Introduction
Acute myeloid leukemia (AML) is an aggressive
malignant disorder of hematopoietic cells that is characterized by
clonal proliferation of myeloid progenitor cells and maturation
arrest (1). It is the most common
acute leukemia in the adult population, with a median age at the
time of diagnosis of 65 years (2).
Previous studies have demonstrated that AML results from mutations
in multiple genes involved in cell proliferation, survival and
apoptosis (3–5). Such changes ultimately result in the
activation of cellular signaling pathways that promote cell growth
and/or maintain survival, leading to leukemogenesis (6,7).
Prognostic factors for AML include: Patient age, preceding
cytotoxic treatments for a primary disorder, antecedent
hematological disease and the presence of specific cytogenetic,
molecular and epigenetic aberrations (8). Treatment of AML has changed little over
the past three decades, with curative therapy consisting of the
nucleoside analog cytarabine in combination with an anthracycline
(9). However, a major complication is
that the current drugs are highly toxic and poorly tolerated,
particularly by older patients (10).
Therefore, the identification of novel strategies for the treatment
of acute myeloid leukemia is urgently required.
Over the previous few years, a novel class of small
RNAs, named microRNAs (miRs), has been demonstrated to be altered
in AML (11,12). miRs are a large family of highly
conserved non-coding RNAs with a short single strand of 19–25
nucleotides in length, which have a role in a wide range of
cellular processes, including cell differentiation, apoptosis,
proliferation and survival signaling pathways (13,14). miRs
have been demonstrated to negatively regulate gene expression by
binding to the 3′untranslated region (UTR) of a target gene's mRNA,
thereby degrading or blocking translation (15). Biological evidence has been observed
that certain upregulated miRs in cancer may act as oncogenes, by
contributing to the transformed phenotype and suppressing tumor
suppressor genes (16). Furthermore,
specific miRs that are downregulated in cancer may act as tumor
suppressors, by allowing the expression of oncogenes (17). In previous years, functional and
prognostic studies have demonstrated that miRs have a significant
role in hematological malignancies, and certain miRs have been
proposed as prognostic markers and therapeutic targets in the
treatment of leukemia (18–21).
The expression of miR-223 has been investigated in
various types of cancer, however, to the best of our knowledge the
expression and function of miR-223 in AML remains to be elucidated.
In the present study, it was demonstrated that miR-223 was
downregulated in AML patients, and overexpression of miR-223
repressed cell proliferation and enhanced cell apoptosis by
directly targeting F-box and WD repeat domain-containing 7 (FBXW7).
The results of the present study have therapeutic implications and
may be exploited for the development of novel treatments for
AML.
Materials and methods
Clinical specimens
The present study included 45 patients with AML (28
males and 17 females; age range, 17–73 years) and 20 healthy
subjects. All patients were enrolled at the Department of
Hematology, Tongji Hospital, Tongji Medical College, Huazhong
University of Science and Technology (Wuhan, China), and provided
written informed consent. The diagnosis and classification of AML
patients were based on French-America-British and World Health
Organization criteria combined to immunophenotyping and cytogenetic
analysis (22–24). All patients were newly diagnosed with
AML between June 2011 and September 2013 and were not taking any
anti-leukemic therapy at the moment of bone marrow aspirates. The
Tongji Hospital's Protection of Human Subjects Committee approved
the present study.
Cell culture and transfection
The human leukemia cell lines, HL-60 and K562, were
purchased from The Cell Bank of Type Culture Collection of Chinese
Academy of Sciences (Shanghai, China). Cells were cultured in
RPMI-1640 (Gibco; Thermo Fisher Scientific, Inc., Waltham, MA, USA)
supplemented with 10% fetal bovine serum (Gibco; Thermo Fisher
Scientific, Inc.), 100 U/ml penicillin G (Gibco; Thermo Fisher
Scientific, Inc.), 100 µg/ml streptomycin (Gibco; Thermo Fisher
Scientific, Inc.) and 2 mM L-glutamine (Gibco; Thermo Fisher
Scientific, Inc.) at 37°C in a humidified atmosphere containing 5%
CO2. Cells were routinely subcultured every 2–3
days.
Mature miR-223 and miR negative control (NC) mimics
were purchased from Shanghai GenePharma Co., Ltd. (Shanghai,
China). The sequence of miR-223 mimic was
5′-UGUCAGUUUGUCAAAUACCCCA-3′. The sequence of NC mimic was
5′-UUCUCCGAACGUGUCACGUTT-3′. For functional analysis, cells were
transfected with miR-223 mimics or NC using Lipofectamine 2000
(Invitrogen; Thermo Fisher Scientific, Inc.) according to the
manufacturer's protocol.
RNA isolation, reverse transcription
and reverse transcription-quantitative polymerase chain reaction
(RT-qPCR)
Mononuclear cells from bone marrow aspirate samples
were isolated by density-gradient centrifugation (400 × g
for 30 min at 20°C and 100 × g for 10 min at 20°C) with the
use of Ficoll-Paque Plus (GE Healthcare Life Sciences, Uppsala,
Sweden) according to the manufacturers protocol. Total RNA was
isolated using TRIzol reagent (Invitrogen; Thermo Fisher
Scientific, Inc.) and reverse transcribed to complementary DNA
using a PrimeScript RT reagent kit (Takara Biotechnology Co., Ltd.,
Dalian, China) according to the manufacturer's protocol. An Applied
Biosystems® 7500 Real-Time PCR System (Thermo Fisher
Scientific, Inc.) and a SYBR Premix Ex Taq™ kit (Takara
Biotechnology Co., Ltd.) were used to perform qPCR. All PCR primers
were purchased from Guangzhou RiboBio Co., Ltd. (Guangzhou, China).
The cycling conditions were as follows: 95°C for 30 sec, followed
by 40 cycles of 95°C for 5 sec and 60°C for 30 sec. Each sample was
analyzed in triplicate. The data were normalized to the endogenous
U6 small nuclear RNA and fold changes were calculated using the
relative quantification method (2−ΔΔCq) (25).
Cell viability assay
Cell viability was determined using the cell
counting kit (CCK)-8 assay (Beyotime Institute of Biotechnology,
Haimen, China) according to the manufacturer's protocol. Cells were
seeded in 96-well culture plates at a density of 4×104
cells per well, a total of 24 h subsequent to transfection with
miR-223 or NC. The viability of the cells was evaluated following
24, 48, 72 and 96 h of incubation at 37°C in humidified air
containing 5% CO2. The absorbance of each well at 450 nm
was measured with an enzyme-linked immunosorbent assay reader
(Bio-Rad Laboratories, Inc., Hercules, CA, USA). All assays were
repeated at least three times.
Cell apoptosis assay
The cells were seeded in 6-well culture plates and
transfected with miR-223 or NC using Lipofectamine 2000. Following
transfection for 48 h, the cells were harvested and washed twice
with phosphate-buffered saline. Following addition of 5 µl of
Annexin V-fluorescein isothiocyanate (FITC) and 10 µl of propidium
iodide (PI), cells were incubated for 15 min in the dark at room
temperature. Cells were subsequently analyzed by flow cytometry (BD
Biosciences, Franklin Lakes, NJ, USA). Apoptotic cells were
classified as those exhibiting a high Annexin V-FITC fluorescence
signal, with a low PI signal. The percentages of apoptotic cells
were calculated by data from fluorescence activated cell-sorting
analysis and the results were presented as a bar chart.
Bioinformatics analysis
TargetScan (http://www.targetscan.org/) was used to identify the
potential targets of miR-223.
Western blotting analysis
Cells were lysed in cold radioimmunoprecipitation
assay buffer (Beyotime Institute of Biotechnology) containing
protease and phosphatase inhibitors and then centrifuged at 12,000
× g for 20 min at 4°C. The samples (20 µg) were then
separated by 10% sodium dodecyl sulfate-polyacrylamide gel
electrophoresis and transferred to polyvinylidene difluoride
membranes (Beyotime Institute of Biotechnology). The membranes were
blocked with Tris-buffered saline containing 0.05% Tween-20 (TBST)
(Beyotime Institute of Biotechnology) containing 5% non-fat dry
milk for 1 h at room temperature, then incubated with primary
rabbit anti-human FBXW7 antibody (1:1,000 dilution; sc-33196; Santa
Cruz Biotechnology, Inc., Dallas, TX, USA) and primary rabbit
anti-human β-actin antibody (1:1,000 dilution; sc-130656; Santa
Cruz Biotechnology, Inc.), according to the manufacturer's
protocol. The membranes were rinsed with TBST at room temperature
and incubated for 1 h with the corresponding horseradish
peroxidase-conjugated secondary antibody (Abcam, Cambridge, MA,
USA) at room temperature. Protein bands were visualized using an
ECL kit (Pierce; Thermo Fisher Scientific, Inc.) and analyzed using
Quantity One software version 4.62 (Bio-Rad Laboratories,
Inc.).
Dual luciferase activity assay
To determine whether FBXW7 is a direct target of
miR-223, the luciferase activity assay vector was used. Cells were
co-transfected with wild-type (WT) or mutant (Mut) 3′-UTR of FBXW7,
miR-223 mimic or NC using Lipofectamine 2000according to the
manufacturers protocol. A total of 48 h subsequent to transfection,
firefly and Renilla luciferase activities were measured
using a Dual-Luciferase® Reporter Assay System (Promega
Corporation, Madison, WI, USA). Each assay was replicated at least
3 times.
Statistical analysis
Data are presented as the mean ± standard deviation,
and compared using Student's t-test in Stata version 10.0
(StataCorp LP, College Station, TX, USA). P<0.05 was considered
to indicate a statistically significant difference.
Results
miR-223 is downregulated in AML
patients
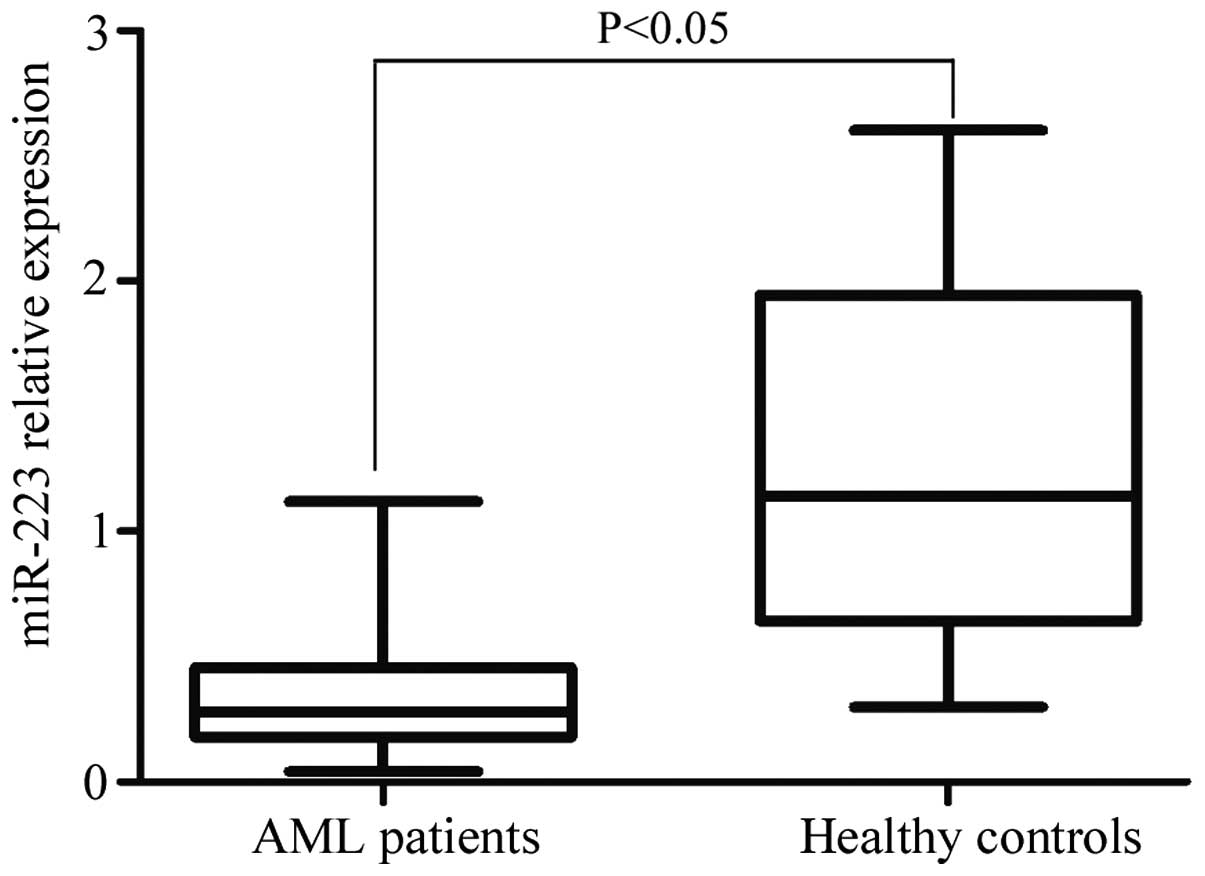
miR-223 expression was detected by RT-qPCR in AML
patients and normal controls. In the present study, it was
demonstrated that miR-223 was significantly downregulated in AML
patients compared with healthy controls (P=0.014; Fig. 1).
miR-223 reduces cell proliferation and
increases cell apoptosis in AML cell lines
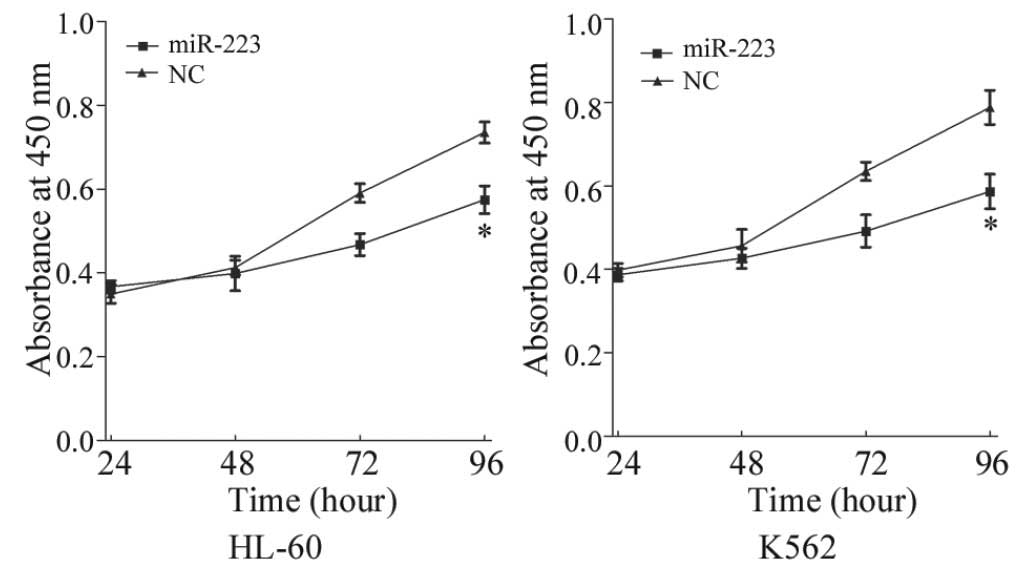
To investigate the effect of miR-223 on cell
proliferation, a CCK-8 assay was performed. The CCK-8 assay
revealed that ectopic expression of miR-223 resulted in
proliferation inhibition in AML cell lines (Fig. 2). CCK-8 assays revealed that following
96 h of treatment, the suppression rate of miR-223 reached
21.82±3.90% (P=0.026) in HL-60 cells and 25.54±4.20% (P=0.020) in
K562 cells. This indicates that miR-223 may be a negative regulator
of AML cell proliferation.
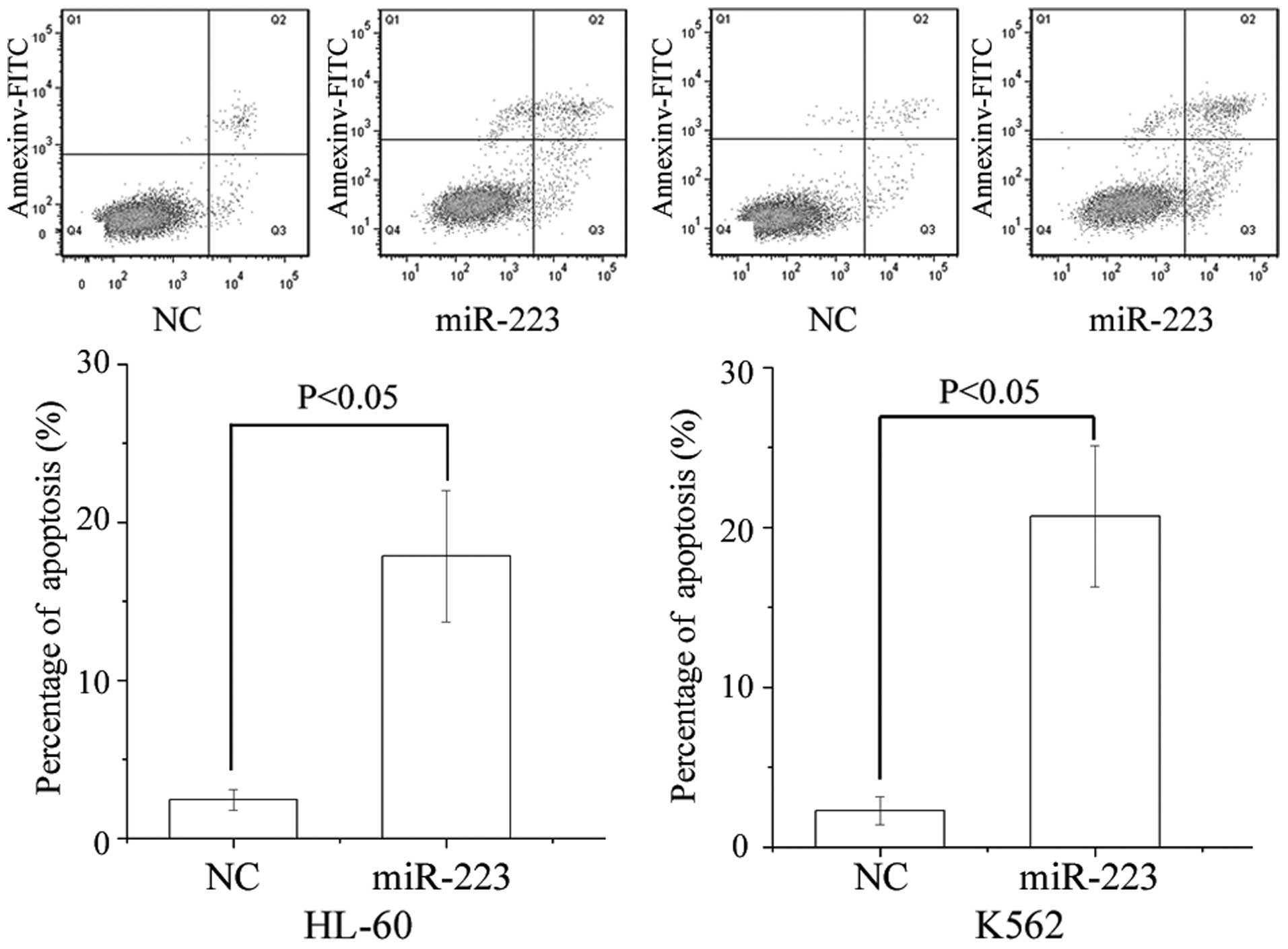
Flow cytometry was performed to evaluate the effect
of miR-223 on apoptosis. As presented in Fig. 3, ectopic expression of miR-223
enhanced apoptosis compared with NC in AML cell lines (P=0.017 for
HL-60 and P=0.010 for K562).
FBXW7 is a direct target gene of
miR-223 in AML
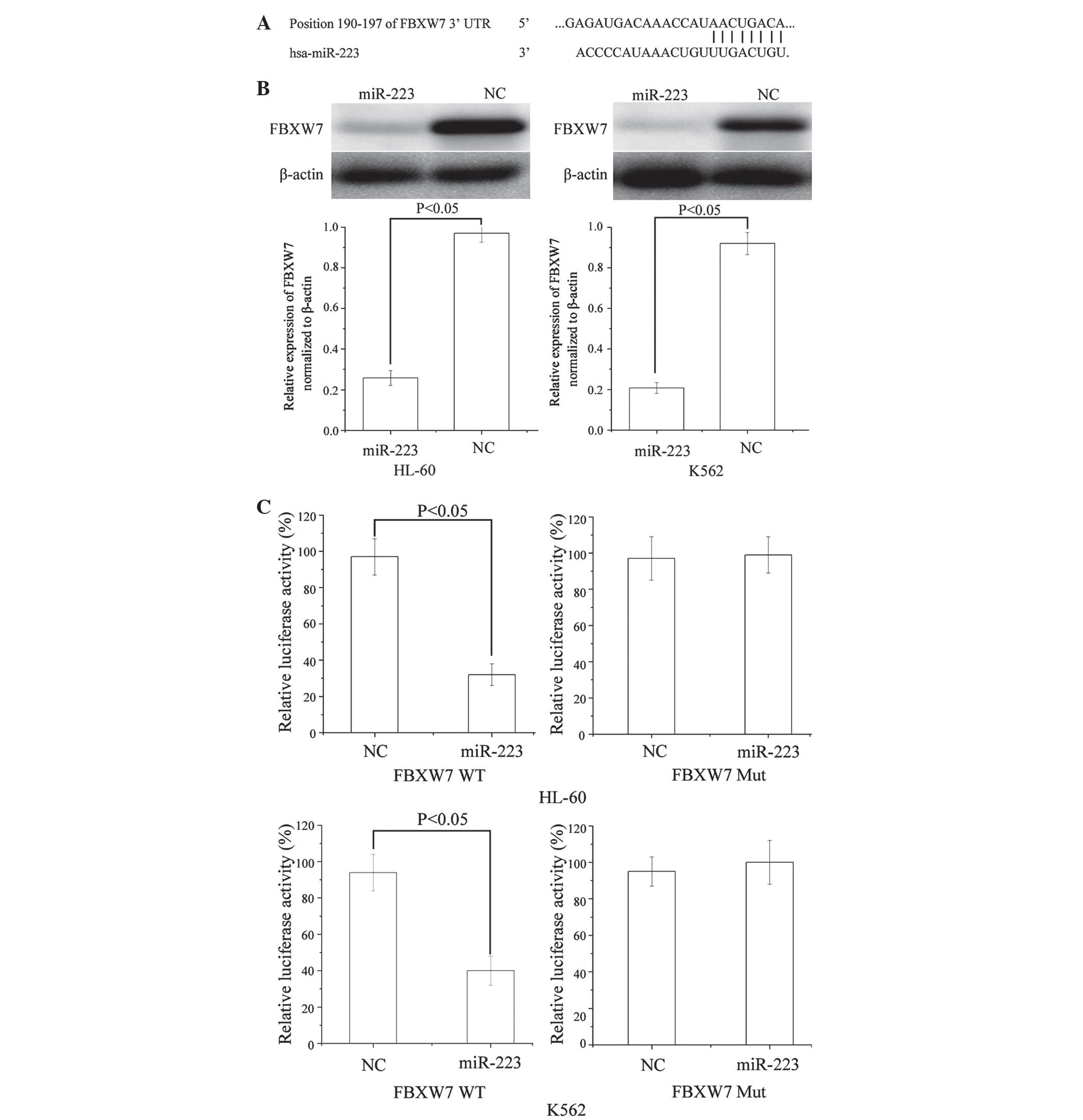
To identify the target of miR-223 in AML, a public
database (TargetScan; http://www.targetscan.org) was used. FBXW7 was
predicted to be a target of miR-223 (Fig.
4A). To verify whether miR-223 directly targeted FBXW7, western
blotting was performed to investigate whether FBXW7 was
downregulated following transfection of miR-223 into AML HL-60 and
K562 cells. As shown in Fig. 4B,
FBXW7 was significantly downregulated in AML HL-60 (P=0.024) and
K562 (P=0.032) cells following transfection of miR-223.
Furthermore, luciferase reporter assays were
conducted. Luciferase reporter assays revealed that miR-223
significantly inhibited the WT (P=0.019 for HL-60 and P=0.270 for
K562), but not the Mut luciferase activity of FBXW7 in AML HL-60
and K562 cells (Fig. 4C). Taken
together, these results appear to indicate that FBXW7 is a direct
target gene of miR-223 in AML.
Discussion
miRNA-223 has a significant role in a number of
types of human cancer. It has been observed to be upregulated in a
number of types of human cancer, including gastric cancer (26), glioblastoma (27), colorectal cancer (28) and lung cancer (29). Furthermore, Li et al found that
gastric cancer patients with lymph node metastasis or organ
metastasis demonstrated significantly increased expression of
miR-223 (30). The expression of
miR-223 was observed to be upregulated in recurrent ovarian cancer
(31). Specifically, patients with
lymph node metastasis or metastatic disease at an advanced
pathological stage exhibited significantly higher expression of
miR-223 compared with patients without lymph node metastasis or
metastatic disease (31). However,
downregulation of miR-223 has been observed in various types of
cancer, including osteosarcoma (32),
hepatocellular carcinoma (33) and
esophageal carcinoma (34). These
previous findings demonstrate that miR-223 may have a vital role as
either a tumor suppressor, or as an oncogenic regulator during
tumor progression. It may also be useful as a potential biomarker
for cancer.
The results of the present study provide evidence
that miR-223 is downregulated in AML, which may indicate that
miR-223 has a suppressive role in AML. By inducing overexpression
of miR-223 in AML cell lines, the present study demonstrated that
miR-223 is able to reduce cell proliferation and promote cell
apoptosis, additionally suggesting a suppressive role for miR-223.
This suggests that miR-223 may be useful in the development of
novel therapies for the treatment of AML.
Identification of miR-223 target genes is critical
for understanding its role in tumorigenesis. Previous studies have
revealed that miR-223 may regulate transcripts in human cells,
including erythrocyte membrane protein band 4.1 like 3 (35), septin 6 (36), FBXW7 (37), ataxia telangiectasia mutated (38), insulin-like growth factor 1 (29), paired box 6 (27) and caprin-1 (27). In the present study, it was
demonstrated that miR-223 may function as a tumor suppressor
through downregulation of FBXW7 in AML. miR-223 transfection
resulted in decreased cell viability and an increase in apoptosis
in human AML cell lines. Therefore, these results may have certain
clinical implications in the future.
In addition, a significant molecular link between
miR-223 and FBXW7 was observed in the present study. Initially,
TargetScan predicted that FBXW7 was a direct target gene of
miR-223. It was demonstrated that FBXW7 mRNA contained a miR-223
eight-nucleotide seed match at position 190–197 of the FBXW7
3′-UTR. Subsequently, in the luciferase dual-activity assay,
miR-223 directly targeted FBXW7 3′-UTR as predicted by
bioinformatics. Finally, it was demonstrated that rescue of miR-223
expression led to downregulation of FBXW7 protein in AML cell
lines. Taken together, the results of the present study revealed
that miR-223 regulated FBXW7 expression in vitro and may act
as a suppressive molecule during AML development and
progression.
FBXW7, a well-known F-box protein in the SCF E3
ligase complex, determines target specificity by recognizing and
binding proteins, leading to their ubiquitination and proteasomal
degradation (39). FBXW7 has been
verified to regulate various cellular processes, including cell
proliferation, differentiation, cell cycle, migration and invasion,
by targeting numerous substrates for degradation (40–42). For
example, downregulation of FBXW7 inhibits cell proliferation by
controlling cell cycle transition via the degradation of crucial
cell-cycle regulators, including c-myc (43), Jun (44), cyclin-E (45) and Notch (46). Consistent with these reports, the
present study demonstrated that overexpression of miR-223 had a
negative effect on FBXW7 and inhibited cell proliferation, as well
as enhancing cell apoptosis. Therefore, the results of the present
study establish a functional link between miR-223 and FBXW7, and
confirm that miR-223 inhibits AML cell proliferation and that
enhanced AML apoptosis is mediated, at least partly, by suppression
of FBXW7.
In conclusion, the present study is the first, to
the best of our knowledge, to demonstrate that miR-223 is
downregulated in AML. It was additionally demonstrated that miR-223
inhibits cell proliferation and enhances cell apoptosis in AML
cells. The identification of candidate target genes of miR-223 may
provide an understanding of potential mechanisms underlying the
development and progression of AML. The results of the present
study have therapeutic implications and may be exploited for the
development of novel treatments for AML. Future work is required to
address the potential of miR-223 as a target for the treatment of
AML.
Acknowledgements
This present study was supported by the Natural
Science Foundation of Hubei Province (grant no. 2012FFB02435) and
the Central University Special Funding (grant no. 2013QN191).
References
|
1
|
Estey E and Döhner H: Acute myeloid
leukaemia. Lancet. 368:1894–1907. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Smith A, Howell D, Patmore R, Jack A and
Roman E: Incidence of haematological malignancy by sub-type: A
report from the Haematological Malignancy Research Network. Br J
Cancer. 105:1684–1692. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Sun Z, Zhang A, Jiang T, Du Z, Che C and
Wang F: MiR-145 suppressed human retinoblastoma cell proliferation
and invasion by targeting ADAM19. Int J Clin Exp Pathol.
8:14521–14527. 2015.PubMed/NCBI
|
|
4
|
Zhou XU, Qi L, Tong S, Cui YU, Chen J,
Huang T, Chen Z and Zu XB: miR-128 downregulation promotes growth
and metastasis of bladder cancer cells and involves VEGF-C
upregulation. Oncol Lett. 10:3183–3190. 2015.PubMed/NCBI
|
|
5
|
Tan G, Wu L, Tan J, Zhang B, Tai WC, Xiong
S, Chen W, Yang J and Li H: MiR-1180 promotes apoptotic resistance
to human hepatocellular carcinoma via activation of NF-κB signaling
pathway. Sci Rep. 6:223282016. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Shih AH, Abdel-Wahab O, Patel JP and
Levine RL: The role of mutations in epigenetic regulators in
myeloid malignancies. Nat Rev Cancer. 12:599–612. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
White BS and DiPersio JF: Genomic tools in
acute myeloid leukemia: From the bench to the bedside. Cancer.
120:1134–1144. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Rommer A, Steinleitner K, Hackl H,
Schneckenleithner C, Engelmann M, Scheideler M, Vlatkovic I,
Kralovics R, Cerny-Reiterer S, Valent P, et al: Overexpression of
primary microRNA 221/222 in acute myeloid leukemia. BMC Cancer.
13:3642013. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
DiNardo CD and Cortes JE: New treatment
for acute myelogenous leukemia. Expert Opin Pharmacother.
16:95–106. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Copsel S, Bruzzone A, May M, Beyrath J,
Wargon V, Cany J, Russel FG, Shayo C and Davio C: Multidrug
resistance protein 4/ ATP binding cassette transporter 4: A new
potential therapeutic target for acute myeloid leukemia.
Oncotarget. 5:9308–9321. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Croce CM: MicroRNA dysregulation in acute
myeloid leukemia. J Clin Oncol. 31:2065–2066. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Erdogan B, Bosompem A, Peng D, Han L,
Smith E, Kennedy ME, Alford CE, Wu H, Zhao Z, Mosse CA, et al:
Methylation of promoters of microRNAs and their host genes in
myelodysplastic syndromes. Leuk Lymphoma. 54:2720–2727. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Alemdehy MF and Erkeland SJ: MicroRNAs:
Key players of normal and malignant myelopoiesis. Curr Opin
Hematol. 19:261–267. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Vasilatou D, Papageorgiou S, Pappa V,
Papageorgiou E and Dervenoulas J: The role of microRNAs in normal
and malignant hematopoiesis. Eur J Haematol. 84:1–16. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Shibayama Y, Kondo T, Ohya H, Fujisawa S,
Teshima T and Iseki K: Upregulation of microRNA-126-5p is
associated with drug resistance to cytarabine and poor prognosis in
AML patients. Oncol Rep. 33:2176–2182. 2015.PubMed/NCBI
|
|
16
|
Jansson MD and Lund AH: MicroRNA and
cancer. Mol Oncol. 6:590–610. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Nana-Sinkam SP and Croce CM: Clinical
applications for microRNAs in cancer. Clin Pharmacol Ther.
93:98–104. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Chen Y, Jacamo R, Konopleva M, Garzon R,
Croce C and Andreeff M: CXCR4 downregulation of let-7a drives
chemoresistance in acute myeloid leukemia. J Clin Invest.
123:2395–2407. 2013. View
Article : Google Scholar : PubMed/NCBI
|
|
19
|
Li X and Zhong H: The diagnosis,
prognosis, and therapeutic application of MicroRNAs in
haematological malignancies. Hematology. 21:263–271. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Wang XX, Wang Y, Wang PP and Li Y:
Expression of microRNA-148/152 family in the mematological
malignancies. Zhongguo Shi Yan Xue Ye Xue Za Zhi. 23:1173–1178.
2015.(In Chinese). PubMed/NCBI
|
|
21
|
Amodio N, Rossi M, Raimondi L, Pitari MR,
Botta C, Tagliaferri P and Tassone P: miR-29s: A family of
epi-miRNAs with therapeutic implications in hematologic
malignancies. Oncotarget. 6:12837–12861. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Sabattini E, Bacci F, Sagramoso C and
Pileri SA: WHO classification of tumours of haematopoietic and
lymphoid tissues in 2008: An overview. Pathologica. 102:83–87.
2010.PubMed/NCBI
|
|
23
|
Lo Coco F and Foa R: Diagnostic and
prognostic advances in the immunophenotypic and genetic
characterization of acute leukaemia. Eur J Haematol. 55:1–9. 1995.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Bennett JM, Catovsky D, Daniel MT,
Flandrin G, Galton DA, Gralnick HR and Sultan C: Proposed revised
criteria for the classification of acute myeloid leukemia. A report
of the French-American-British cooperative group. Ann Intern Med.
103:620–625. 1985. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Livak and Schmittgen, . Analysis of
relative gene expression data using real-time quantitative PCR and
the 2-ΔΔCt method. Methods. 25:402–408. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Li BS, Zhao YL, Guo G, Li W, Zhu ED, Luo
X, Mao XH, Zou QM, Yu PW, Zuo QF, et al: Plasma microRNAs, miR-223,
miR-21 and miR-218, as novel potential biomarkers for gastric
cancer detection. PLoS One. 7:e416292012. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Huang BS, Luo QZ, Han Y, Li XB, Cao LJ and
Wu LX: microRNA-223 promotes the growth and invasion of
glioblastoma cells by targeting tumor suppressor PAX6. Oncol Rep.
30:2263–2269. 2013.PubMed/NCBI
|
|
28
|
Wu L, Li H, Jia CY, Cheng W, Yu M, Peng M,
Zhu Y, Zhao Q, Dong YW, Shao K, et al: MicroRNA-223 regulates FOXO1
expression and cell proliferation. FEBS Lett. 586:1038–1043. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Nian W, Ao X, Wu Y, Huang Y, Shao J, Wang
Y, Chen Z, Chen F and Wang D: miR-223 functions as a potent tumor
suppressor of the Lewis lung carcinoma cell line by targeting
insulin-like growth factor-1 receptor and cyclin-dependent kinase
2. Oncol Lett. 6:359–366. 2013.PubMed/NCBI
|
|
30
|
Li J, Guo Y, Liang X, Sun M, Wang G, De W
and Wu W: MicroRNA-223 functions as an oncogene in human gastric
cancer by targeting FBXW7/hCdc4. J Cancer Res Clin Oncol.
138:763–774. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Laios A, O'Toole S, Flavin R, Martin C,
Kelly L, Ring M, Finn SP, Barrett C, Loda M, Gleeson N, et al:
Potential role of miR-9 and miR-223 in recurrent ovarian cancer.
Mol Cancer. 7:352008. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Namløs HM, Meza-Zepeda LA, Barøy T,
Østensen IH, Kresse SH, Kuijjer ML, Serra M, Bürger H,
Cleton-Jansen AM and Myklebost O: Modulation of the osteosarcoma
expression phenotype by microRNAs. PLoS One. 7:e480862012.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Karakatsanis A, Papaconstantinou I,
Gazouli M, Lyberopoulou A, Polymeneas G and Voros D: Expression of
microRNAs, miR-21, miR-31, miR-122, miR-145, miR-146a, miR-200c,
miR-221, miR-222, and miR-223 in patients with hepatocellular
carcinoma or intrahepatic cholangiocarcinoma and its prognostic
significance. Mol Carcinog. 52:297–303. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Li S, Li Z, Guo F, Qin X, Liu B, Lei Z,
Song Z, Sun L, Zhang HT, You J and Zhou Q: miR-223 regulates
migration and invasion by targeting Artemin in human esophageal
carcinoma. J Biomed Sci. 18:242011. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Liang H, Yan X, Pan Y, Wang Y, Wang N, Li
L, Liu Y, Chen X, Zhang CY, Gu H and Zen K: MicroRNA-223 delivered
by platelet-derived microvesicles promotes lung cancer cell
invasion via targeting tumor suppressor EPB41L3. Mol Cancer.
14:582015. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Wei Y, Yang J, Yi L, Wang Y, Dong Z, Liu
Z, Ou-yang S, Wu H, Zhong Z, Yin Z, et al: MiR-223-3p targeting
SEPT6 promotes the biological behavior of prostate cancer. Sci Rep.
4:75462014. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Zhou X, Jin W, Jia H, Yan J and Zhang G:
MiR-223 promotes the cisplatin resistance of human gastric cancer
cells via regulating cell cycle by targeting FBXW7. J Exp Clin
Cancer Res. 34:282015. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Liang L, Zhu J, Zaorsky NG, Deng Y, Wu X,
Liu Y, Liu F, Cai G, Gu W, Shen L and Zhang Z: MicroRNA-223
enhances radiation sensitivity of U87MG cells in vitro and in vivo
by targeting ataxia telangiectasia mutated. Int J Radiat Oncol Biol
Phys. 88:955–960. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Wang Z, Inuzuka H, Zhong J, Wan L,
Fukushima H, Sarkar FH and Wei W: Tumor suppressor functions of
FBW7 in cancer development and progression. FEBS Lett.
586:1409–1418. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Lau AW, Fukushima H and Wei W: The Fbw7
and betaTRCP E3 ubiquitin ligases and their roles in tumorigenesis.
Front Biosci (Landmark Ed). 17:2197–2212. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Welcker M and Clurman BE: FBW7 ubiquitin
ligase: A tumour suppressor at the crossroads of cell division,
growth and differentiation. Nat Rev Cancer. 8:83–93. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Minella AC and Clurman BE: Mechanisms of
tumor suppression by the SCF(Fbw7). Cell Cycle. 4:1356–1359. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Yada M, Hatakeyama S, Kamura T, Nishiyama
M, Tsunematsu R, Imaki H, Ishida N, Okumura F, Nakayama K and
Nakayama KI: Phosphorylation-dependent degradation of c-Myc is
mediated by the F-box protein Fbw7. EMBO J. 23:2116–2125. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Nateri AS, Riera-Sans L, Da Costa C and
Behrens A: The ubiquitin ligase SCFFbw7 antagonizes apoptotic JNK
signaling. Science. 303:1374–1378. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Koepp DM, Schaefer LK, Ye X, Keyomarsi K,
Chu C, Harper JW and Elledge SJ: Phosphorylation-dependent
ubiquitination of cyclin E by the SCFFbw7 ubiquitin ligase.
Science. 294:173–177. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Fryer CJ, White JB and Jones KA:
Mastermind recruits CycC: CDK8 to phosphorylate the Notch ICD and
coordinate activation with turnover. Mol Cell. 16:509–520. 2004.
View Article : Google Scholar : PubMed/NCBI
|