Introduction
An unstable genome is an important characteristic of
cancer cells, and gene expression between cancerous and
non-cancerous tissues is significantly different (1). Approximately one half of microRNA (miR)
genes are distributed at unstable sites of the genome, including
fragile sites of chromatin and cleavage sites (2,3). There is
a significant difference in the expression level of miRs in
cancerous tissue and normal tissue (4). Based on their diversity and wide
distribution throughout the genome, miRs may serve as an important
tumor diagnostic method (5).
Lung cancer has the highest mortality rate worldwide
at 92%, and 80% of lung cancer cases are non-small-cell lung cancer
(NSCLCs) (6). Although early-stage
diagnostic techniques, chemotherapy and other targeted therapies
are improving, the 5-year survival rate of NSCLC remains low at 6%
(7). The incidence and progression of
lung cancer is a multi-factor and multi-step complex process
(8). Numerous miRs have roles as
oncogenes or cancer suppressor genes in lung cancer cells, and
there has been a great deal of investigation concerning the role of
miRs in lung cancer; miR17-29 cluster, miR-31, miR-26a, miR-107,
miR-185, miR-let-7 and miR-29a are all associated with the onset of
lung cancer (9–15). These findings indicate that miRs are
critical in the incidence and progression of lung cancer.
Currently, antitumor drugs specific to the cell
cycle, including colchicine and taxol, demonstrate poor specificity
to tumor cells and possess side-effects, including toxicity
(16). In addition, antitumor drugs
are liable to the development of drug resistance, which restricts
clinical treatment (17). Therefore,
treatment methods that target specific cell signaling pathways
require investigation, and studies have identified novel targets
for targeted therapy of tumor molecules, including the regulation
of miRs (18,19). Therefore, miRs provide a novel method
for treating malignant tumors, and it has been observed that
treating cancer by regulating the level of miRs in the cancerous
cells is possible (20). The present
study investigated miRs in human lung cancer and paracarcinoma
tissue using the miRchip technique (21), and investigated if the expression
levels of miRs were associated with lung cancer cells in
vitro using quantitative polymerase chain reaction (qPCR). In
addition, the present study investigated the biological functions
of the potential target gene of miR-4458 in combination with
cellular proliferation and cell cycle assays, and therefore
preliminarily explained the role of miR-4458 in the onset of lung
cancer.
Materials and methods
General data and patient tissue
specimens
The present study selected 94 NSCLC patients with
complete clinical and follow-up visit data, who underwent excision
of NSCLC tissue at the Inner Mongolia Medical University (Hohhot,
China) between January 2007 and June 2014. FOr each patient,
specimens were taken of normal lung tissue from the lung cancer and
normal paracarcinoma lung tissue (excised 5 cm from the edge of the
tumor). No patient underwent radiotherapy or chemotherapy prior to
surgery. The tissue was cryopreserved immediately following
surgery, when all patients were pathologically diagnosed with
NSCLC. All histological diagnoses were demonstrated by hematoxylin
and eosin staining (Beyotime Institute of Biotechnology, Haimen,
China). The information and specimen collection for all patients
and the experiment specification were in accordance with standard
operating procedures of the Ethics Committee of Inner Mongolia
Medical University (Hohhot, China). The experimental content
involving design of the medical ethics was approved by the Ethics
Committee of the Inner Mongolia Medical University (22,23).
Written informed consent was obtained from all patients.
General characteristics of the
patients
There were 94 patients that underwent NSCLC surgery,
aged 49–86 years (median age, 54 years). There were 69 male
patients (73.4%) and 25 female patients (26.6%).
Tumor-node-metastasis (TNM) stages (24) for the patients was as follows: T1N0M0,
32 patients; T2N2M0, 25 patients; T2N0M0, 14 patients; T2N1M0, 11
patients; other stage, 12 patients. Histological stage: Ia, 17
patients; Ib, 13 patients; IIa, 20 patients; IIb, 9 patients; IIIa,
31 patients; and IV, 4 patients (25). The final follow-up appointment for all
patients was on June 30, 2014.
Cell lines and culture
Human lung carcinoma A549 and H460 cell lines were
purchased from Sinovac Biotech Ltd. (Shanghai, China), and human
lung fibroblast HFL1 and human embryonic epithelial HEK293T cell
lines were obtained from the Molecular Biology Experimental Center
of Inner Mongolia Medical University (Hohhot, China). These two
cells lines were cultured with Dulbecco's modified Eagle's medium
(DMEM; Beyotime Institute of Biotechnology) containing 10% fetal
bovine serum (Beyotime Institute of Biotechnology) in an atmosphere
of 5% CO2 at 37°C. Cells in the logarithmic phase
(rapidly growing for 24 h) were used for the following
experiments.
miR hybrid chip
The present study selected 15 patients from the
original 94 patients at random. Total RNA was extracted from the
lung cancer tissues using TRIzol® Reagent (Invitrogen™;
Thermo Fisher Scientific, Inc., Waltham, MA, USA), according to the
manufacturer's protocol. The hybrid chip used in the present study
was miRCURY™ LNA™ microRNA Array kit (Exiqon, Vedbaek, Denmark).
The chip used the Sanger microRNAs sequence database version 20.0
(www.mirbase.org/). An RNA fluorescent marker and
chip hybridization was performed using the miRCURY™LNA™ microRNA
Array Hi-Power Labeling kit (Exiqon). The marker enzyme Hy3TM
fluorophore (Shanghai Biotechnology Corp., Shanghai, China) was
used to mark the RNA for the chip hybridization fluorescence probe,
according to the manufacturer's protocol. Each probe experiment was
repeated 3 times. The green fluorescence signal was scanned with
the Gene Pix 4000B Microarray Scanner (Molecular Devices LLC,
Sunnyvale, CA, USA). The green fluorescence intensity was analyzed
using Gene Pix Pro version 6.0 (Axon Instruments; Molecular
Devices, LLC). The median value correction method was used to
obtain the corrected value (26). The
expression was regarded as upregulated if the ratio of the miR
fluorescence corrected values of the NSCLC and paracarcinoma
specimens was ≥1.5. The expression was regarded as downregulated if
the ratio of the miR fluorescence corrected values of the two
specimens was ≤0.67. The database miRGen version 3.0
(carolina.imis.athena-innovation.gr/index.php?r=mirgenv3) was used
for bioinformatic analysis, which integrated various target
prediction tools, including PicTar, miRanda, DIANA-micro T and
Target Scan S. DAVID software version 2.4 (david.abcc.ncifcrf.gov/) was used for functional
classification analysis of the target genes (27).
Detecting cellular proliferation using
CellTiter
A549, H460 and HFLl cells in the logarithmic phase
were selected, seeded in a 96-well microplate (4×103
cells/well; Beyotime Institute of Biotechnology) with DMEM
containing 10% fetal bovine serum and placed in an incubator (5%
CO2; 37°C) for culture overnight. The culture medium was
discarded following 1 h of incubation. The cells were agitated for
10 min at room temperature following the addition of 35 µl fresh
culture medium and 35 µl CellTiter 96 detection reagent (Promega
Corporation, Madison, WI, USA). The cells (50 µl) were transferred
to an opaque white plate (Beyotime Institute of Biotechnology) at
24, 48 and 72 h. The fluorescence value was tested using a
fluorescence luminometer (F97Pro; Shanghai Lengguang Technology Co.
Ltd, Shanghai, China). Relative proliferative activity =
appreciation rate of fluorescence value of treatment group /
appreciation rate of fluorescence value of control group (28).
qPCR assay
Small RNAs (<100 nt) were extracted from the lung
cancer cells and tissues using the Universal MicroRNA kit (Qiagen,
Hilden, Germany). In total, 5 g tissue = 1 ml water extract.
Complementary (c)DNA was reverse transcribed using TaqMan MicroRNA
Reverse Transcription kit (Applied Biosystems; Thermo Fisher
Scientific, Inc.) on ice. The cDNA synthesis reaction mixture was
15 µl, which consisted of the following: 0.15 µl dNTPs (100 mM),
1.00 µl MultiScribe™ Reverse Transcriptase, 1.50 µl 10X Reverse
Transcription Buffer, 0.19 µl RNase Inhibitor, 4.16 µl
nuclease-free water, 3 µl hsa-miR-4458 or U6 5X RT Primer and 5 µl
RNA sample (10 ng total RNA). Primers were synthesized by Shanghai
Yanjing Biotechnology Co., Ltd (Shanghai, China). Primer Design
software version 5 (PREMIER Biosoft, Palo Alto, CA, USA) and DNAMAN
gene analysis software demo version 5.2.9 (Lynnon Biosoft, San
Ramon, CA, USA) were used in order to detect the primer location
and to screen the appropriate primers. The primer sequences were as
follows: miR-4458, 5′-ACCTACAATGTGTGCTGGCTTTC-3′; miR-4458
inhibitor, 5′-GATCCCAGCAGCCAAGGCTATGTTTCTACCGAAC-3′; and internal
reference U6, 5′-CACCACGTTTATACGCCGGTG-3′. The reaction conditions
were 16°C for 30 min, 42°C for 30 min, and 85°C for 5 min. The cDNA
synthesized by reverse transcription was stored at −20°C for later
use. A relative qPCR analysis was conducted with U6 as the internal
reference. The PCR reaction occurred in a qPCR amplifier
(Quantstudio™ DX Real-Time PCR Instrument; Thermo Fisher
Scientific, Inc.). The reaction mixture was 20 µl and consisted of
the following: 1.33 µl cDNA, 1 µl hsa-miR-4458, or U6 20X Real Time
primer, 10 µl TaqMan 2X Universal PCR Master Mix and 7.67 µl
nuclease-free water, taken from the Platinum®
Quantitative PCR SuperMix (Invitrogen; Thermo Fisher Scientific,
Inc.). The reaction conditions were 95°C for 10 min, 95°C for 15
sec and 60°C for 1 min for a total of 40 cycles (29). qPCR data analysis was performed using
Sequence Detection System version 2.3 software (ABI
PRISM® 7900 HT; Applied Biosystems; Thermo Fisher
Scientific, Inc.). The expression level of miRs was expressed using
the ΔCq value (Cq miR relative to Cq U6) (30). Each experiment was repeated 3
times.
Plasmid cell transfection
miR-4458 mimic and inhibitor and miR-Scramble were
synthesized by miR-Ribo (Guangzhou RiboBio Co., Ltd., Guangzhou,
China). The sequences were as follows: miR-4458 mimic,
5′-AGAGGUAGGUGUGGAAGAA-3′; miR-4458 inhibitor,
5′-UCGCACUGCUAGCUACGCUAGC-3′; miR-Scramble,
5′-CGAGUAGACUCCAACUGUGAUC-3′. The miRs were transfected into
plasmids, and these plasmids were transfected into A549 and H460
cells. The A549 and H460 cells were cultured for 24 h in a 24-well
microplate (5×104 cells/well) prior to transfection. The
cell transfection process was in accordance with the manufacturer's
protocol for Oligofectamine™ Transfection Reagent (Invitrogen;
Thermo Fisher Scientific, Inc.). miRs were transfected into
riboFECT™ CP plasmids (Guangzhou RiboBio Co., Ltd.). DMEM was added
to each well at 4 h following transfection. The cells were cultured
for 48 and 72 h at 37°C in an atmosphere of 5% CO2. The
experiment included a negative control (miR-Scramble), blank
control (no miR mimic) and lipidosome (Guangzhou RiboBio Co., Ltd.)
groups with 3 duplicated wells for each group.
Luciferase reporter gene
experiment
The 3′-UTRs of CCND1 were amplified by PCR using the
following primers: 5′-GTACTGGAGAATGTGCCCTGCGGCA-3′ and
5′-AGCTCGAGTAAGGCTCTGCACCCGC-3′ (Primer Premier version 6.0;
PrimerDesign Ltd., Chandler's Ford, UK). MUT-CCND1–3′UTR was
designed to completely inhibit the 9 consistent base sequences of
miR-4458 and WT-CCND1–3′UTR. DNAMAN gene analysis software demo
version 5.2.9 was used to identify the consistent base sequences.
The luciferase reporter vector pGL3M-CCND1–3′UTR (50 ng) (Promega
Corporation) and the control plasmid pcDNA3.1 (10 ng) (GeneCopoeia,
Inc., Rockville, MD, USA) were co-transfected into 293T cells using
Lipofectamine® 2000 Reagent (Thermo Fisher Scientific,
Inc.) and the lipidosome mediation transfection method (31). The cells were named the experimental
group pGL3M-WT-CCND1–3′UTR, positive control group
pGL3M-MUT-CCND1–3′UTR and negative control group pGL3 M. The PCR
products were then inserted into pGL3-basic vector. Target site
mutations were generated using the PCR products with the
appropriate primers containing point substitutions (MUT-CCND1
5′-GUUGCUGCAACACAACUAUAUAU-3′). The sequences were verified by DNA
sequencing. HEK293T cells were co-transfected with 0.1 mg reporter
plasmid with 0.65 pmol miRNA mimic or control miRNA in 96-well
plates. Luciferase activity was detected 48 h later using a
dual-luciferase reporter assay system and normalized to
Renilla activity.
Investigating the cell cycle using
flow cytometry
A549 and H460 cells were digested with pancreatic
enzymes (Beytime Institute of Biotechnology) to obtain a cell
suspension (following culturing for 48 h at 37°C in an atmosphere
of 5% CO2), washed with phosphate-buffered saline (PBS;
Beyotime Institute of Biotechnology) and resuspended in 700 µl
ethanol and 300 µl 10% fetal bovine serum. The cells were mixed
with 1 ml of −20°C absolute ethyl alcohol (Beyotime Institute of
Biotechnology) and fixed at 4°C overnight using P0020 fixing
reagent (Beyotime Institute of Biotechnology). The cells were
centrifuged at 560 × g for 5 min, washed once or twice with PBS
following discarding of the ethanol and resuspended in PBS.
Following addition of the PBS staining solution containing
propidium iodide (PI; final concentration, 50 µg/ml; Beyotime
Institute of Biotechnology) and RNase A (final concentration 100
µg/ml; Shanghai Biotechnology Corp.), the cells were stained for 1
h in the dark. Subsequently, the cells were detected using the
MoFlo® Astrios™ EQ Flow Cytometer (Beckman Coulter,
Inc., Bream, CA, USA) (29). The
experiment was repeated 3 times.
Investigating CCND1 protein expression
using western blot analysis
A549 and H460 cells were transferred to a
centrifugal tube. The cells were subjected to protein extraction
(ProteoPrep® Total Extraction Sample kit; Sigma-Aldrich,
St. Louis, MO, USA) and quantification in accordance with the
manufacturer's protocol. Subsequently, 30 µg protein underwent 10%
sodium dodecyl sulfate-polyacrylamide gel electrophoresis. The gel
was electrotransferred onto a polyvinylidene difluoride (PVDF)
membrane and blocked in 5% skim milk powder for 2 h at room
temperature. The blocked PVDF membrane was placed in the primary
antibody (rabbit anti-CCND1 polyclonal antibody; dilution, 1:200;
catalog no., BA0873; Boster Wuhan Biological Engineering, Co.,
Ltd., Wuhan, China) solution diluted with Tris-HCl-buffered saline
and Tween (TBST) and slowly agitated at 4°C overnight. The membrane
was washed at room temperature with western wash buffer (Beyotime
Institute of Biotechnology). The PVDF membrane was incubated with
secondary antibodies (fluorescein isothiocyanate-conjugated donkey
anti-rabbit; dilution, 1:200; catalog no., 711-475-152; and cyanine
3-conjugated donkey anti-rabbit; dilution, 1:200; catalog no.,
711-475-205; Jackson Immuno Research Laboratories, Inc., West
Grove, PA, USA) diluted with TBST (1:10,000) for 2 h at room
temperature. Enhanced chemiluminescence X-ray imaging (Beyotime
Institute of Biotechnology) was used to detect the signals. The
signal intensity was subject to a relative quantitative analysis
using imaging analysis software (Image-Pro Plus version 6; Media
Cybernetics, Rockville, MD, USA). The gel analysis was expressed
with integrated optical density value. All reagents for the western
blot were purchased from Sigma-Aldrich.
Immunohistochemistry
The patient tissue specimens were fixed in 4%
formaldehyde (Beyotime Institute of Biotechnology) at 4°C, embedded
in paraffin (Beyotime Institute of Biotechnology), sectioned (5 µm
thickness) with a paraffin slicing machine (YD-202A; Zhengzhou
Nanbei Instrument Equipment Co., Ltd., Zhengzhou, China), dried in
an incubator at 37°C, subjected to antigen retrieval following
dewaxing with xylene (Beyotime Institute of Biotechnology), blocked
in 5% bovine serum albumin (diluted with PBS; Beyotime Institute of
Biotechnology), incubated with primary antibody (rabbit polyclonal
CCND1 antibody; dilution, 1:200; Leica Microsystems GmbH, Wetzlar,
Germany) at 4°C overnight followed by washing with PBS 3 times for
3 min each. The sections were subsequently incubated with goat IgG
secondary antibody (dilution, 1:200; Leica Microsystems GmbH) for
30 min at room temperature, washed with PBS 3 times for 3 min each
and stained with the color reagent 3,3′-diaminobenzidine, followed
by rinsing in water and counterstaining with hematoxylin (Beyotime
Institute of Biotechnology). A CX31-LV320 light microscope (Olympus
Corp., Tokyo, Japan; magnification, ×400) was used to observe the
samples.
Statistical analysis
The miR chip expression profile used the SAM and
TIGR Multiple Array Viewer version 4.0 software (www.tm4.org/index.html) for analysis. The data were
expressed as the mean ± standard deviation. Inter-group differences
were subject to Student's t-test. Inter-group enumeration data were
compared with the χ2 test or Fisher's exact probability
method. P<0.05 was considered to indicate a statistically
significant difference. P<0.01 was considered to indicate a
distinctively statistically significant difference. SPSS version
18.0 software (SPSS, Inc., Chicago, IL, USA) was used for
statistical analysis.
Results
Expression of miR-4458 in NSCLC and
paracarcinoma tissues
Based on the chip data, it was observed that there
was a significant difference in the expression of 58 miRs in 15
pairs of NSCLC and paracarcinoma tissues
(upregulated/downregulated, >1.5 times; Fig. 1A). There were differences in 26 miRs
in the NSCLC tissue compared with the paracarcinoma tissue with
variation >2 times, and 20 downregulated and 6 upregulated miRs
(Fig. 1B). miR-4458 had the most
significant alteration in its expression level between NSCLC and
paracarcinoma tissue (0.27 times in the paracarcinoma tissue). qPCR
was conducted to investigate the miR-4458 expression level in 94
patients with NSCLC. It was demonstrated that the median expression
level of miR-4458 in the paracarcinoma tissue of the 94 patients
decreased from 2.38 to 0.65 (P<0.001; Fig. 1C). The median downregulation of
miR-4458 expression in the 94 patients between non-cancerous and
cancerous tissue was 0.65, and the patients were divided into two
groups based on this cut-off value: miR-4458 low-expression group
(52 patients) and miR-4458 high-expression group (42 patients). The
1, 2, 3, 4 and 5-year total survival rates were 72.6, 48.2, 28.9,
16.1 and 14.4%, respectively, in the miR-4458 low-expression group,
compared with 69.7, 51.5, 44.9, 32.2 and 29.1%, respectively, in
the miR-4458 high-expression group (P=0.025; Fig. 1D). The 1, 2, 3, 4 and 5-year
relapse-free survival rates were 58.8, 33.9, 13.4, 7.8 and 7.8%,
respectively, in the miR-4458 low-expression group, compared with
70.6, 38.2, 34.4, 21.3 and 17.6% in the miR-4458 high-expression
group (P=0.019; Fig. 1E).
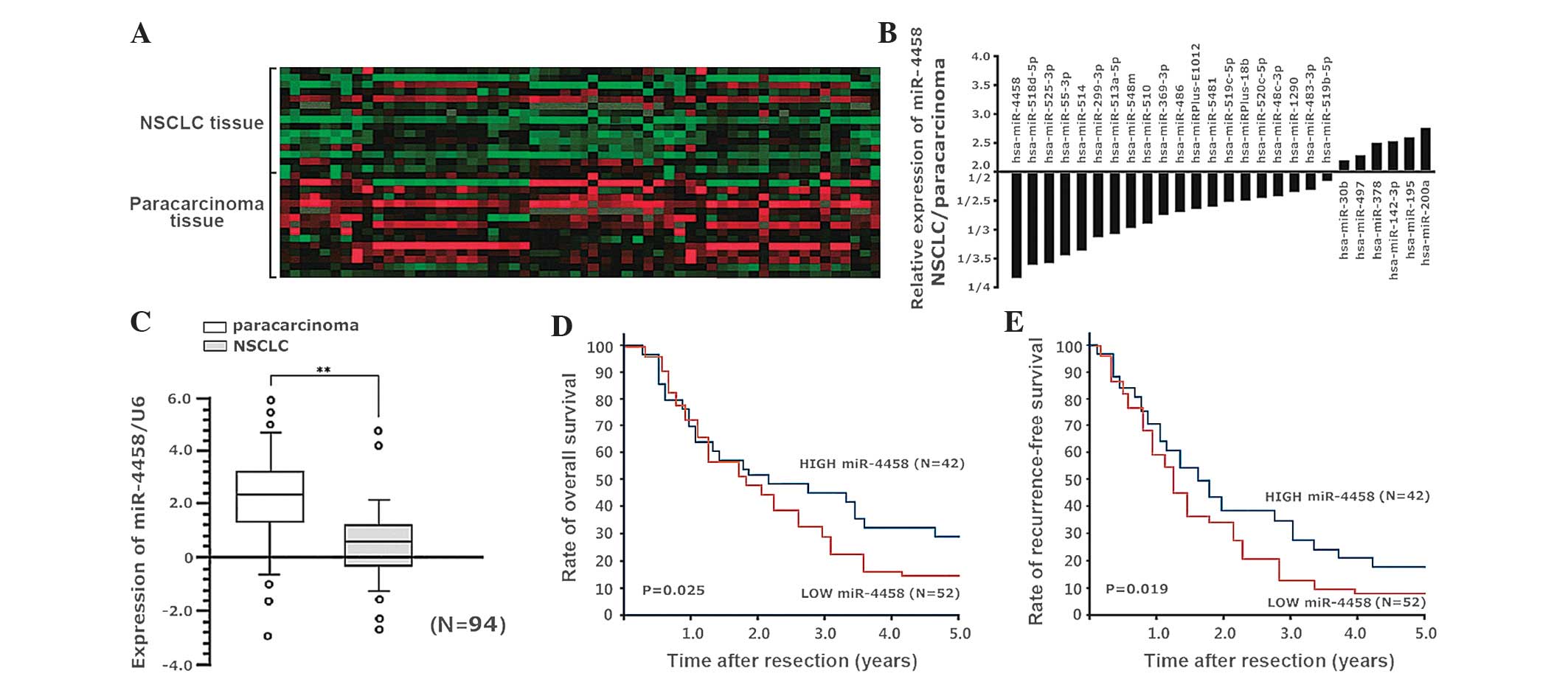 | Figure 1.(A) miR chip analysis demonstrated
that there was a significant difference in the expression of 58
miRs in 15 pairs of NSCLC and paracarcinoma tissues (variation
>1.5 times). (B) Compared with the paracarcinoma tissue, there
were 26 miRs (downregulated, 20; upregulated, 6) in NSCLC tissue
with variation >2 times. (C) Quantitative polymerase chain
reaction was conducted to investigate the miR-4458 level of 94
patients with NSCLC (U6 used as the internal reference). The level
of miR-4458 in the cancerous tissue was significantly reduced
compared with the paracarcinoma tissue (P<0.001). (D-E) The
patients were divided into miR-4458 low-expression group (52
patients) and miR-4458 high-expression group (42 patients), based
on the average downregulation of miR-4458 as a division point
(0.65). (D) The 1, 2, 3, 4 and 5-year overall survival rates in the
miR-4458 low-expression group and high-expression group (P=0.025)
and (E) the 1, 2, 3, 4 and 5-year relapse-free survival rates in
the miR-4458 low-expression and high-expression groups (P=0.019).
miR, microRNA; NSCLC, non-small cell lung cancer. |
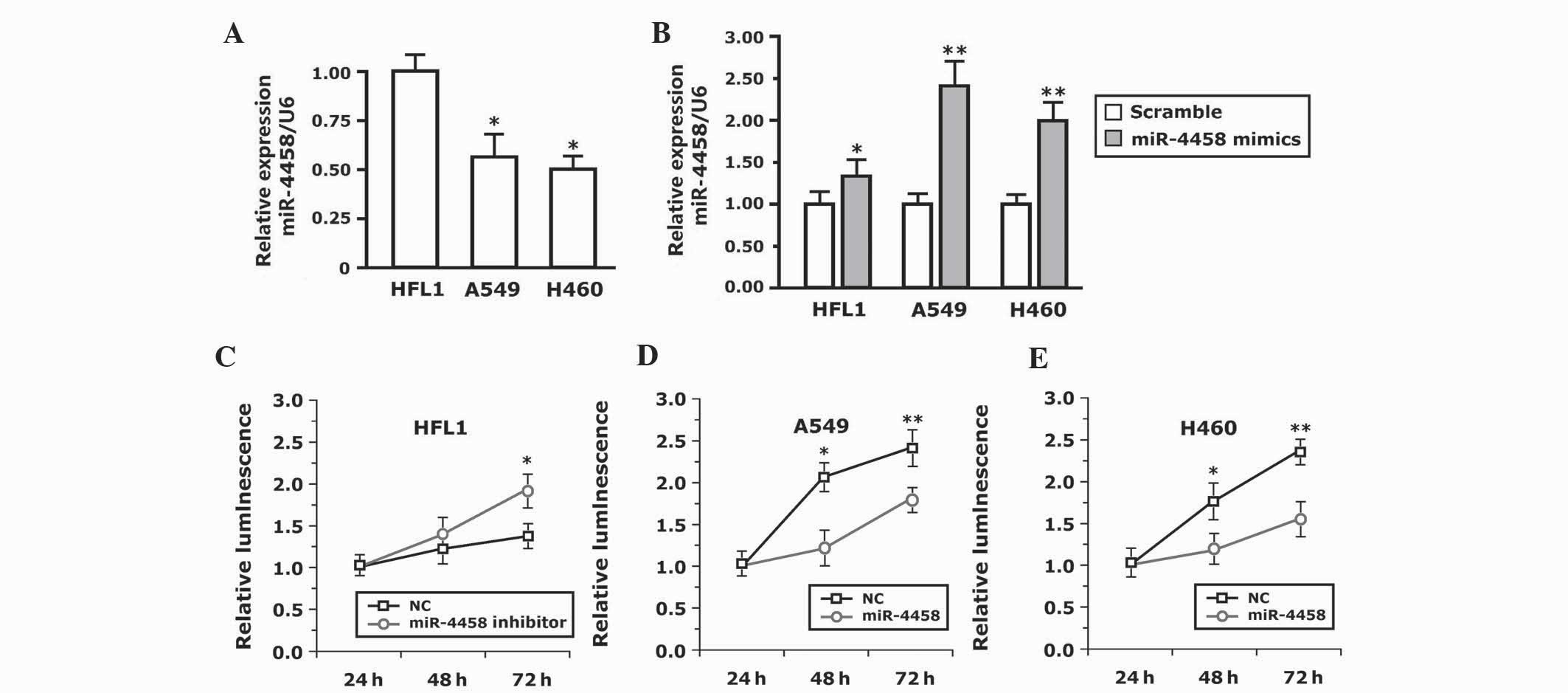
Association of expression of miR-4458
in lung carcinoma cell lines and cell proliferation
To study the biological functions of miR-4458, the
present study initially investigated the expression of miR-4458 in
human lung carcinoma A549 and H460 cell lines and the human lung
fibroblast HFL1 cell line. The results demonstrated that the
expression level of miR-4458 in A549 and H460 cell lines was
downregulated significantly compared with normal HFL1 cells
(P=0.017; Fig. 2A). The expression
level of miR-4458 in the A549 and H460 cell lines increased
significantly compared with HFL1 cells (P<0.001; Fig. 2B). Therefore, transfection of cells
with miR-4458 mimics was able to successfully increase the
expression of endogenous miR-4458.
The normal lung HFL1 cells were first transfected
with the miR-4458 inhibitor. A CellTiter kit was used to detect the
cell proliferation at 24, 48 and 72 h. There was no difference in
cell proliferation within the initial 48 h (data not shown). The
proliferation of the cells transfected with the miR-4458 inhibitor
was increased compared with the cells transfected with the negative
control miRNA (NC) at 72 h (P=0.025; Fig.
2C). To additionally verify the effect of miR-4458 on cellular
proliferation, A549 and H460 cells were transfected with miR-4458
and NC. Fig. 2D and E demonstrates
that the A549 and H460 cells transfected with miR-4458 proliferated
slowly compared with NC. There was no difference in cellular
proliferation within the initial 24 h. The cells transfected with
NC proliferated significantly more at 48 h compared with the cells
transfected with miR-4458 (P=0.011). The viability of the cells
transfected with miR-4458 decreased significantly compared with
that of the cells transfected with NC (P<0.001). Therefore,
miR-4458 was able to inhibit cellular proliferation in A549 and
H460 cells.
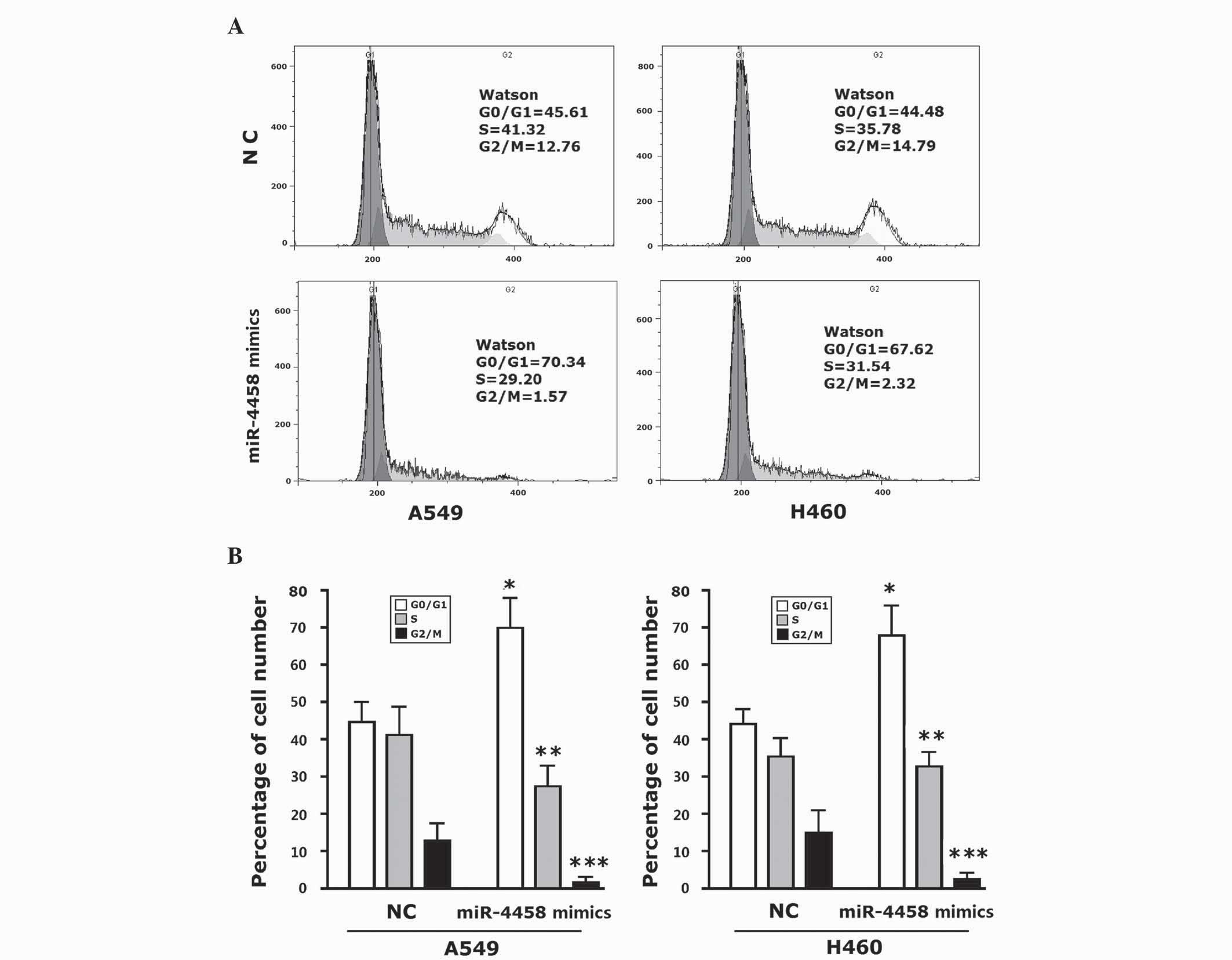
miR-4458 arrests lung carcinoma at
cell cycle stage G0/G1
Inhibition of cellular proliferation indicated the
possibility of miR-4458 inhibiting the cell cycle. Therefore, the
present study investigated the distribution of various phases of
the cells using flow cytometery. The cell cycle was analyzed using
PI-stained DNA. A549 cells transfected with miR-4458 mimics at
stage G0/G1 accounted for 69.94±8.05%. H460 cells at stage G0/G1
accounted for 68.15±7.75% (Fig. 3A and
B). These percentages were increased significantly compared
with the control group (P<0.001). The percentage of A549 cells
at stage S was significantly decreased compared with the control
cells (P=0.028), and the growth rate of H460 cells was also
observed to have slowed down. A549 and H460 cells at stage G2/M
were significantly decreased compared with the control group
(P<0.001). There were more A549 and H460 miR-4458-transfected
cells at stage G0/G1.
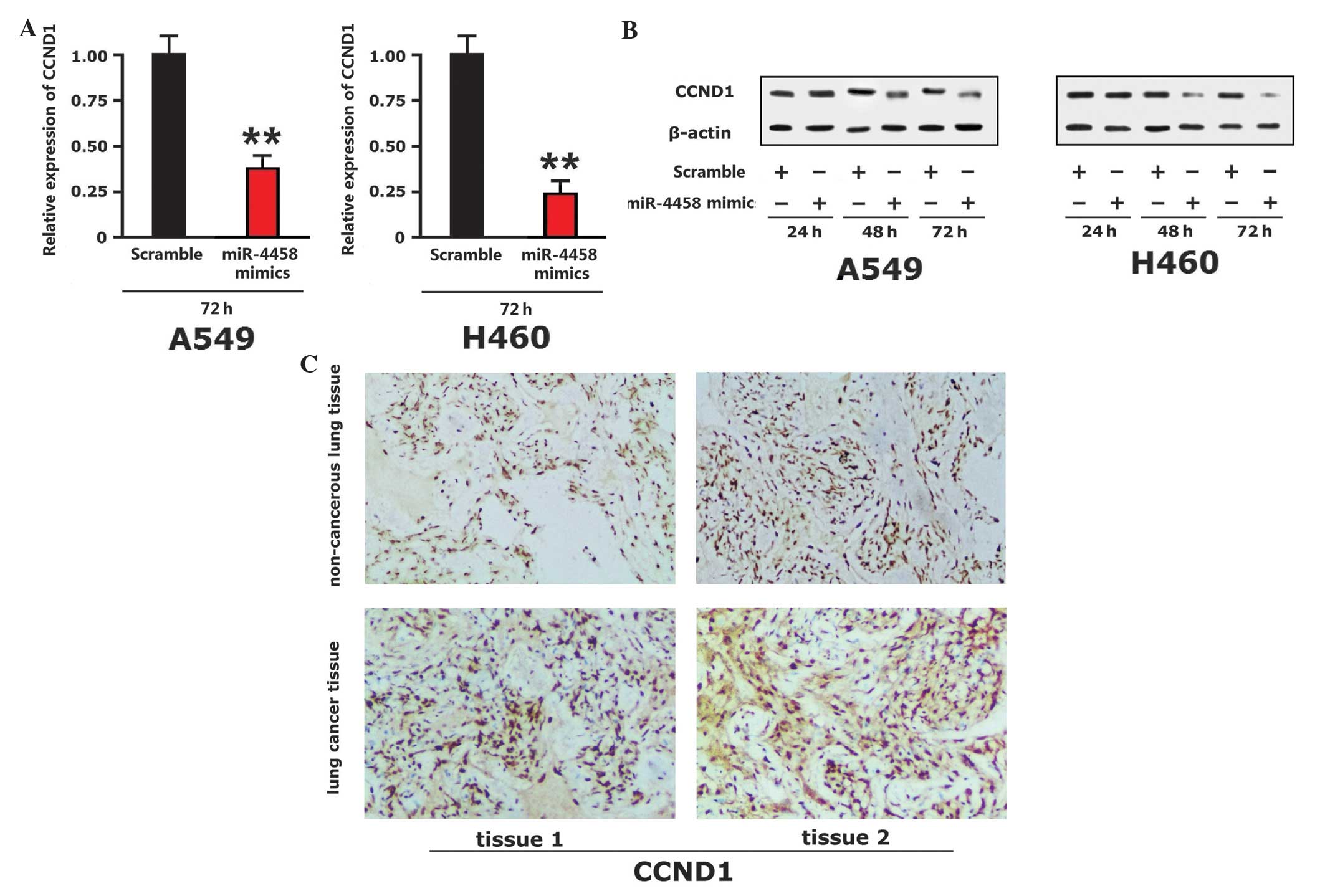
miR-4458 is capable of inhibiting
CCND1 protein expression in human NSCLC cells
Western blot analysis indicated that there was no
significant alteration in the levels of CCDN1 24 h subsequent to
the addition of A549 and H460 cells to miR-4458 mimics. CCND1
protein expression was downregulated at 48 h. CCND1 protein
expression was significantly downregulated at 72 h (P<0.001;
Fig. 4A and B). The present study
observed the expression of CCND1 protein in lung cancer tissue and
paracarcinoma tissue of lung cancer patients using
immunohistochemistry. The expression level of CCND1 in lung cancer
tissues was increased compared with the paracarcinoma tissue
(Fig. 4C). Therefore, miR-4458 was
capable of inhibiting the expression of CCND1 in lung cancer
tissue.
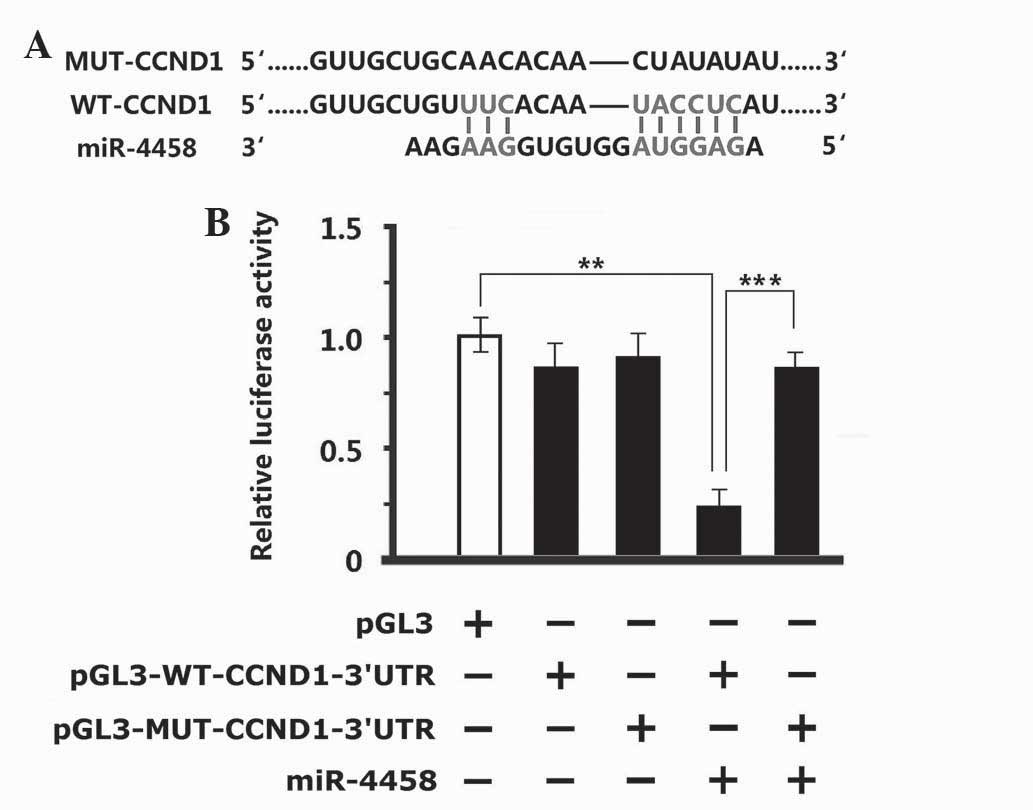
miR-4458 had 9 sequences completely consistent with
WT-CCND1–3′UTR (Fig. 5A). After
MUT-CCND1 was used to generate target site mutations at the seed
sequence, the reporter result of the luciferase assay conducted in
293T cells indicated that there were no significant changes in
pGL3M-MUT-CCND1–3′UTR and pGL3M-WT-CCND1–3′UTR in the negative
control group compared with pGL3M in the vacant plasmid group.
There were no significant changes in viability of the cells in the
MUT group while the fluorescence intensity in the WT group
decreased significantly following the addition of miR-4458 mimics
(Fig. 5B; P<0.001). These results
indicate that miR-4458 was capable of binding with the specific
sequence in the promoter of WT-CCND1–3′UTR. miR-4458 did not
function following an alteration in the specific sequence of the
promoter.
Discussion
There have been numerous studies concerning the gene
expression difference between cancerous and non-cancerous cells,
and they have demonstrated that metabolic signaling pathways alter
as a result of the difference in cancerous cells and normal
physiological processes (32). miRs
are diverse and are widely distributed in the genome; therefore,
alterations in the genomes of cancerous cells may be observed by
studying alterations in the expression of miRs (33). Consequently, miRs are molecules that
may serve as diagnostic markers for cancers.
The present study demonstrated that there is a low
expression of miR-4458 in lung cancer tissue based on the
difference in the expression of miRs between lung cancer and
paracarcinoma tissue in combination with qPCR verification. These
results indicate that miR-4458 may have a specific role in the
proliferation and progression of lung cancer. The present study
additionally revealed that miR-4458 inhibits the proliferation of
lung cancer cells to a greater extent in the lung carcinoma A549
and H460 cell lines. The present study demonstrated, using cell
cycle assays, that miR-4458 is capable of causing arrest of the
cell cycle at stage G0/G1, and therefore, inhibiting the
proliferation of cells (34). The
present study used TargetScan software to predict the target gene
of miR-4458; the software provided several hundred results.
Considering that miRs function by inhibiting target genes, the
present study focused on genes relevant to cell cycle and
apoptosis. Based on western blot analysis, the present results
demonstrated that miR-4458 is capable of inhibiting the expression
of CCND1 at the protein level, which confirms that CCND1 is a
target gene of miR-4458 (35). In the
present study, double reporter experiments have also demonstrated
this result. Overall, miR-4458 may participate in inhibiting the
onset of lung cancer, as a cancer suppressor gene.
CCND1 protein is encoded by the human CCND1 gene
(36). CCND1 is important in
controlling the cell growth cycle, and numerous types of cancer
abnormally express CCND1 proteins at a high level to stimulate cell
growth (37). Previous studies have
indicated that these proteins may be a fatal characteristic of
cancerous cells, since CCND1 inhibitors halt tumor growth and cause
cancer cell death (38,39). In addition, studies have revealed that
the inhibition of cyclin D1 induces aging of breast cancer cells in
mice and inhibition of CCND3 induces apoptosis of cancerous cells
of leukemic mice (40). Scientists
have identified that CCND inhibitor drugs also have similar effects
on human blood tumor cells (41).
This protein controls the cell cycle and regulates cell growth and
division. In numerous types of cancer, excessive cell cycle
proteins allow for fast growth of tumor cells (42,43). It
has been demonstrated that abnormal CCND1 is present in breast,
lung, endometrial, pancreatic and testicular cancer, multiple
myeloma and other types of blood cancer (44–48).
Mutation, amplification and over-expression of CCND1 may alter the
cell cycle process. These phenomena frequently occur in numerous
types of cancer and may cause the development of tumors (49). The present study hypothesizes that
miR-4458 may be developed and utilized in the future as a miR drug
for the treatment of cancer.
In conclusion, the present study indicates that
miR-4458 effectively inhibits migration, proliferation and the cell
cycle of lung tumor cells, as well as inhibiting CCND1 expression.
The present results provide a novel clue for a deeper understanding
of the association between miRs and the incidence of lung cancer,
and provide a novel hypothesis and basis for seeking potential
molecular targets. The cell cycle specificity of miR-4458 provides
a novel direction for developing drugs for blocking cell cycle
stage G0/G1 of tumor cells.
Acknowledgements
The present study was supported by the Project of
Incentive Funds for Guidance of Scientific and Technical Innovation
in Inner Mongolia Autonomous Region (grant no. 2014CZTCXYD), the
National Natural Science Foundation of China (grant no. 81260571),
the Public Health Project of The State Administration of
Traditional Chinese Medicine (grant no. GJZYYGLJ 11 AGL NZD) and
the Public Health Project of The State Administration of
Traditional Chinese Medicine (grant no. GJZYYGLJ 11 AGL).
References
|
1
|
Stepanenko AA and Dmitrenko VV: HEK293 in
cell biology and cancer research: Phenotype, karyotype,
tumorigenicity, and stress-induced genome-phenotype evolution.
Gene. 569:182–190. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Ma MZ, Li CX, Zhang Y, Weng MZ, Zhang MD,
Qin YY, Gong W and Quan ZW: Long non-coding RNA HOTAIR, a c-Myc
activated driver of malignancy, negatively regulates miRNA-130a in
gallbladder cancer. Mol Cancer. 13:1562014. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Verma M, Khoury MJ and Ioannidis JP:
Opportunities and challenges for selected emerging technologies in
cancer epidemiology: Mitochondrial, epigenomic, metabolomic and
telomerase profiling. Cancer Epidemiol Biomarkers Prev. 22:189–200.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Zhang L, Wei P, Shen X, Zhang Y, Xu B,
Zhou J, Fan S, Hao Z, Shi H, Zhang X, et al: MicroRNA expression
profile in penile cancer revealed by next-generation small RNA
sequencing. PloS One. 10:e01313362015. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Mo W, Zhang J, Li X, Meng D, Gao Y, Yang
S, Wan X, Zhou C, Guo F, Huang Y, et al: Identification of novel
AR-targeted microRNAs mediating androgen signalling through
critical pathways to regulate cell viability in prostate cancer.
PLoS One. 8:e565922013. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Hsiao SH, Chung CL, Lee CM, Chen WY, Chou
YT, Wu ZH, Chen YC and Lin SE: Suitability of computed
tomography-guided biopsy specimens for subtyping and genotyping of
non-small-cell lung cancer. Clin Lung Cancer. 14:719–725. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Ramnath N, Dilling TJ, Harris LJ, Kim AW,
Michaud GC, Balekian AA, Diekemper R, Detterbeck FC and Arenberg
DA: Treatment of stage III non-small cell lung cancer: Diagnosis
and management of lung cancer, 3rd ed: American College of Chest
Physicians evidence-based clinical practice guidelines. Chest.
143(Suppl 5): e314S–e340S. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Zhong KZ, Chen WW, Hu XY, Jiang AL and
Zhao J: Clinicopathological and prognostic significance of
microRNA-107 in human non small cell lung cancer. Int J Clin Exp
Pathol. 7:4545–4551. 2014.PubMed/NCBI
|
|
9
|
Chatterjee A, Chattopadhyay D and
Chakrabarti G: miR-17-5p downregulation contributes to paclitaxel
resistance of lung cancer cells through altering beclin1
expression. PLoS One. 9:e957162014. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Okudela K, Tateishi Y, Umeda S, Mitsui H,
Suzuki T, Saito Y, Woo T, Tajiri M, Masuda M, Miyagi Y and Ohashi
K: Allelic imbalance in the miR-31 host gene locus in lung
cancer-its potential role in carcinogenesis. PLoS One.
9:e1005812014. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Liu B, Wu X, Liu B, Wang C, Liu Y, Zhou Q
and Xu K: miR-26a enhances metastasis potential of lung cancer
cells via AKT pathway by targeting PTEN. Biochim Biophys Acta.
1822:1692–1704. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Zhang Z, Zhang L, Yin ZY, Fan XL, Hu B,
Wang LQ and Zhang D: miR-107 regulates cisplatin chemosensitivity
of A549 non small cell lung cancer cell line by targeting cyclin
dependent kinase 8. Int J Clin Exp Pathol. 7:7236–7241.
2014.PubMed/NCBI
|
|
13
|
Takahashi Y, Forrest AR, Maeno E,
Hashimoto T, Daub CO and Yasuda J: miR-107 and miR-185 can induce
cell cycle arrest in human non small cell lung cancer cell lines.
PLoS One. 4:e66772009. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Zhou C, Chen H, Han L, Wang A and Chen LA:
Identification of featured biomarkers in different types of lung
cancer with DNA microarray. Mol Biol Rep. 41:6357–6363. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Jakopovic M, Thomas A, Balasubramaniam S,
Schrump D, Giaccone G and Bates SE: Targeting the epigenome in lung
cancer: Expanding approaches to epigenetic therapy. Front Oncol.
3:2612013. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Gray BP, McGuire MJ and Brown KC: A
liposomal drug platform overrides peptide ligand targeting to a
cancer biomarker, irrespective of ligand affinity or density. PloS
One. 8:e729382013. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Gao CZ, Zhang Y, Chen J, Fei F, Wang TS,
Yang B, Dong P and Zhang YJ: Research progress of the drug delivery
system of antitumor platinum drugs with macrocyclic compounds. Yao
Xue Xue Bao. 50:650–657. 2015.(In Chinese). PubMed/NCBI
|
|
18
|
Lee WH, Liu HE, Chang JY, Liou JP and
Huang HM: MPT0B169, a new tubulin inhibitor, inhibits cell growth
and induces G2/M arrest in nonresistant and paclitaxel-resistant
cancer cells. Pharmacology. 92:90–98. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Gailhouste L, Gomez-Santos L and Ochiya T:
Potential applications of miRNAs as diagnostic and prognostic
markers in liver cancer. Front Biosci (Landmark Ed). 18:199–223.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Song T, Zhang X, Yang G, Song Y and Cai W:
Decrement of miR-199a-5p contributes to the tumorigenesis of
bladder urothelial carcinoma by regulating MLK3/NF-κB pathway. Am J
Transl Res. 7:2786–2794. 2015.PubMed/NCBI
|
|
21
|
Fesler A, Xu X, Zheng X, Li X, Jiang J,
Russo JJ and Ju J: Identification of miR-215 mediated
targets/pathways via translational immunoprecipitation expression
analysis (TrIP-chip). Oncotarget. 6:24463–24473. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Ochieng J, Ecuru J, Nakwagala F and
Kutyabami P: Research site monitoring for compliance with ethics
regulatory standards: Review of experience from Uganda. BMC Med
Ethics. 14:232013. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Marchetti D, Spagnolo A, Cicerone M,
Cascini F, La Monaca G and Spagnolo AG: Research ethics committee
auditing: The experience of a university hospital. HEC Forum.
25:257–268. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Jhun BW, Lee KJ, Jeon K, Suh GY, Chung MP,
Kim H, Kwon OJ, Sun JM, Ahn JS, Ahn MJ, et al: Clinical
applicability of staging small cell lung cancer according to the
seventh edition of the TNM staging system. Lung Cancer. 81:65–70.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Lee DS, Kim YS, Kay CS, Kim SH, Yeo CD,
Kim JW, Kim SJ, Kim YK, Ko YH, Kang JH and Lee KY: Distinctive
patterns of initially presenting metastases and clinical outcomes
according to the histological subtypes in stage IV non-small cell
lung cancer. Medicine (Baltimore). 95:e27952016. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Sokolenko S and Aucoin MG: A correction
method for systematic error in (1)H-NMR time-course data validated
through stochastic cell culture simulation. BMC Syst Biol.
9:512015. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Ying H, Lyu J, Ying T, Li J, Jin S, Shao
J, Wang L and Xu H: Risk miRNA screening of ovarian cancer based on
miRNA functional synergistic network. J Ovarian Res. 7:92014.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Kumarathasan P, Breznan D, Das D, Salam
MA, Siddiqui Y, Mackinnon-Roy C, Guan J, de Silva N, Simard B and
Vincent R: Cytotoxicity of carbon nanotube variants: A comparative
in vitro exposure study with A549 epithelial and J774 macrophage
cells. Nanotoxicology. 9:148–161. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Bao L, Zhao J, Dai X, Wang Y, Ma R, Su Y,
Cui H, Niu J, Bai S, Xiao Z, et al: Correlation between miR-23a and
onset of hepatocellular carcinoma. Clin Res Hepatol Gastroenterol.
38:318–330. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Ji H, Yang Z, Jiang W, Geng C, Gong M,
Xiao H, Wang Z and Cheng L: Antiviral activity of nano carbon
fullerene lipidosome against influenza virus in vitro. J Huazhong
Univ Sci Technolog Med Sci. 28:243–246. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Livak and Schmittgen, . Analysis of
relative gene expression data using real-time quantitative PCR and
the 2-ΔΔCt method. Methods. 25:402–408. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Liu XH, Sun M, Nie FQ, Ge YB, Zhang EB,
Yin DD, Kong R, Xia R, Lu KH, Li JH, et al: Lnc RNA HOTAIR
functions as a competing endogenous RNA to regulate HER2 expression
by sponging miR-331-3p in gastric cancer. Mol Cancer. 13:922014.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Paul S and Giri AK: Epimutagenesis: A
prospective mechanism to remediate arsenic-induced toxicity.
Environ Int. 81:8–17. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Yu X, Song H, Xia T, Han S, Xiao B, Luo L,
Xi Y and Guo J: Growth inhibitory effects of three miR-129 family
members on gastric cancer. Gene. 532:87–93. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Du B, Wang Z, Zhang X, Feng S, Wang G, He
J and Zhang B: MicroRNA-545 suppresses cell proliferation by
targeting cyclin D1 and CDK4 in lung cancer cells. PLoS One.
9:e880222014. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Li Z, Li X, Li C, Su Y, Fang W, Zhong C,
Ji W, Zhang Q and Su C: Transcription factor OCT4 promotes cell
cycle progression by regulating CCND1 expression in esophageal
carcinoma. Cancer Lett. 354:77–86. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Wong L, Power N, Miles A and Tropepe V:
Mutual antagonism of the paired-type homeobox genes, vsx2 and
dmbx1, regulates retinal progenitor cell cycle exit upstream of
ccnd1 expression. Dev Biol. 402:216–228. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Komori T: Regulation of Rb family proteins
by Cdk6/Ccnd1 in growth plates. Cell Cycle. 12:2161–2162. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Junk DJ, Cipriano R, Stampfer M and
Jackson MW: Constitutive CCND1/CDK2 activity substitutes for p53
loss, or MYC or oncogenic RAS expression in the transformation of
human mammary epithelial cells. PLoS One. 8:e537762013. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Jensen LB, Bartlett JM, Witton CJ,
Kirkegaard T, Brown S, Müller S, Campbell F, Cooke TG and Nielsen
KV: Frequent amplifications and deletions of G1/S-phase transition
genes, CCND1 and MYC in early breast cancers: A potential role in
G1/S escape. Cancer Biomark. 5:41–49. 2009.PubMed/NCBI
|
|
41
|
Choi YJ, Li X, Hydbring P, Sanda T,
Stefano J, Christie AL, Signoretti S, Look AT, Kung AL, von Boehmer
H and Sicinski P: The requirement for cyclin D function in tumor
maintenance. Cancer Cell. 22:438–451. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Liu X, Lv XB, Wang XP, Sang Y, Xu S, Hu K,
Wu M, Liang Y, Liu P, Tang J, et al: MiR-138 suppressed
nasopharyngeal carcinoma growth and tumorigenesis by targeting the
CCND1 oncogene. Cell Cycle. 11:2495–2506. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Cao L, Li C, Shen S, Yan Y, Ji W, Wang J,
Qian H, Jiang X, Li Z, Wu M, et al: OCT4 increases BIRC5 and CCND1
expression and promotes cancer progression in hepatocellular
carcinoma. BMC Cancer. 13:822013. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Zhong Z, Yeow WS, Zou C, Wassell R, Wang
C, Pestell RG, Quong JN and Quong AA: Cyclin D1/cyclin-dependent
kinase 4 interacts with filamin A and affects the migration and
invasion potential of breast cancer cells. Cancer Res.
70:2105–2114. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Achiwa Y, Hasegawa K and Udagawa Y: Effect
of ursolic acid on MAPK in cyclin D1 signaling and RING-type E3
ligase (SCF E3s) in two endometrial cancer cell lines. Nutr Cancer.
65:1026–1033. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Wang JC, Thiere M, Henne-Bruns D,
Knippschild U and Kornmann M: Inhibition of pancreatic cancer cell
growth in vivo using a tetracycline-inducible cyclin D1 antisense
expression system. Pancreas. 42:141–148. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Schonbrunn E, Betzi S, Alam R, Martin MP,
Becker A, Han H, Francis R, Chakrasali R, Jakkaraj S, Kazi A, et
al: Development of highly potent and selective diaminothiazole
inhibitors of cyclin-dependent kinases. J Med Chem. 56:3768–3782.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Sewify EM, Afifi OA, Mosad E, Zaki AH and
El Gammal SA: Cyclin D1 amplification in multiple myeloma is
associated with multidrug resistance expression. Clin Lymphoma
Myeloma Leuk. 14:215–222. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Ikeda Y, Oda K, Hiraike-Wada O, Koso T,
Miyasaka A, Kashiyama T, Tanikawa M, Sone K, Nagasaka K, Maeda D,
et al: Cyclin D1 harboring the T286I mutation promotes oncogenic
activation in endometrial cancer. Oncol Rep. 30:584–588.
2013.PubMed/NCBI
|