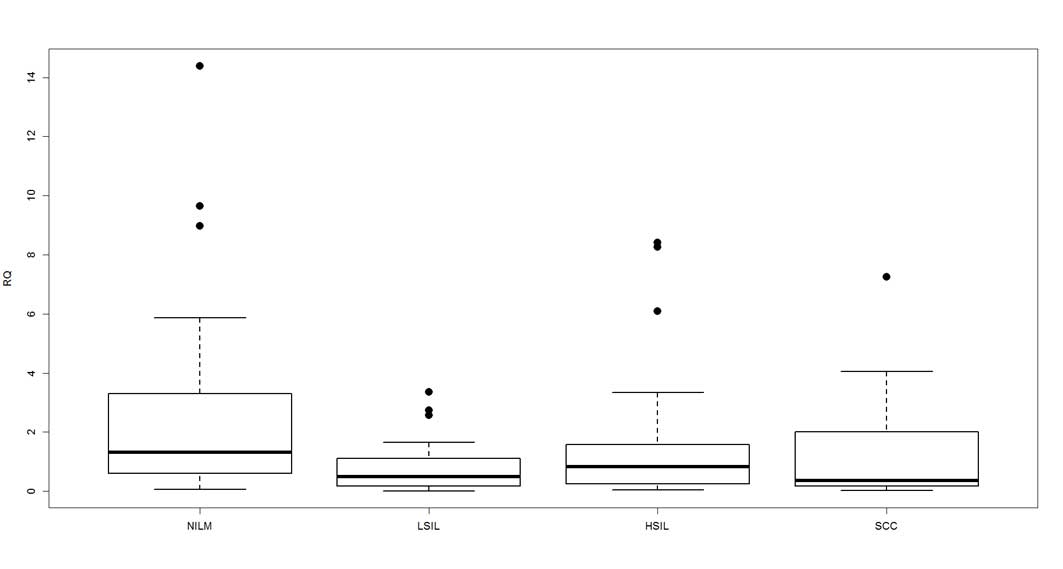
dysplasia), 0.7944 in LSIL, 1.3402 in HSIL and 1.5110 in SCC
(Fig. 2). The Kruskal-Wallis rank sum
test determined that the mean expression values were significantly
different among the groups (P=0.0162).
The present study also investigated the association
between the mean level of CDH1 gene expression and the
presence of HPV infection. It was observed that HPV infection had
no significant effect on CDH1 gene expression (P=0.8117; 95%
CI, -∞ to 1.0659). In HPV16 and 18-positive samples, information
about the oncogene E6 expression was also available. Overall, E6
expression did not have a significant effect on CDH1 gene
expression (P=0.3299; 95% CI, -∞ to 0.4519), although the presence
of E6 expression was significantly associated with decreased
CDH1 gene expression in the LSIL group (P=0.03596; 95% CI,
-∞ to 0.0445).
By means of multivariate regression model, the
association between CDH1 gene expression and methylation of
the nucleotides was explored separately for cervical lesions and
for normal cervical epithelium. Upon model building (exploratory
analysis, outlier detection, treatment of multicollinearity by
ridge regression, model diagnostics, robust regression and model
selection by AIC), it was observed that the expression of the
CDH1 gene was decreased in cervical lesions with a high
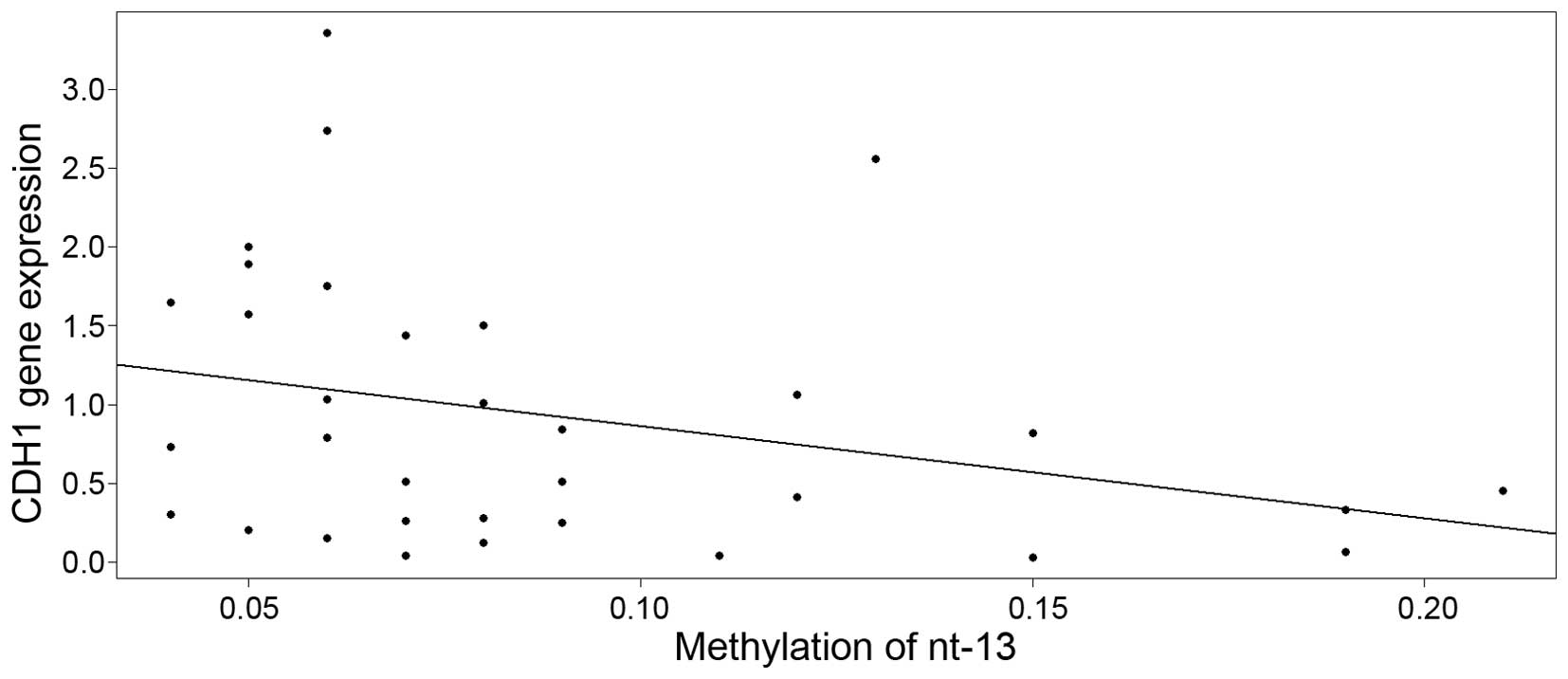
methylation level at nucleotide −13 (Fig.
3), with an estimate of descent of −5.8735 [95% CI, −12.4992 to
0.7522; standard error (SE)=3.2487] and intercept equal to 1.4494
(95% CI, −12.4992 to 0.7522; SE=0.3224). Although the slope was
only weakly significant (P=0.0803), the P-value cannot be taken at
face value due to the well-known post-model selection inferences
issues (27).
In control samples, model building with ridge
regression revealed that nucleotides +103 and +107 are the only
significant predictors. When fed into the multivariate regression
analysis, solely nucleotide +107 was revealed to be statistically
significant (P=0.04815), with a negative slope of −30.1142
(SE=14.7210) and an intercept of 2.5474 (SE=0.5961). As an
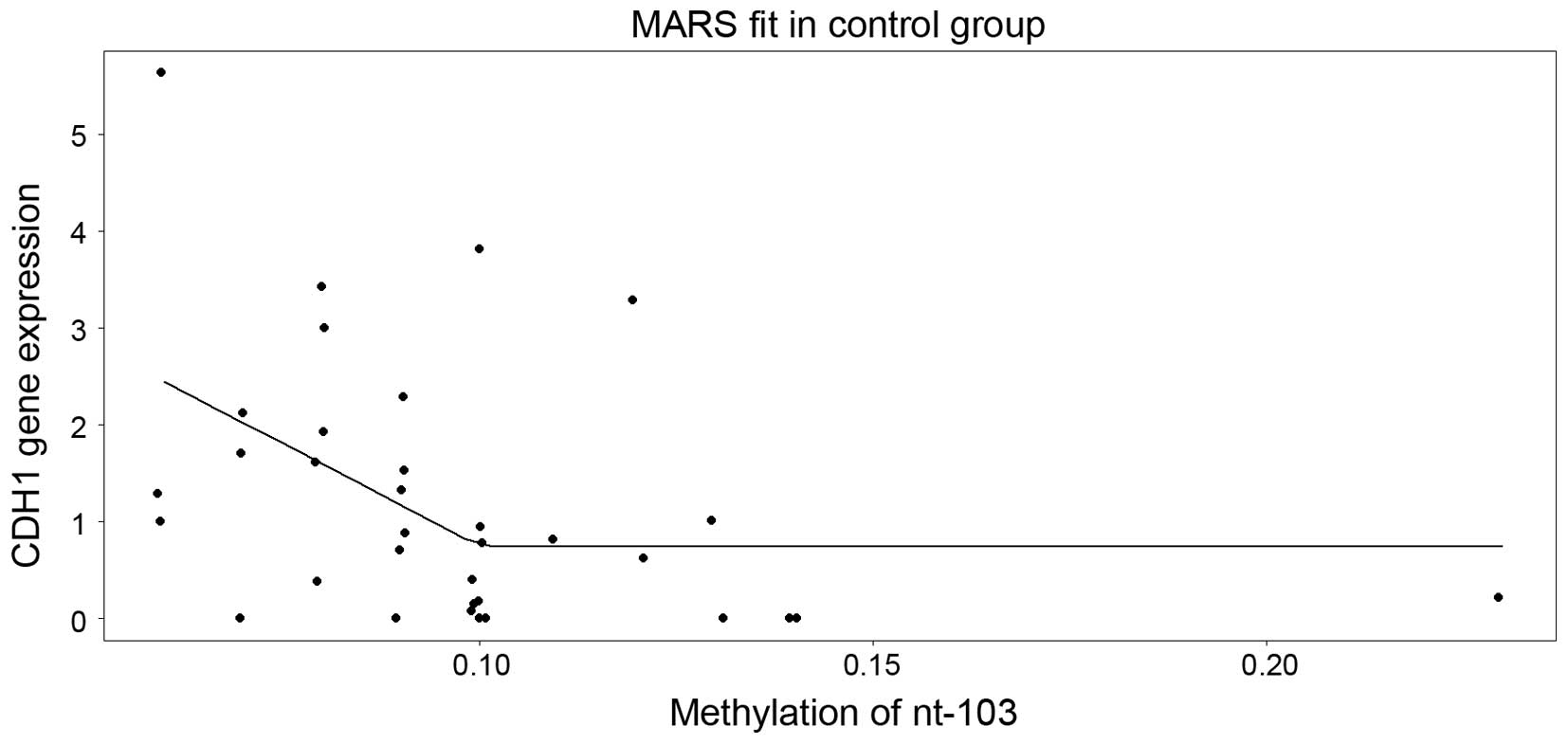
alternative to the general linear model, the multivariate adaptive
regression splines model was used, which among all the nucleotides,
it selected solely nucleotide +103 (Fig.
4). Of note, this model suggested that the level of expression
decreases as the methylation in nucleotide +103 increases up to
~10%, and above the cut-off, a saturation (plateau) appears.
Discussion
In present study, the methylation status of the
CDH1 promoter was investigated in cervical cells from
precursor lesions, which represent a source for detecting
biomarkers of relevance to cervical carcinogenesis. The CDH1
promoter was frequently reported to be methylated in numerous types
of gynecological cancer, including breast (28), ovarian (29), endometrial (30) and cervical cancer. CDH1 gene
hypermethylation is also detectable in the serum of patients with
cervical cancer (31). The majority
of studies to date have examined promoter methylation in tissue
sections or cell lines, while studies on cervical cytology
specimens were less frequent and had more different results for
CDH1 methylation (21–23). The selected promoter was observed to
be methylated in 58% of cervical cancer specimens and in 29% of
HSIL, although, these results, in a survey of 51 studies, were
observed to be dependent on the analysis method and the type of
clinical material used (23).
Similarly, the present study detected CDH1 methylation in
21.7% of HSIL, 46.2% of SCC and 20.6% of LSIL specimens. Previous
studies investigating the CDH1 methylation status of LSIL
specimens are uncommon; CDH1 methylation was not present
(32,33) or was only detected in a small
percentage of samples (11.3% of LSIL and 13.3% of normal cervical
epithelium) (34). The unequal
probability of CDH1 methylation in LSIL and HSIL was
statistically significant (P=0.0012 and P=0.0005, respectively),
which suggests that methylation in this lesions is not random and
may represent an early carcinogenesis event, in contrast to the
random methylation of CDH1 in SCC patients. The presence of
CDH1 methylation in cervical lesions represents just one of
all possible methylated targets. As it is generally known,
different genes can be methylated in cervical cancer, while the
combination of various methylated genes can act as a co-driver of
carcinogenesis (21–24). As a consequence, and due to its
specific role in cell adhesion, CDH1 should be
considered.
It has been postulated that DNA methylation is
age-related and usually occurs at age-related sites of the human
genome (35). Cells have a lower
threshold for malignant transformation and are more susceptible to
cancer when acquire methylation at age-related sites (35). In the present study, CDH1
methylation was more frequent in patients older than 50 years
(P=0.01085), indicating that the presence of promoter
hypermethylation could be age-related, as it has been reported for
other genes (36).
Cervical cancer is also associated with long-term
persistence of HPV infection, which may induce progression of
high-grade cervical dysplasia to cervical cancer, together with
aberrant DNA methylation in the host genome (37). HPV infection with high-risk HPV types
causes changes in the methylation status of cellular genes through
upregulation of DNA methyltransferases (37). Viral oncogenes can induce tumor
suppressor gene methylation (38), as
well as expression of E7 and E6, which results in a further
reduction in surface E-cadherin levels (39). A previous study by Flatley et
al (40) reported that high-risk
HPV infection may influence folate status and the frequency of
promoter methylation of three tumor suppressor genes (CDH1,
death-associated protein kinase and hypermethylated in cancer 1),
which increased with the progression of cervical neoplasia. In the
present study, HPV infection was not demonstrated to affect the
methylation pattern of the CDH1 gene (P=0.627; 95% CI, −1.00
to 0.2596) or the E6 expression of high-risk HPV genotypes
(P=0.3299; 95% CI, -∞ to −0.4519; data not shown). No correlation
between the promoter methylation status of the CDH1 gene and
the patients' clinicopathological parameters, including HPV
infection, phenotypic distribution or stage of the disease, was
observed in other studies (41,42). Thus,
the impact of HPV infection on DNA methylation of the host genome
still remains controversial.
To distinguish samples with methylated and
unmethylated CDH1 promoter, MSP was used as the first step
of the current study. Quantitative pyrosequencing was used for
monitoring the ratio of methylated CpG islands in a nucleotide
sequence. This provided additional information concerning the
methylation of CpG islands. The level of DNA methylation within the
CDH1 promoter was measured in normal cervical specimens and
in cervical lesions with various stages of dysplasia. MSP is a
widely used method to assess the methylation status of any group of
CpG sites within a CpG island without the requirement for
restriction enzymes, and exhibits a sensitivity of methylation
detection of 0.1% of alleles (43).
By pyrosequencing, the degree of methylation at several CpGs in
close proximity can be quantitatively measured (44). The methylation at each CpG position in
a sequence is determined from the ratio of T and C (44). However, the methylation values
measured by pyrosequencing in the present study ranged from 0 to 6%
in certain CpGs analyzed in unmethylated control DNA samples (data
not shown). Various authors recommend the use of a ≥10% threshold
of methylation to classify samples as methylated (45). However, the present study used
statistical analysis to differentiate between significantly
methylated and unmethylated CpGs in various types of cervical
dysplasia compared with the mean methylation level at each CpG
position in the control group (Table
II). The results confirmed that certain CpGs in MSP-methylated
samples had significantly higher methylation levels than those
detected in the unmethylated controls. Following a detailed
analysis of the CDH1 promoter sequence, two CpGs in the
internal forward primer (nucleotides −13 and +9) and three CpGs in
the internal reverse primer sequence (nucleotides +70, +75 and +80)
were observed to have a significant influence on the result of MSP.
When these nucleotides were used in the logistic regression for MSP
an AUC of 61.18% was achieved. If, instead, a logistic regression
model for MSP was constructed with use of all CpGs, the nucleotides
−45, +70, +90 and +107 appeared to be most important, leading to an
AUC of 79.6%. ROC curves for each nucleotide had weak AUC, and due
to the low number of samples in the study, no analysis for
prediction of high-grade lesion or cancer was performed. However,
studies evaluating the ability of DNA methylation levels to
identify cervical cancer cases usually use combination of genes and
clinicopathological features to improve AUC and to increase the
sensitivity and specificity of the test to identify cancer
(46).
In the last part of the present study, the level of
CDH1 gene expression was measured, and the association
between CDH1 hypermethylation at each CpG island and
CDH1 gene expression level was evaluated. The expression of
E-cadherin, as a major adhesion component of epithelial cells, has
been observed to be reduced or lost in epithelial tumor types by
promoter hypermethylation mechanisms (47). It has been also reported that the
presence and localization of cytoplasmic E-cadherin correlated with
CIN grade (48). In other types of
cancer, the degree of CpG methylation increased as the precancerous
conditions progressed (49). The
present study detected significantly reduced expression of the
CDH1 gene in SILs or cancers (P=0.0162) compared with
control samples. According to our observations, HPV infection had
no effect on the relative quantity of E-cadherin (P=0.8117).
However, E6 oncogene expression decreased the CDH1 gene
expression only in HPV16 or 18-positive LSILs (P=0.0359). The HPV
E6 protein has been shown to interact with cellular proteins; it
creates a complex with p53 and mediates its degradation by the
ubiquitin system (50). The E6
protein reduces the expression of cell-surface E-cadherin, and thus
has also a function in the control of cell-surface E-cadherin
expression and in the regulation of the cutaneous immune response
in virus-infected skin (51). A
previous study noticed that E-cadherin transcription regulation by
E6 is independent of direct methylation of the E-cadherin promoter
(39). Therefore, further studies are
required.
In statistical analysis investigating the influence
of methylation at each CpG position of the CDH1 promoter on
CDH1 gene expression, the present study revealed that
nucleotides +103 and +107 were the most influential ones on
CDH1 gene expression in control samples. These nucleotides
are localized near the CTCF binding site, and were observed to be
methylated also in the control cell line HCT116 (39). In the present study in SILs, the main
effect on CDH1 gene expression was exerted by nucleotide
−13, which is localized near the Snail binding site. Regulation of
E-cadherin gene expression in metastatic and non-metastatic cancer
cells demonstrated that methylation states, chromatin constraint
and Snail family transcription factors are important in the
downregulation of E-cadherin gene expression (52). The presence of DNA methylation sites
near the transcription factor binding sites could be required for
efficient transcriptional regulation of E-cadherin and other tumor
suppressor genes. However, further studies are required to
elucidate the molecular interaction of E-cadherin promoter
methylation and transcription factor binding (53).
In summary, E-cadherin expression during tumor
progression was observed to be downregulated by several mechanisms,
including genetic, epigenetic and transcriptional changes. The
present study confirmed that epigenetic changes such as DNA
methylation of the CDH1 promoter are frequent in LSILs.
Similarly, CDH1 methylation was observed to be present in
HSILs and in ~50% of cervical cancer specimens. CDH1 gene
expression was reduced during SIL progression in the present study;
however, the influence of HPV infection or HPV E6 expression on the
methylation pattern of the CDH1 gene or its gene expression
could not be confirmed. It was established that not all HPV
infected cervical dysplasia develop into cancer, indicating that
factors other than HPV viral proteins contribute to the progression
to cervical cancer (54). The current
findings support also the claim that methylation of the CDH1
gene is age-related, and therefore, older patients could be more
susceptible to cancer than younger ones. The important methylation
of the CDH1 promoter occurred near the transcription factor
binding sites, which suggests that methylation at these sites may
be an important event in the transcriptional regulation of
E-cadherin and other genes, although additional studies are
required to confirm this hypothesis. Inactivation of the E-cadherin
system by multiple mechanisms, including genetic and epigenetic
events, plays a significant role in both the early and late stages
of multistep carcinogenesis (49).
Acknowledgements
The present study was supported by the projects
‘Increasing Opportunities for Career Growth in Research and
Development in the Field of Medical Sciences’ of the Institute of
Experimental Pharmacology and Toxicology (ITMS; Bratislava,
Slovakia; grant no. 26110230067), VEGA (grant no., 1/0102/15) and
‘Molecular diagnosis of cervical cancer’ (ITMS; grant no.
26220220113), which were co-funded by the European Union and the
European Social Fund (Brussels, Belgium).
References
|
1
|
Arbyn M, Castellsagué X, de Sanjosé S,
Bruni L, Saraiya M, Bray F and Ferlay J: Worldwide burden of
cervical cancer in 2008. Am Oncol. 22:2675–2686. 2011.
|
|
2
|
Dürst M, Gissmann L, Ikenberg H and zur
Hausen H: A papillomavirus DNA from a cervical carcinoma and its
prevalence in cancer biopsy samples from different geographic
regions. Proc Natl Acad Sci. 80:3812–3815. 1983. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
zurHausen H: Papillomaviruses causing
cancer: Evasion from host-cell control in early events in
carcinogenesis. J Natl Cancer Inst. 92:690–698. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Korzeniewski N, Spardy N, Duensing A and
Duensing S: Genomic instability and cancer: Lessons learned from
human papillomaviruses. Cancer Lett. 305:113–122. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Visnovsky J, Kudela E, Farkasova A,
Balharek T, Krkoska M and Danko J: Amplification of TERT and TERC
genes in cervical intraepithelial neoplasia and cervical cancer.
Neuro Endocrinol Lett. 35:518–522. 2014.PubMed/NCBI
|
|
6
|
McCredie MR, Sharples KJ, Paul C, Baranyia
J, Medley G, Jones RW and Skegg DC: Natural history of cervical
neoplasia and risk of invasive cancer in women with cervical
intraepithelial neoplasia 3: A retrospective cohort study. Lancet
Oncol. 9:425–434. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Boone JD, Erickson BK and Huh WK: New
insights into cervical cancer screening. J Gynecol Oncol.
23:282–287. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Dunne EF and Markowitz LE: Genital human
papillomavirus infection. Clin Infect Dis. 43:624–629. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Tjalma WA: The ideal cervical cancer
screening recommendation for Belgium, an industrialized country in
Europe. Eur J Gynaecol Oncol. 35:211–218. 2014.PubMed/NCBI
|
|
10
|
Solomon D, Davey D, Kurman R, Moriarty A,
O'Connor D, Prey M, Raab S, Sherman M, Wilbur D and Wright T Jr:
The 2001 Bethesda system: Terminology for reporting results of
cervical cytology. JAMA. 287:2114–2119. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Bentley J, Bertrand M, Brydon L, Gagne H,
Hauck B, Mayrand MH, McFaul S, Power P, Schepansky A and
Straszak-Suri M: Colposcopic management of abnormal cervical
cytology and histology. J Obstet Gynaecol Can. 34:1188–1202. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Mayrand MH, Duarte-Franco E, Rodrigues I,
Walter SD, Hanley J, Ferenczy A, Ratnam S, Coutlee F and Franco EL:
Canadian Cervical Cancer Screening Trial Study Group: Human
papillomavirus DNA versus Papanicolaou screening tests for cervical
cancer. N Engl J Med. 357:1579–1588. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Agramunt S, Checa MÁ, Gonzáles-Comadrán M,
Larrazabai F, Arbós A, Alameda F, Mancebo G and Carreras R:
High-grade squamous intraepithelial lesion could be managed
conservatively in women up to 25 years: Results from a
retrospective cohort study. J Low Genit Tract Dis. 17:459–462.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Tindle RW: Immune evasion in human
papillomavirus-associated cervical cancer. Nat Rev Cancer. 2:59–65.
2002. View
Article : Google Scholar : PubMed/NCBI
|
|
15
|
Tang A, Amagai M, Granger LG, Stanley JR
and Udey MC: Adhesion of epidermal Langerhans cells to
keratinocytes mediated by E-cadherin. Nature. 361:82–85. 1993.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Hubert P, Caberg JH, Gilles C, Bousarghin
L, Franzen-Detrooz E, Boniver J and Delvenne P:
E-cadherin-dependent adhesion of dendritic and Langerhans cells to
keratinocytes is defective in cervical human
papillomavirus-associated (pre)neoplastic lesions. J Pathol.
206:346–355. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Oda H and Takeichi M: Evolution:
Structural and functional diversity of cadherin at the adherens
junction. J Cell Biol. 193:1137–1146. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Pećina-Slaus N: Tumor suppressor gene
E-cadherin and its role in normal and malignant cells. Cancer Cell
Int. 3:172003. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Sebova K, Zmetakova I, Bella V, Kajo K,
Stankovicova I, Kajabova V, Krivulcik T, Lasabova Z, Tomka M,
Galbavy S and Fridrichova I: RASSF1A and CDH1 hypermethylation as
potential epimarkers in breast cancer. Cancer Biomark. 10:13–26.
2011–2012.
|
|
20
|
Wijnhoven BP, Dinjens WN and Pignatelli M:
E-cadherin-catenin cell-cell adhesion complex and human cancer. Br
J Surg. 87:992–1005. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Wajed SA, Laird PW and DeMeester TR: DNA
methylation: An alternative pathway to cancer. Ann Surg. 234:10–20.
2001. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Baylin SB and Herman JG: DNA
hypermethylation in tumorigenesis: Epigenetics joins genetics.
Trends Genet. 16:168–174. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Wentzensen N, Sherman ME, Schiffman M and
Wang SS: Utility of methylation markers in cervical cancer early
detection: Appraisal of the state-of-the-science. Gynecol Oncol.
112:293–299. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Narayan G, Arias-Pulido H, Koul S, Vargas
H, Zhang FF, Villella J, Schneider A, Terry MB, Mansukhani M and
Murty VV: Frequent promoter methylation of CDH1, DAPK, RARB, and
HIC1 genes in carcinoma of cervix uteri: Its relationship to
clinical outcome. Mol Cancer. 2:242003. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Janusicova V, Mendelova A, Zubor P,
Kapustova I, Svecova I, Kudela E, Burjanivova T, Lasabova Z and
Danko J: mRNA expression in cervical specimens for determination of
severe dysplasia or worse in HPV-16/18-positive squamous lesions. J
Low Genit Tract Dis. 18:273–280. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
House MG, Guo M, Iacobuzio-Donahue C and
Herman JG: Molecular progression of promoter methylation in
intraductal papillary mucinous neoplasms (IPMN) of the pancreas.
Carcinogenesis. 24:193–198. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Leeb H and Pötscher BM: Model selection
and inference: Facts and fiction. Econometric Theory. 1:21–59.
2005.
|
|
28
|
Caldeira JR, Prando EC, Quevedo FC, Neto
FA, Rainho CA and Rogatto SR: CDH1 promoter hypermethylation and
E-cadherin protein expression in infiltrating breast cancer. BMC
Cancer. 6:482006. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Dhillon VS, Young AR, Husain SA and Aslam
M: Promoter hypermethylation of MGMT, CDH1, RAR-beta and SYK tumour
suppressor genes in granulosa cell tumours (GCTs) of ovarian
origin. Br J Cancer. 90:874–881. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Visnovsky J, Fiolka R, Kudela E, Slavik P,
Krkoska M, Lasabová Z and Danko J: Hypermethylation of selected
genes in endometrial carcinogenesis. Neuro Endocrinol Lett.
34:675–680. 2013.PubMed/NCBI
|
|
31
|
Abudukadeer A, Bakry R, Goebel G,
Mutz-Dehbalaie I, Widschendter A, Bonn GK and Fiegl H: Clinical
relevance of CDH1 and CDH13 DNA-methylation in serum of cervical
cancer patients. Int J Mol Sci. 13:8353–8363. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Gustafson KS, Furth EE, Heitjan DF,
Fansler ZB and Clark DP: DNA methylation profiling of cervical
squamous intraepithelial lesions using liquid-based cytology
specimens: An approach that utilizes receiver-operating
characteristic analysis. Cancer. 102:259–268. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Kim JH, Choi YD, Lee JS, Lee JH, Nam JH
and Choi C: Assessment of DNA methylation for the detection of
cervical neoplasia in liquid-based cytology specimens. Gynecol
Oncol. 116:99–104. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
McCormick TM, Canedo NH, Furtado YL,
Silveira FA, de Lima RJ, Rosman AD, Filho GL Almeida and Mda G
Carvalho: Association between human papillomavirus and Epstein-Barr
virus DNA and gene promoter methylation of RB1 and CDH1 in the
cervical lesions: A transversal study. Diagn Pathol. 10:592015.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Xu Z and Taylor JA: Genome-wide
age-related DNA methylation changes in blood and other tissues
relate to histone modification, expression and cancer.
Carcinogenesis. 35:356–364. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Jung M and Pfeifer GP: Aging and DNA
methylation. BMC Biol. 13:72015. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Leonard SM, Wei W, Collins SI, Pereira M,
Divaf A, Constandinou-Williams C, Young LS, Roberts S and Woodman
CB: Oncogenic human papillomavirus imposes an instructive pattern
of DNA methylation changes which parallel the natural history of
cervical HPV infection in young women. Carcinogenesis.
33:1286–1293. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Woodman CB, Collins SI and Young LS: The
natural history of cervical HPV infection: Unresolved issues. Nat
Rev Cancer. 7:11–22. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
D'Costa ZJ, Jolly C, Androphy EJ, Mercer
A, Matthews CM and Hibma MH: Transcriptional repression of
E-cadherin by human papillomavirus type 16 E6. PLoS One.
7:e489542012. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Flatley JE, McNeir K, Balasubramani L,
Tidy J, Stuart EL, Young TA and Powers HJ: Folate status and
aberrant DNA methylation are associated with HPV infection and
cervical pathogenesis. Cancer Epidemiol Biomarkers Prev.
18:2782–2789. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Attaleb M, El hamadani W, Khyatti M,
Benbacer L, Benchekroun N, Benider A, Amrani M and El Mzibbri M:
Status of p16(INK4a) and E-cadherin gene promoter methylation in
Moroccan patients with cervical carcinoma. Oncol Res. 18:185–192.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Kahn SL, Ronnett BM, Gravitt PE and
Gustafson KS: Quantitative methylation-specific PCR for the
detection of aberrant DNA methylation in liquid-based Pap tests.
Cancer. 114:57–64. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Herman JG, Graff JR, Myöhänen S, Nelkin BD
and Baylin SB: Methylation-specific PCR: A novel PCR assay for
methylation status of CpG islands. Proc Natl Acad Sci USA.
93:9821–9826. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Tost J and Gut IG: DNA methylation
analysis by pyrosequencing. Nat Protoc. 2:2265–2275. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Colella S, Shen L, Baggerly KA, Issa JP
and Krahe R: Sensitive and quantitative universal Pyrosequencing
methylation analysis of CpG sites. Biotechniques. 35:146–150.
2003.PubMed/NCBI
|
|
46
|
Siegel EM, Riggs BM, Delmas AL, Koch A,
Hakam A and Brown KD: Quantitative DNA methylation analysis of
candidate genes in cervical cancer. PLoS One. 10:e01224952015.
View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Esteller M, Corn PG, Baylin SB and Herman
JG: A gene hypermethylation profile of human cancer. Cancer Res.
61:3225–3229. 2001.PubMed/NCBI
|
|
48
|
Branca M, Giorgi C, Ciotti M, Santini D,
Di Bonito L, Costa S, Benedetto A, Bonifacio D, Di Bonito P, Paba
P, et al: HPV-PathogenISS Study Group: Down-regulation of
E-cadherin is closely associated with progression of cervical
intraepithelial neoplasia (CIN), but not with high-risk human
papillomavisrus (HPV) or disease outcome in cervical cancer. Eur J
Gynaecol Oncol. 27:215–223. 2006.PubMed/NCBI
|
|
49
|
Strathdee G: Epigenetic versus genetic
alterations in the inactivation of E-cadherin. Semin Cancer Biol.
12:373–379. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Thomas M, Pim D and Banks L: The role of
the E6-p53 interaction in the molecular pathogenesis of HPV.
Oncogene. 18:7690–7700. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Hirohashi S: Inactivation of the
E-cadherin-mediated cell adhesion system in human cancers. Am J
Pathol. 153:333–339. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Liu YN, Lee WW, Wang CY, Chao TH, Chen Y
and Chen JH: Regulatory mechanisms controlling human E-cadherin
gene expression. Oncogene. 24:8277–8290. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Kwon O, Jeong SJ, Kim SO, He L, Lee HG,
Jang KL, Osada H, Jung M, Kim BY and Ahn JS: Modulation of
E-cadherin expression by K-Ras; involvement of DNA
methyltransferase-3b. Carcinogenesis. 31:1194–1201. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Syrjänen K, Kataja V, Yliskoski M, Chang
F, Syrjänen S and Saarikoski S: Natural history of cervical human
papillomavirus lesions does not substantiate the biologic relevance
of the Bethesda system. Obstet Gynecol. 79:675–682. 1992.PubMed/NCBI
|