Introduction
Gastric cancer is one of the most common human
malignancies, globally accounting for ~1 million novel cases and
>700,000 cancer-associated mortalities annually (1). Surgical resection remains the primary
treatment for gastric cancer; however, ~60% of patients have
locally advanced and metastatic disease at the time of surgery,
leading to a relatively low therapeutic efficacy (2). Therefore, nonsurgical methods have
attracted increasing attention (3).
Iodine-125 (125I) seeds have a long
half-life and low energy with excellent stability, which has
resulted in their extensive clinical use (4). Brachytherapy with low-dose
125I seeds has been demonstrated to be an effective
salvage therapy for gastric cancer and other malignant carcinomas
(5–10). Although 125I seed
implantation has been successfully utilized in clinics, its
radiobiological effect and underlying molecular mechanisms have not
been fully elucidated. Recent evidence has indicated that altered
DNA methylation patterns have a critical role in tumor inhibition
resulting from 125I irradiation (3). In addition, consecutive low-energy
125I irradiation significantly inhibits the expression
of DNA methyltransferases (DNMTs) in cancer cells (11). Following a decrease in DNMT
expression, the irradiation-induced DNA demethylation contributes
to tumor inhibition by reactivating tumor suppressor genes
(11).
MicroRNAs (miRs) are small non-coding RNAs that
function as endogenous silencers of numerous target genes.
Downregulation of miRs is a common characteristic observed in
various types of cancer, suggesting that these molecules may act as
a novel class of tumor suppressors (12–18).
Previous studies have revealed that a growing number of tumor
suppressor miRs are inactivated by promoter hypermethylation in
cancers (19–27). One of these miRs, miR-181c, is
silenced in gastric cancer by promoter hypermethylation.
Furthermore, when miR-181c expression is upregulated, it has been
demonstrated to induce growth inhibition of gastric cancer cells,
suggesting its role as a potential tumor suppressor in gastric
cancer (20). Based on these studies,
the present study hypothesized that tumor suppressor miRs, which
are epigenetically silenced in cancer, may be activated by
irradiation-inducing DNA demethylation and contribute to the
anticancer effects of 125I irradiation. The aim of the
present study was to evaluate whether miR-181c is regulated by
125I irradiation and is involved in
irradiation-triggering tumor inhibition in gastric cancer
cells.
Materials and methods
Cell culture
Human gastric cancer KATO-III and MKN45 cell lines
were purchased from the Shanghai Institute of Cytobiology of the
Chinese Academy of Sciences (Shanghai, China). The cells were
cultured in RPMI-1640 (Thermo Fisher Scientific, Inc., Waltham, MA,
USA) supplemented with 10% fetal bovine serum (FBS;
Gibco®; Thermo Fisher Scientific, Inc.), 100 U/ml
penicillin and 100 µg/ml streptomycin. All cell lines were
maintained at 37°C in a humidified incubator containing 5%
CO2.
125I irradiation treatment
of gastric cancer cells
In-house 125I seeds were obtained from
Beijing Atom and High Technique Industries, Inc. (Beijing, China).
An in vitro irradiation model was constructed as previously
described (28). The absorbed dose
was measured and verified as follows: 44, 92, 144 and 204 h were
required for doses of 2, 4, 6 and 8 Gy, respectively.
MTT assay
Cell viability was determined by measuring the
ability of the cells to transform thiazolyl blue tetrazolium
bromide (MTT) to a purple formazan dye as previously described
(28,29). Briefly, cells were irradiated and 20
µl MTT solution was added to the cells in each well of a 96-well
plate and incubated for 5 h. The medium was replaced with dimethyl
sulfoxide to dissolve the purple formazan. The color intensity of
the formazan solution, which is positively associated with cell
viability, was measured with a microplate spectrophotometer
(VersaMax™; Molecular Devices, LLC, Sunnyvale, CA, USA) at 570
nm.
Transwell invasion assay
Invasiveness of gastric cancer cells were measured
using a modified Boyden chamber (BD Bioscience, Franklin Lakes, NJ,
USA). Briefly, the gastric cancer cell suspensions were obtained 92
h following irradiation at a total dose of 4 Gy. In total, 104
cells were plated in 200 µl RPMI-1640 containing 10% FBS in the
upper chambers of the Boyden chamber. The lower chambers were
filled with 500 µl RPMI-1640 containing 10% FBS. Subsequently,
cells were incubated for 48 h at 37°C, and the membrane was stained
with crystal violet and viewed under a microscope to calculate the
average number of invasive cells.
Western blotting
Total protein from gastric cancer cells was
extracted using RIPA lysis buffer (Cloud-Seq, Inc., Shanghai,
China) and quantified using a BCA assay (Pierce™; Thermo Fisher
Scientific, Inc.). Protein extracts were separated using sodium
dodecyl sulfate-polyacrylamide gel electrophoresis and transferred
onto a nitrocellulose membrane using RapidBlot Transfer Buffer
(Cloud-Seq, Inc.). The membranes were hybridized with the following
primary antibodies overnight at 4°C: Mouse monoclonal anti-DNA
(cytosine-5)-methyltransferase 1 (DNMT1; catalog no., sc-271729;
dilution, 1:200); and mouse monoclonal anti-glyceraldehyde
3-phosphate dehydrogenase (GAPDH; catalog no., sc-365062; dilution,
1:200) (Santa Cruz Biotechnology, Inc., Dallas, TX, USA).
Subsequently, membranes were probed with horseradish
peroxidase-conjugated goat anti-mouse secondary antibody (catalog
no., 115-035-003; dilution, 1:10,000; Jackson ImmunoReserach, West
Grove, PA, USA) for 1 h at room temperature, and the immunoreactive
signals were detected using a SuperSen Enhanced Chemiluminescence
kit (Cloud-Seq, Inc.). The DNMT1 protein expression levels were
normalized to GAPDH. The signal intensities on western blots were
semi-quantified using ImageJ version 1.43 software (National
Institutes of Health, Bethesda, MA, USA).
Methylated DNA immunoprecipitation
(MeDIP)-quantitative polymerase chain reaction (qPCR) assay
The MeDIP assay coupled with qPCR was used to
quantitatively evaluate the methylation status of miR-181c in the
cells treated without (control) or with 4 Gy of 125I
irradiation. MeDIP was performed as described previously (3). Briefly, genomic DNA was extracted from
cells using DNeasy Blood and Tissue kit (Qiagen, Inc., Hilden,
Germany), according to the manufacturer's protocol, and sonicated
to produce random fragments ~200–600 bp in size. Following
denaturation at 95°C for 10 min, immunoprecipitation was performed
using a mouse monoclonal anti-5-methylcytidine antibody (catalog
no., 39649; dilution 1:10; incubation, 2 h at 4°C; Active Motif,
Carlsbad, CA, USA). Immunoprecipitated complexes were collected
with Dynabeads® Protein A (Thermo Fisher Scientific,
Inc.), and the methylated DNA fragments were purified by
phenol-chloroform extraction and isopropanol precipitation.
Subsequently, methylated DNA fragments and input DNA were analyzed
by qPCR using an ABI 7900 Real-Time PCR System (Applied
Biosystems™; Thermo Fisher Scientific, Inc.) and Rapid SYBR Green
PCR Master Mix from Cloud-Seq, Inc. The following cycling
conditions were used: 95°C for 1 min, followed by 40 cycles of 95°C
for 15 sec, then 60°C for 1 min, concluding with melt curve
analysis. The relative changes in the extent of gene methylation
were determined by measuring the amount of detected genes in
methylated DNA fragments following normalization to the input DNA
(30). The primer sequences targeting
the promoter of miR-181c were synthesized by Sangon Biotech Co.,
Ltd. (Shanghai, China) as follows: Forward,
5′-GAGGGATGAGGAAATGGA-3′ and reverse, 5′-TCACAACAGCGTGAGTGG-3′. The
experiment was performed in triplicate.
Transfection of cells with miR-181c
mimic and inhibitor
miR-181c was repressed or overexpressed by
transfection of cells with miR inhibitor or miR mimics using
Lipofectamine® 2000 (Invitrogen™; Thermo Fisher
Scientific, Inc.), according to the manufacturer's protocol.
miR-181c mimic and scrambled control miR sequences were designed
and synthesized by GenePharama Co., Ltd. (Shanghai, China) as
follows: miR-181c mimic, sense 5′-AACAUUCAACCUGUCGGUGAGUUA-3′ and
antisense 5′-ACUCACCGACAGGUUGAAUGUUUU-3′; miR-181c inhibitor, sense
5′-TAACUCACCGACAGGUUGAAUGUU-3′; scrambled negative control, sense
5′-UUGUACUACACAAAAGUACUG-3′. All of the oligonucleotides were
transfected at a final concentration of 100 nM. Following
transfection, the expression of miR-181c was confirmed by qPCR.
Reverse transcription-qPCR
In total, ~5×106 cells were treated without
(control) or with 4 Gy of 125I irradiation. Total RNA
was extracted from cells using ExRNA Reagent (Cloud-Seq, Inc.),
according to manufacturer's protocol. Total RNA from each sample
was quantified using NanoDrop ND-1000 (NanoDrop; Thermo Fisher
Scientific, Inc., Wilmington, DE, USA), and RNA integrity was
assessed by standard denaturing agarose gel electrophoresis. miRs
and mRNAs were processed into cDNA using a FlashScript Reverse
Transcription kit (Cloud-Seq, Inc.) with a miR-181c-specific stem
loop primer and oligo dT (18)
primer. qPCR was performed with Rapid SYBR PCR Master Mix
(Cloud-Seq, Inc.) on an ABI 7900 Real-Time PCR System. Primers were
synthesized by Sangon Biotech Co., Ltd. as follows: U6, forward
5′-CTCGCTTCGGCAGCACA-3′ and reverse 5′-AACGCTTCACGAATTTGCGT-3′;
GAPDH, forward 5′-CGGAGTCAACGGATTTGGTCGTAT-3′ and reverse
5′-AGCCTTCTCCATGGTGGTGAAGAC-3′; miR-181c, forward
5′-AACATTCAACCTGTCGGTG-3′ and reverse
5′-GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACACTCAC-3′; miRNA
universal, reverse 5′-CCAGTGCAGGGTCCGAGGTAT-3′. For normalization,
GAPDH and U6 were used to normalize mRNA and miRNA, respectively.
Cycles were as follows: 95°C for 10 min, followed by 40 cycles at
95°C for 10 sec and annealing/extension at 60°C for 30 sec. Each
experiment was repeated at least three times, and the results were
calculated according to the 2−ΔΔCq method (31).
Statistical analysis
Statistical analysis was performed with SPSS version
11.0 software (SPSS, Inc., Chicago, IL, USA). Each experiment was
repeated three times. Student's t-test was used to assess the
statistical significance of differences between groups. Data are
presented as the mean ± standard error of the mean. P<0.05 was
considered to indicate a statistically significant difference.
Results
125I irradiation inhibits
viability and invasiveness of gastric cancer cells
To investigate the effects of 125I
irradiation on the viability of gastric cancer cells, KATO-III and
MKN45 cells were treated with continuous 125I
irradiation at a dose between 0 and 8 Gy. Subsequently, MTT assay
was performed to determine the effects of 125I
treatment. MTT assays indicated that 125I treatment
significantly decreased the viability of KATO-III and MKN45 cells
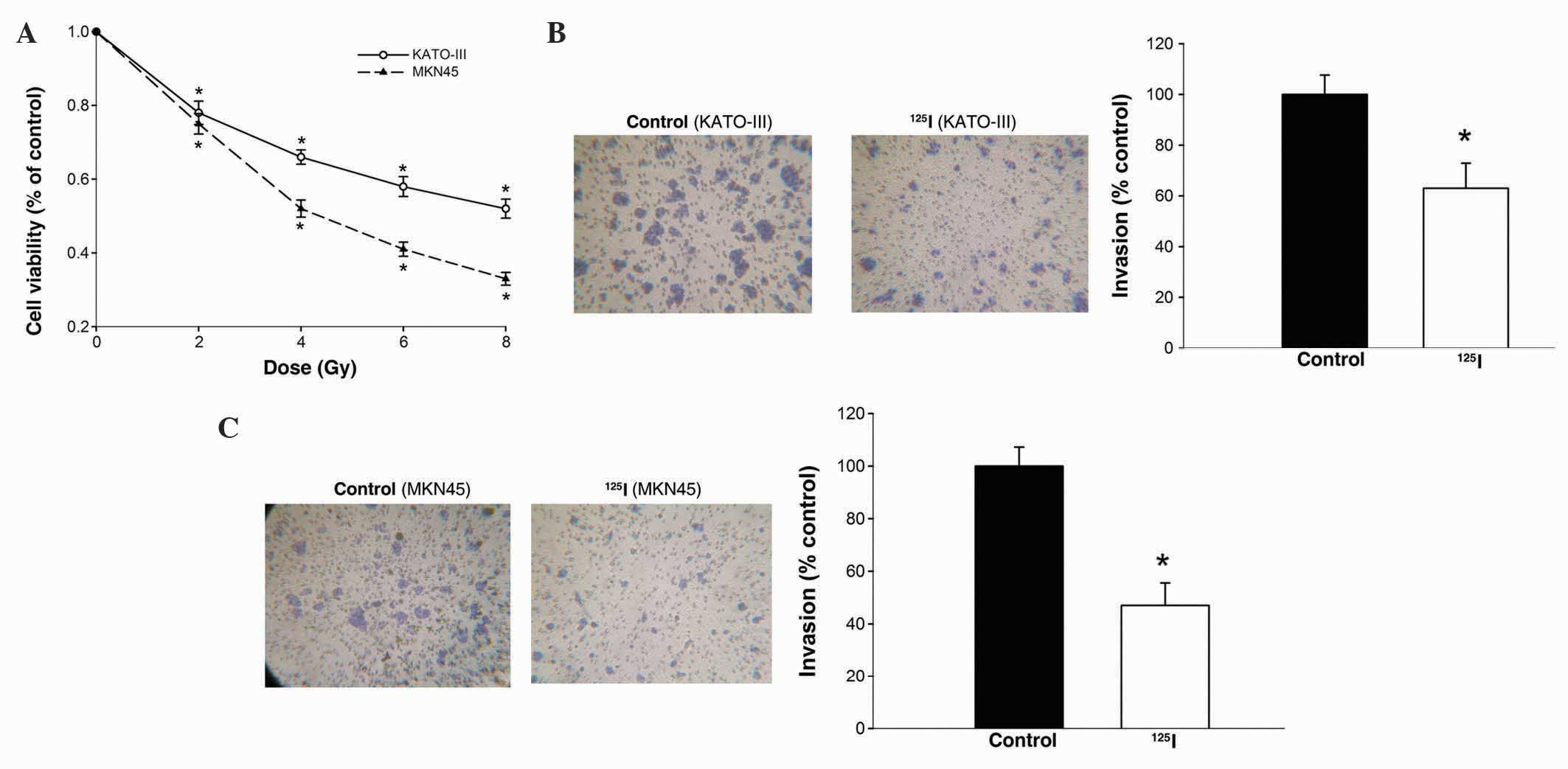
at a low dose (P<0.05; Fig. 1A).
On the basis of these results, 4 Gy was used for subsequent
experiments.
To investigate the effects of 125I
irradiation on the invasion of gastric cancers, KATO-III and MKN45
cells were treated with 125I irradiation at 0 (control)
or 4 Gy. In total ~48 h later, the invasive ability of the cells
was analyzed using Transwell assays. As shown in Fig. 1B, the number of KATO-III cells
invading through the Matrigel following 125I irradiation
was clearly attenuated by 53% compared with the control group. A
similar reduction in cell invasive activity was observed in MKN45
cells treated with 125I irradiation (P=0.043; Fig. 1C). These data indicate a negative
regulatory effect of 125I irradiation on the viability
and invasiveness of gastric cancer cells.
125I irradiation modulates
miR-181c activity by inducing DNA demethylation at its promoter
region
Previous evidence has demonstrated that
125I irradiation alters the expression of DNMT1, leading
to epigenetic alterations, which reactivates silenced tumor
suppressor genes (11). miR-181c is a
potential tumor suppressor that is epigenetically silenced by
promoter hypermethylation in gastric cancers (12). Thus, the present study hypothesizes
that 125I irradiation may modulate the activity of the
miR-181c gene by affecting its methylation status.
To confirm this hypothesis, the present study
primarily examined the effects of 125I irradiation on
the expression of DNMT1 protein in KATO-III and MKN45 cells. DNMT1
and GAPDH proteins were detected by western blotting, and protein
expression levels of DNMT1 were calculated by normalization to
GAPDH density. As indicated in Fig.
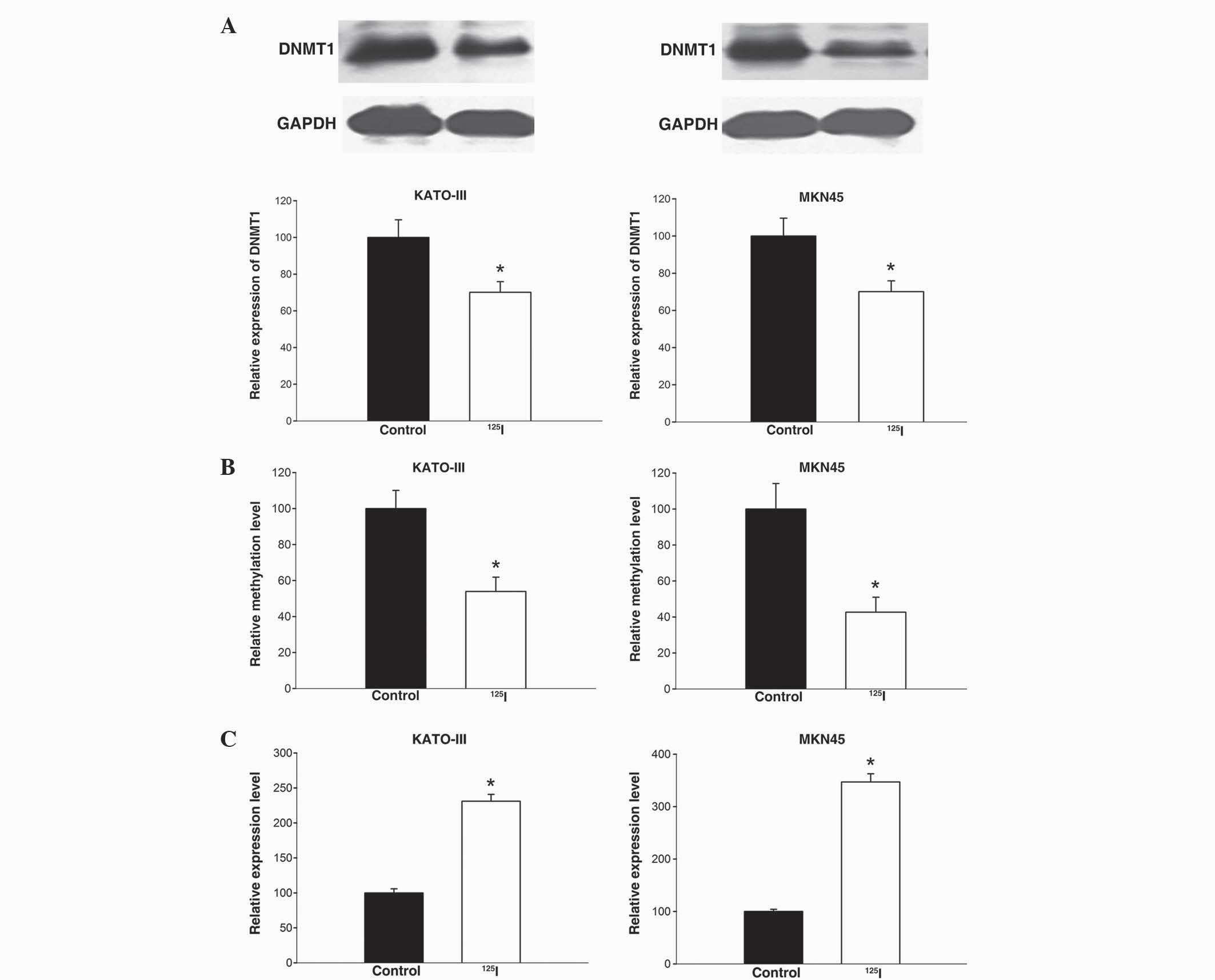
2A, 125I treatment lead to a significant reduction
of DNMT1 protein expression in KATO-III (P=0.040) and MKN45 cells
(P=0.025). This decrease of DNMT1, which is responsible for the
maintenance of DNA methylation patterns, may result in methylation
alterations of target genes.
To determine whether 125I irradiation
affects the methylation status of the miR-181c gene, a MeDIP-PCR
assay was performed in gastric cancer cells treated with and
without 125I irradiation (4 Gy). As indicated in
Fig. 2B, 125I-irradiated
cells exhibited a significantly decrease in the methylation status
of miR-181c compared with control cells (KATO-III, P=0.032; MKN45,
P=0.025); there was a 53.9% and 42.7% decrease in KATO-III and
MKN45 cells, respectively. These results demonstrate that
125I irradiation induces DNA demethylation at the
promoter region of the miR-181c gene.
Finally, alterations in the expression of miR-181c
in gastric cancer cells following 125I treatment were
determined using qPCR. As indicated in Fig. 2C, 125I treatment caused
upregulation of miR-181c expression in KATO-III and MKN45 cells,
which was significant compared with the control group (P=0.018 and
P=0.014, respectively). Overall, these data suggest that
125I irradiation upregulates miR-181c expression in
gastric cancer cells, which may be partially attributed to the
irradiation inducing DNA demethylation.
miR-181c exerts a functional role as a
tumor suppressor in gastric cancer cells
A previous study reported that there was a decreased
expression of miR-181c in gastric cancer, and revealed that
miR-181c suppresses cell growth in KATO-III and MKN45 cells
(20). To further determine the
functional role of miR-181c in gastric cancer cells, the
invasiveness of KATO-III and MKN45 cells transfected with miR-181c
mimics and scrambled negative control was evaluated by the present
study.
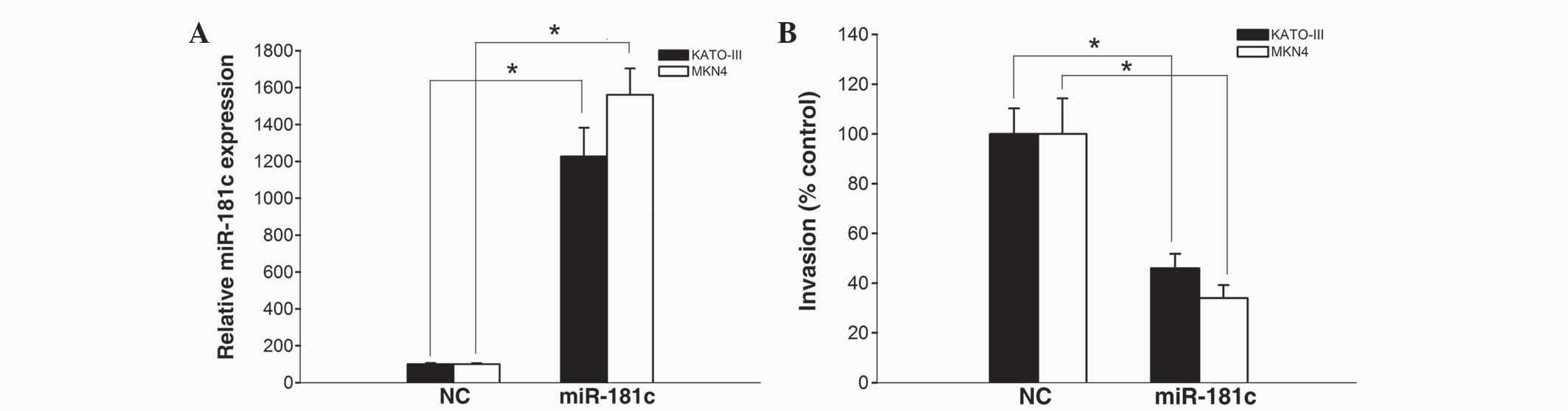
The results of a qPCR revealed that miR-181c mimics
significantly increased miR-181c expression in KATO-III and MKN45
cells (P=0.016 and P=0.009, respectively; Fig. 3A), suggesting that miR-181c mimics
were efficiently introduced into the cells and upregulated miR-181c
expression. Furthermore, the number of gastric cancer cells
invading through Matrigel following transfection with miR-181c
mimics was remarkably attenuated compared with KATO-III and MKN45
cells transfected with scrambled negative control (P=0.027 and
P=0.021, respectively; Fig. 3B).
These data indicate a potential negative regulatory effect of
miR-181c on the invasion of gastric cancer cells.
Downregulation of miR-181c compromises
the anticancer effects of 125I irradiation
To evaluate the role of miR-181c on the inhibitory
effect of 125I irradiation, KATO-III and MKN45 cells
were treated with 4 Gy 125I irradiation followed by
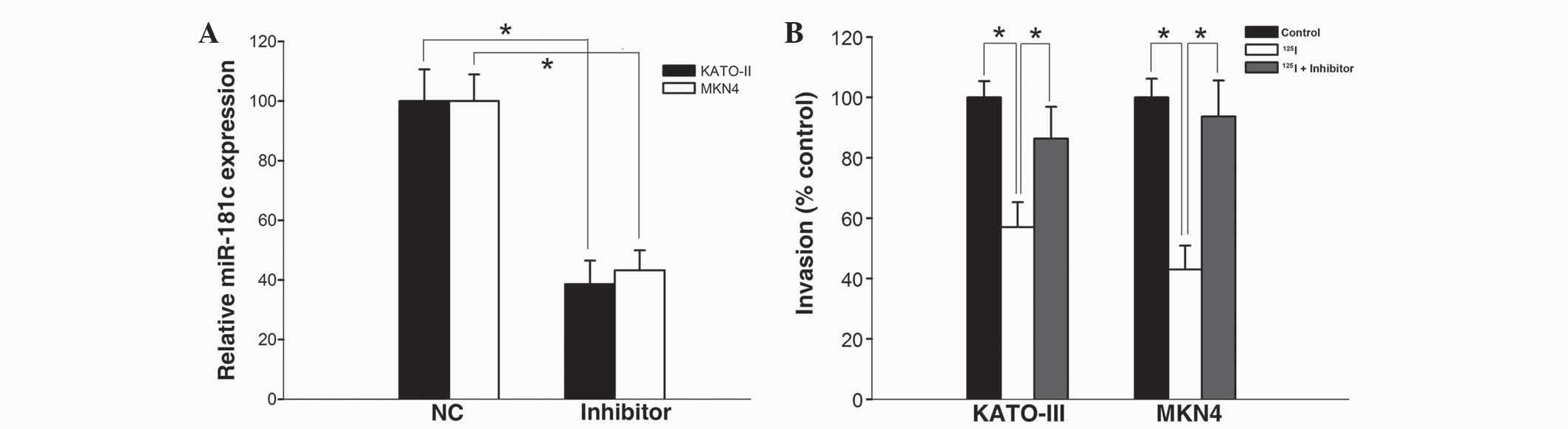
transfection with miR-181c inhibitors. As shown in Fig. 4A, miR-181c inhibitors significantly
decreased the expression of miR-181c in KATO-III and MKN45 cells
compared with the scrambled normal control-transfected cells
(P=0.037 and P=0.030, respectively). In addition, the
125I-irradiation-induced inhibition on cell invasion was
significantly attenuated in KATO-III and MKN45 cells transfected
with the miR-181c inhibitor (P<0.05; Fig. 4B). These data implied that miR-181c
was responsible for the suppression of cell invasion, which was
induced by 125I irradiation.
Discussion
125I seed implantation has been widely
used to treat various types of cancer, including gastric cancer,
due to its high precision, low trauma to patients, strong lethality
and few side effects (9,10). Although 125I seed
implantation has been successfully used in clinics, its
radiobiological effects and underlying molecular mechanisms are not
fully elucidated.
Several studies have indicated that 125I
seed irradiation is effective in inducing cell apoptosis in
pancreatic and colonic cancer cells (4,11,32). More recently, Tian et al
(28) revealed that 125I
seeds effectively inhibit cell growth and invasion of
nasopharyngeal carcinoma. However, the effects of 125I
seed irradiation in gastric cancer cell lines have not yet been
investigated. The present study evaluated the efficacy of
125I irradiation on the death of KATO-III and MKN45
cells, as assessed by cell viability assays, and demonstrated that
a low dose of 125I irradiation (4 Gy) effectively kills
gastric cancer cells.
Previous studies have revealed that altered DNA
methylation patterns may be critical in tumor inhibition resulting
from consecutive low-energy 125I irradiation (11). Ma et al (11) demonstrated that low-dose (4 Gy)
125I irradiation causes a significant decrease in DNMT
expression in pancreatic cancer cells. Consistent with the present
study, a significant decrease in DNMT1 expression was observed in
gastric cancer xenografts implanted with 125I seeds
(3). In addition, there is a clear
and positive association between DNA methylation and expression of
DNMTs, since DNMTs maintain DNA methylation patterns (11). As the result of decreased DNMT
expression induced by 125I seed irradiation, tumor
suppressor genes, including BCL2/adenovirus E1B 19kDa interacting
protein 3, Wnt family member 9A and germ cell associated 2, are
extensively reactivated by promoter demethylation (3).
miRs are known as a class of small noncoding RNAs,
which are critical in cancer progression as oncogenes and tumor
suppressor genes. It has been reported that numerous tumor
suppressor miRs, including miR-9, miR-124 and miR-200 family, are
epigenetically silenced by DNA methylation in cancer (33–35). It is
reasonable to hypothesize that these miRs may be reactivated by
125I-irradiation inducing promoter demethylation,
thereby contributing to anticancer effects. Among these tumor
suppressor miRs, miR-181c has been reported to be epigenetically
silenced in gastric carcinogenesis (20). In the present study, MeDIP-PCR assays
were performed to examine the methylation status at the promoter
region of miR-181c. As expected, the present results revealed that
125I irradiation significantly induced DNA demethylation
at the promoter of the miR-181c gene in KATO-III and MKN45 cells.
Additionally, a notable upregulation of miR-181c in gastric cancer
cells was observed following continuous low-dose 125I
irradiation. These data suggested that 125I irradiation
induces DNA demethylation at the promoter region of miR-181c,
resulting in the reactivation of miR-181c. Further experiments by
the present study demonstrated that overexpression of miR-181c
effectively decreased cell invasiveness. In addition,
downregulation of miR-181c compromised the anticancer effects
observed with 125I irradiation. According to these
results, it may be concluded that miR-181c is reactivated by
125I irradiation through DNA demethylation, and thus, is
involved in 125I-irradiation induced tumor
inhibition.
In summary, the present study provides, to the best
of our knowledge, the first demonstration that miRs are involved in
the therapeutic effect of 125I seed irradiation. The
present study provides an illustrative example in gastric cancer
with the reactivation of miR-181c by 125I irradiation,
as well as its functional consequences for tumor inhibition. In
addition, the present study has revealed that miR-181c reactivation
may be mediated through 125I-induced demethylation,
emphasizing the critical role of epigenetic regulation underlying
the anticancer effects of 125I irradiation.
Acknowledgements
The present study was supported by the Foundation of
Applied Basic Research Program of Yunnan Province (Kunming, China;
grant nos. 2011FB150 and 2013FB183).
References
|
1
|
Wang Z, Wang J, Yang Y, Hao B, Wang R, Li
Y and Wu Q: Loss of has-miR-337-3p expression is associated with
lymph node metastasis of human gastric cancer. J Exp Clin Cancer
Res. 32:762013. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Chu D, Zhu S, Li J, Ji G, Wang W, Wu G and
Zheng J: CD147 expression in human gastric cancer is associated
with tumor recurrence and prognosis. PloS One. 9:e1010272014.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Ma ZH, Yang Y, Zou L and Luo KY: 125I seed
irradiation induces up-regulation of the genes associated with
apoptosis and cell cycle arrest and inhibits growth of gastric
cancer xenografts. J Exp Clin Cancer Res. 31:612012. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Zhuang HQ, Wang JJ, Liao AY, Wang JD and
Zhao Y: The biological effect of 125I seed continuous low dose rate
irradiation in CL187 cells. J Exp Clin Cancer Res. 28:122009.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Yu Y, Anderson LL, Li Z, Mellenberg DE,
Nath R, Schell MC, Waterman FM, Wu A and Blasko JC: Permanent
prostate seed implant brachytherapy: Report of the american
association of physicists in medicine task group No. 64. Med Phys.
26:2054–2076. 1999. View
Article : Google Scholar : PubMed/NCBI
|
|
6
|
Wang JJ, Yuan HS, Li JN, Jiang WJ, Jiang
YL and Tian SQ: Interstitial permanent implantation of 125I seeds
as salvage therapy for re-recurrent rectal carcinoma. Int J
Colorectal Dis. 24:391–399. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Zhao E, Xiao S, Qin Y, Wang Q, Guo M and
Wang R: Permanent interstitial implantation of 125I seeds for 5
cases of advanced head and neck cancer. Lin Chuang Er Bi Yan Hou Ke
Za Zhi. 18:348–349, 352. 2004.PubMed/NCBI
|
|
8
|
Wang ZM, Wu J, Wang GC, Ren JL and Kjelle
D: Efficacy of permanent interstitial implantation of 125I seeds
for solitary brain metastasis from non-small cell lung carcinoma.
Ai Zheng. 21:1145–1148. 2002.PubMed/NCBI
|
|
9
|
Wang J, Sui A, Jia Y, Xu B, Wei L, Chen J
and Shen W: Treatment of unresectable advanced gastric cancer using
lodine-125 brachytherapy. Chinese Journal of Clinical Oncology.
3:212–215. 2006. View Article : Google Scholar
|
|
10
|
Joyce F, Burcharth F, Holm HH and Strøyer
I: Ultrasonically guided percutaneous implantation of iodine-125
seeds in pancreatic carcinoma. Int J Radiat Oncol Biol Phys.
19:1049–1052. 1990. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Ma JX, Jin ZD, Si PR, Liu Y, Lu Z, Wu HY,
Pan X, Wang LW, Gong YF, Gao J and Zhao-shen L: Continuous and
low-energy 125I seed irradiation changes DNA methyltransferases
expression patterns and inhibits pancreatic cancer tumor growth. J
Exp Clin Cancer Res. 30:352011. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Suzuki H, Maruyama R, Yamamoto E and Kai
M: DNA methylation and microRNA dysregulation in cancer. Mol Oncol.
6:567–578. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Zhang B, Pan X, Cobb GP and Anderson TA:
microRNAs as oncogenes and tumor suppressors. Dev Biol. 302:1–12.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Yu S, Lu Z, Liu C, Meng Y, Ma Y, Zhao W,
Liu J, Yu J and Chen J: miRNA-96 suppresses KRAS and functions as a
tumor suppressor gene in pancreatic cancer. Cancer Res.
70:6015–6025. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Lopez-Serra P and Esteller M: DNA
methylation-associated silencing of tumor-suppressor microRNAs in
cancer. Oncogene. 31:1609–1622. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Lee YS and Dutta A: The tumor suppressor
microRNA let-7 represses the HMGA2 oncogene. Genes Dev.
21:1025–1030. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Noguchi S, Mori T, Hoshino Y, Maruo K,
Yamada N, Kitade Y, Naoe T and Akao Y: MicroRNA-143 functions as a
tumor suppressor in human bladder cancer T24 cells. Cancer Lett.
307:211–220. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Nan Y, Han L, Zhang A, Wang G, Jia Z, Yang
Y, Yue X, Pu P, Zhong Y and Kang C: MiRNA-451 plays a role as tumor
suppressor in human glioma cells. Brain Res. 1359:14–21. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Saito Y, Suzuki H, Tsugawa H, Nakagawa I,
Matsuzaki J, Kanai Y and Hibi T: Chromatin remodeling at Alu
repeats by epigenetic treatment activates silenced microRNA-512-5p
with downregulation of Mcl-1 in human gastric cancer cells.
Oncogene. 28:2738–2744. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Hashimoto Y, Akiyama Y, Otsubo T, Shimada
S and Yuasa Y: Involvement of epigenetically silenced microRNA-181c
in gastric carcinogenesis. Carcinogenesis. 31:777–784. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Zhang Y, Yan LX, Wu QN, Du ZM, Chen J,
Liao DZ, Huang MY, Hou JH, Wu QL, Zeng MS, et al: miR-125b is
methylated and functions as a tumor suppressor by regulating the
ETS1 proto-oncogene in human invasive breast cancer. Cancer Res.
71:3552–3562. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Ng EK, Tsang WP, Ng SS, Jin HC, Yu J, Li
JJ, Röcken C, Ebert MP, Kwok TT and Sung JJ: MicroRNA-143 targets
DNA methyltransferases 3A in colorectal cancer. Br J Cancer.
101:699–706. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Tsai KW, Liao YL, Wu CW, Hu LY, Li SC,
Chan WC, Ho MR, Lai CH, Kao HW, Fang WL, et al: Aberrant
hypermethylation of miR-9 genes in gastric cancer. Epigenetics.
6:1189–1197. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Tsai KW, Wu CW, Hu LY, Li SC, Liao YL, Lai
CH, Kao HW, Fang WL, Huang KH, Chan WC and Lin WC: Epigenetic
regulation of miR-34b and miR-129 expression in gastric cancer. Int
J Cancer. 129:2600–2610. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Tsuruta T, Kozaki K, Uesugi A, Furuta M,
Hirasawa A, Imoto I, Susumu N, Aoki D and Inazawa J: miR-152 is a
tumor suppressor microRNA that is silenced by DNA hypermethylation
in endometrial cancer. Cancer Res. 71:6450–6462. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Vogt M, Munding J, Grüner M, Liffers ST,
Verdoodt B, Hauk J, Steinstraesser L, Tannapfel A and Hermeking H:
Frequent concomitant inactivation of miR-34a and miR-34b/c by CpG
methylation in colorectal, pancreatic, mammary, ovarian, urothelial
and renal cell carcinomas and soft tissue sarcomas. Virchows
Archiv. 458:313–322. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Vrba L, Jensen TJ, Garbe JC, Heimark RL,
Cress AE, Dickinson S, Stampfer MR and Futscher BW: Role for DNA
methylation in the regulation of miR-200c and miR-141 expression in
normal and cancer cells. PloS One. 5:e86972010. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Tian Y, Xie Q, Tian Y, Liu Y, Huang Z, Fan
C, Hou B, Sun D, Yao K and Chen T: Radioactive 125I seed
inhibits the cell growth, migration and invasion of nasopharyngeal
carcinoma by triggering DNA damage and inactivating VEGF-A/ERK
signaling. PloS One. 8:e740382013. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Yang TS, Yang XH, Wang XD, Wang YL, Zhou B
and Song ZS: MiR-214 regulate gastric cancer cell proliferation,
migration and invasion by targeting PTEN. Cancer Cell Int.
13:682013. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Clark C, Palta P, Joyce CJ, Scott C,
Grundberg E, Deloukas P, Palotie A and Coffey AJ: A comparison of
the whole genome approach of MeDIP-seq to the targeted approach of
the Infinium HumanMethylation450 BeadChip(®) for
methylome profiling. PLoS One. 7:e502332012. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) Method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Ma Z, Yang Y, Yang G, Wan J, Li G, Lu P
and Du L: Iodine-125 induces apoptosis via regulating p53,
microvessel density, and vascular endothelial growth factor in
colorectal cancer. World J Surg Oncol. 12:2222014. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Wilting SM, van Boerdonk RA, Henken FE,
Meijer CJ, Diosdado B, Meijer GA, le Sage C, Agami R, Snijders PJ
and Steenbergen RD: Methylation-mediated silencing and tumour
suppressive function of hsa-miR-124 in cervical cancer. Mol Cancer.
9:1672010. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Wiklund ED, Bramsen JB, Hulf T, Dyrskjøt
L, Ramanathan R, Hansen TB, Villadsen SB, Gao S, Ostenfeld MS,
Borre M, et al: Coordinated epigenetic repression of the miR-200
family and miR-205 in invasive bladder cancer. Int J Cancer.
128:1327–1334. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Neves R, Scheel C, Weinhold S, Honisch E,
Iwaniuk KM, Trompeter HI, Niederacher D, Wernet P, Santourlidis S
and Uhrberg M: Role of DNA methylation in miR-200c/141 cluster
silencing in invasive breast cancer cells. BMC Res Note. 3:2192010.
View Article : Google Scholar
|