Introduction
Lung cancer is a life-threatening malignant tumor
that is associated with the highest morbidity and mortality rates
globally (1). Lung cancer can be
classified into two major pathological categories: Small cell lung
cancer (SCLC) and non-SCLC (NSCLC) (2). NSCLC, as the predominant form of lung
cancer, accounts for nearly 85% of all lung cancer cases (3). Due to the increasing rate of cigarette
smoking and air pollution, it has become a common malignancy in
developing countries (4). Although
the survival rate for lung cancer has increased due to improvements
in diagnosis and treatment, the prognosis of NSCLC remains poor.
The 5- and 10-year survival rates remain at <15 and <7%,
respectively (5). Metastasis is the
major cause of mortality among patients with NSCLC (6). Tumor metastasis is a complex biological
process, which involves cell migration, invasion, matrix
degradation and angiogenesis (7).
Therefore, the identification of predictive molecular factors and
an understanding of the underlying molecular mechanisms during
tumor metastasis are critical to further improve the survival of
NSCLC patients.
Over the past few years, microRNAs (miRNAs/miRs)
have been demonstrated to be involved in the pathogenesis of human
lung cancer (8). miRNAs, a class of
small non-coding RNAs (~22 nucleotides long), are widely expressed
in eukaryotes and regulate the expression of their target mRNAs by
base pairing with mRNAs in the 3′-untranslated region (UTR),
leading to mRNA cleavage or translation repression (9–12). Thus,
they are known to have essential roles in a variety of
physiological and pathological processes, including the cell cycle,
proliferation, migration, invasion, apoptosis, differentiation and
development (13–15). Recent studies have demonstrated that
miRNAs could also function as tumor suppressors or oncogenes, and
that they are aberrantly expressed in numerous types of human
cancer (16). Upregulated miRNAs
function as oncogenes through the negative regulation of tumor
suppressor genes. By contrast, downregulated miRNAs function as
tumor suppressor genes and inhibit cancer through the regulation of
oncogenes (17,18). One miRNA can regulate a number of
target genes, while one gene can also be regulated by multiple
miRNAs. It has been estimated that >60% of all human genes are
regulated by miRNAs (19). Therefore,
the identification of miRNA targets is essential in order to
understand the function of miRNAs in cancer development and
progression.
miR-181b has been reported to be frequently
downregulated in various tumors (20–22).
However, the expression and functions of miR-181b in NSCLC are yet
to be investigated. The objective of the present study was to
elucidate the expression and effect of miR-181b in NSCLC, and to
investigate its underlying mechanisms.
Materials and methods
Clinical specimens and cell
culture
The protocol of the present study was approved by
the Protection of Human Subjects Committee of Yichang Central
People's Hospital (Yichang, Hubei, China). Written informed consent
was provided by all patients in this study. Primary cancer tissues
and matched normal adjacent tissues (NATs; located >3 cm from
the tumor) were collected from 62 NSCLC patients, including 24 male
and 38 female patients (age range, 28–74 years), who had undergone
surgical resection at Yichang Central People's Hospital between
January 2012 and July 2014. None of the patients had received any
pre-operative cancer treatment, such as radiotherapy or
chemotherapy. Tissues were rapidly snap-frozen in liquid nitrogen
and stored at −80°C refrigerator for subsequent experiments. The
clinical data of the NSCLC patients were also collected.
The human NSCLC cell lines, H23 and H522, were
purchased from the Shanghai Institute of Biochemistry and Cell
Biology (Shanghai, China). The H23 and H522 cells were cultured in
RPMI-1640 medium supplemented with 10% fetal bovine serum (FBS;
Gibco; Thermo Fisher Scientific, Inc., Waltham, MA, USA) in a 5%
CO2 cell incubator at 37°C.
RNA isolation and reverse
transcription-quantitative polymerase chain reaction (RT-qPCR)
Total RNA was extracted with TRIzol reagent
(Invitrogen; Thermo Fisher Scientific Inc.) according to the
manufacturer's protocols. The concentration of the extracted RNA
was determined using the ND-2000 spectrophotometer (NanoDrop
Technologies; Thermo Fisher Scientific, Inc., Pittsburgh, PA, USA).
RNA (1 µg) was reverse transcribed into cDNA using a reverse
transcription kit (Tiangen Biotech Co., Ltd., Beijing, China). qPCR
was performed using a standard SYBR PrimeScript miRNA RT-PCR kit
(Takara Bio, Inc., Otsu, Japan) on an Applied Biosystems 7500
Real-Time PCR system (Thermo Fisher Scientific, Inc., Waltham, MA,
USA). U6 was used as an internal control. The primer sequences were
as follows: miR-181b forward, 5′-AACATTCATTGCTGTCGGTG-3′ and
reverse, 5′-GCTGTCAACGATACGCTACGT-3′; and U6 forward,
5′-GCTTCGGCAGCACATATACTAAA-3′ and reverse,
5′-GCTTCACGAATTTGCGTGTCAT-3′. The cycling conditions were as
follows: 42°C for 5 min; 95°C for 10 sec; and 40 cycles of 95°C for
5 sec, 55°C for 30 sec and 70°C for 30 sec. Each sample was
analyzed in triplicate. The relative expression of miR-181b was
analyzed using the 2−ΔΔCq method (23).
Cell transfection
Mature miR-181b mimic, miRNA mimic negative control
(NC) and the luciferase reporter plasmid were purchased from
GenePharma (Shanghai, China). The sequence of the miR-181b mimic
was 5′-AACAUUCAUUGCUGUCGGUGGGU-3′. The sequence of the NC mimic was
5′-UUCUCCGAACGUGUCACGUTT-3′. Transfection with miR-181b mimic, NC
or luciferase reporter plasmid was performed using Lipofectamine
2000 (Invitrogen; Thermo Fisher Scientific, Inc., Waltham, MA, USA)
according to the manufacturer's protocols.
Cell migration and invasion assay
Migration and invasion analysis of the NSCLC cells
was performed using Transwell chambers with an 8-µm pore
polycarbonate membrane (Costar; Corning Incorporated, Corning, NY,
USA). For the migration assays, transfected cells
(5×104) in 200 µl RPMI-1640 medium with 0.1% serum were
placed into the upper chamber. For the invasion assays, transfected
cells (5×104) in 200 µl RPMI-1640 medium with 0.1% serum
were placed into the upper chamber coated with Matrigel (BD
Biosciences, San Jose, CA, USA). In each assay, a volume of 0.5 ml
RPMI-1640 medium with 20% FBS was then added to the lower chamber
as a chemoattractant. The cells were then incubated for another 12
h for the migration assay and for 24 h for the invasion assay. The
cells that did not migrate or invade through the pores were
carefully wiped away with cotton wool. Next, the inserts were fixed
with 100% methanol (Shanghai Macklin Biochemical Co., Ltd.,
Shanghai, China), stained with 0.5% crystal violet (Beyotime
Institute of Biotechnology, Haimen, China) and the number of cells
was counted with an inverted microscope (CKX41; Olympus, Tokyo,
Japan).
Western blotting
Total cellular protein was isolated using
radioimmunoprecipitation assay lysis buffer [50 mM Tris-HCl (pH
7.4), 1% NP-40, 0.25% Na-deoxycholate, 150 mM NaCl, 1 mM EDTA, 1 mM
phenylmethylsulfonyl fluoride, aprotinin, leupeptin and pepstatin
(1 µg/ml each), 1 mM Na3VO4 and 1 mM NaF) at 72 h
post-transfection. Protein concentration was measured using a
bicinchoninic acid assay kit (Nanjing KeyGen Biotech. Co., Ltd.,
Nanjing, China). Subsequently, equal amounts of protein were
separated by 10% SDS-PAGE and electrotransferred onto
polyvinylidene fluoride membranes (EMD Millipore, Billerica, MA,
USA). The membranes were blocked with 5% non-fat milk in 0.1% TBST
and incubated overnight at 4°C with rabbit anti-high-mobility group
box-1 protein (HMGB1) monoclonal antibody (1:1,000; cat. no.
ab92310; Abcam, Cambridge, UK) and mouse anti-β-actin monoclonal
antibody (1:1,000; cat. no. sc-130301; Santa Cruz Biotechnology,
Inc., Dallas, TX, USA). After washing with TBST, the membranes were
incubated with horseradish peroxidase-conjugated goat anti-rabbit
and goat anti-mouse secondary antibodies (1:5,000; cat. nos.
sc-2004 and sc-2005, respectively; Santa Cruz Biotechnology, Inc.)
for 1 h at room temperature. The protein bands were developed with
enhanced chemiluminescence reagents (Pierce; Thermo Fisher
Scientific, Inc., Waltham, MA, USA) and images were captured using
a FluorChem imaging system (Alpha Innotech, San Leandro, CA,
USA).
Luciferase assay
To determine whether HMGB1 is a direct target of
miR-181b, luciferase reporter assays were performed. The luciferase
reporter plasmids, pmirGLO-HMGB1–3′UTR wild-type (WT) and
pmirGLO-HMGB1–3′UTR mutant (MUT), were synthesized and purified by
GenePharma. The H23 and H522 cells were transfected with 0.5 µg
reporter plasmid, 40 nmol miR-181b mimic or NC in a 12-well plate
using Lipofectamine 2000 according to the manufacturer's
instructions. At 48 h post-transfection, the activities of the
firefly and renilla luciferases were determined with the
Dual-Luciferase Reporter Assay System (Promega, Manheim, Germany).
Each assay was replicated 3 times. The firefly luciferase activity
was normalized to the renilla luciferase activity for each
transfected well.
Statistical analysis
Data are presented as the mean ± standard deviation
and compared using Student's t-tests in Stata 10.0 (StataCorp LP,
College Station, Texas, USA). Two-tailed P-values of <0.05 were
considered to indicate a statistically significant difference.
Results
miR-181b expression in NSCLC tissues
and its association with clinicopathological factors
A total of 62 NSCLC tissues were included in this
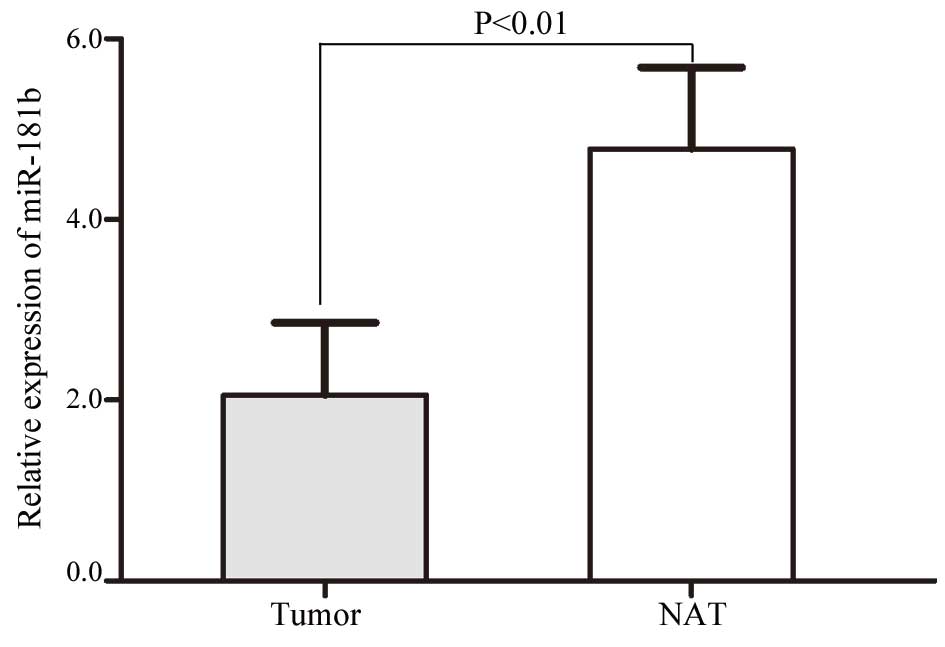
study. As shown in Fig. 1, miR-181b
was significantly downregulated in the NSCLC tissues compared with
the NATs (P=0.004). These results indicated that miR-181b may have
a suppressive role in NSCLC.
The study then examined whether the expression level
of miR-181b is associated with clinicopathological factors. The
statistical analysis showed that miR-181b expression was
significantly associated with the tumor-node-metastasis (TNM)
classification and metastasis (Table
I).
 | Table I.A comparison between miR-181b
expression in non-small cell lung cancer and clinicopathological
features. |
Table I.
A comparison between miR-181b
expression in non-small cell lung cancer and clinicopathological
features.
|
|
| miR-181b expression,
n |
|
|---|
|
|
|
|
|
|---|
| Clinical
features | Cases, n | Low | High | P-value |
|---|
| Gender |
|
|
| 0.734 |
| Male | 24 | 18 | 6 |
|
|
Female | 38 | 27 | 11 |
|
| Age, years |
|
|
| 0.359 |
|
<57 | 35 | 27 | 8 |
|
| ≥57 | 27 | 18 | 9 |
|
| Smoking history,
years |
|
|
| 0.407 |
|
<10 | 24 | 16 | 8 |
|
| ≥10 | 38 | 29 | 9 |
|
| Tumor
differentiation |
|
|
| 0.098 |
| I–II | 36 | 29 | 7 |
|
|
III–IV | 26 | 16 | 10 |
|
| TNM
classification |
|
|
| 0.026 |
| I | 27 | 15 | 12 |
|
| II | 19 | 17 | 2 |
|
|
III+IV | 16 | 13 | 3 |
|
| Metastasis |
|
|
| 0.035 |
| No | 34 | 21 | 13 |
|
|
Yes | 28 | 24 | 4 |
|
miR-181b inhibits cell migration and
invasion in NSCLC H23 and H522 cells
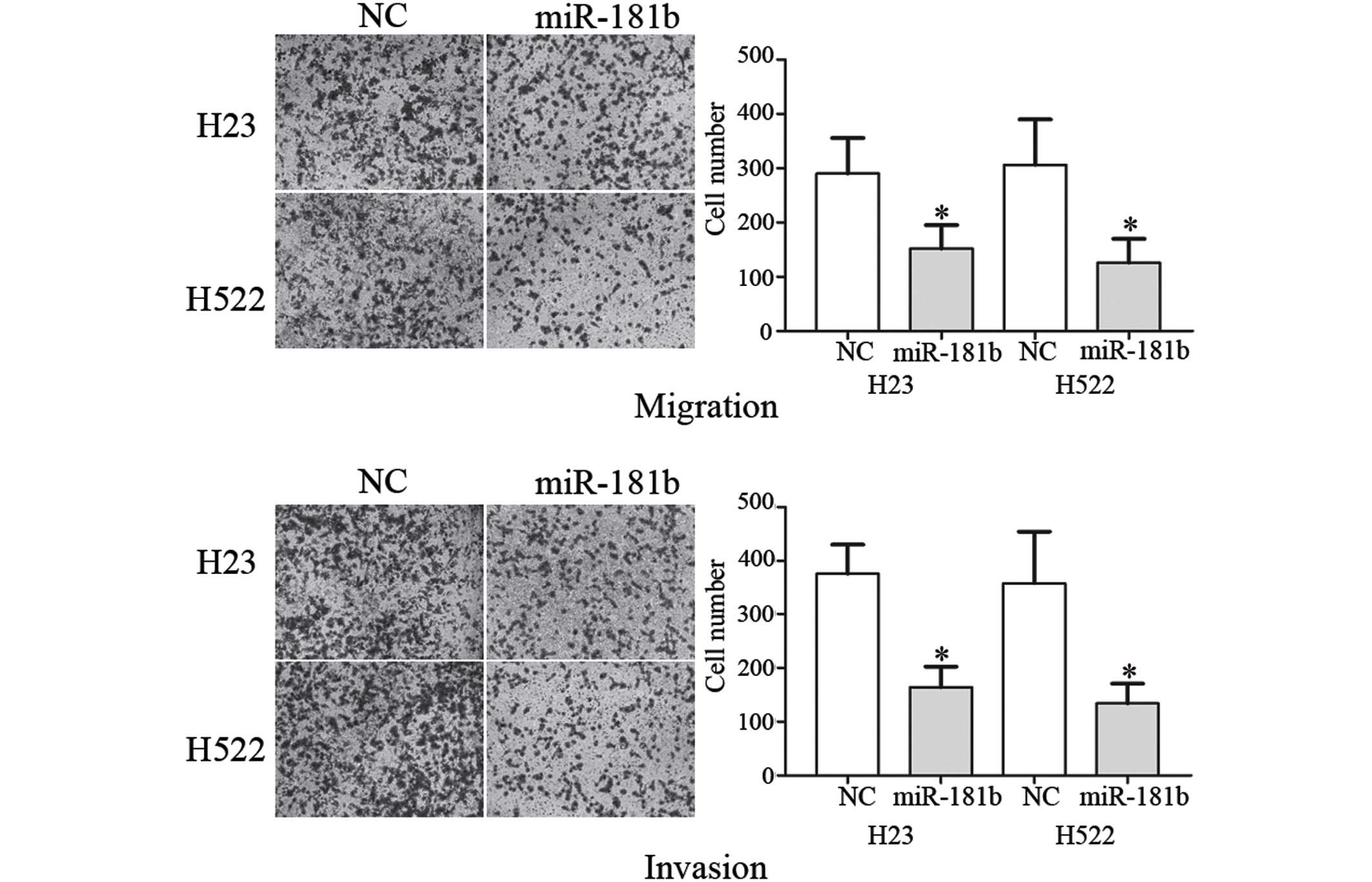
Next, the suppressive role of miR-181b on cell
migration and invasion was investigated using the Transwell assay.
As shown in Fig. 2, the migration and
invasion of the NSCLC H23 (P=0.032 for migration; P=0.280 for
invasion) and H522 (P=0.020 for migration; P=0.017 for invasion)
cells transfected with miR-181b was markedly decreased compared
with the NC. These results indicated that miR-181b decreased the
migration and invasion in the NSCLC cells.
HMGB1 is a direct target gene of
miR-181b in NSCLC
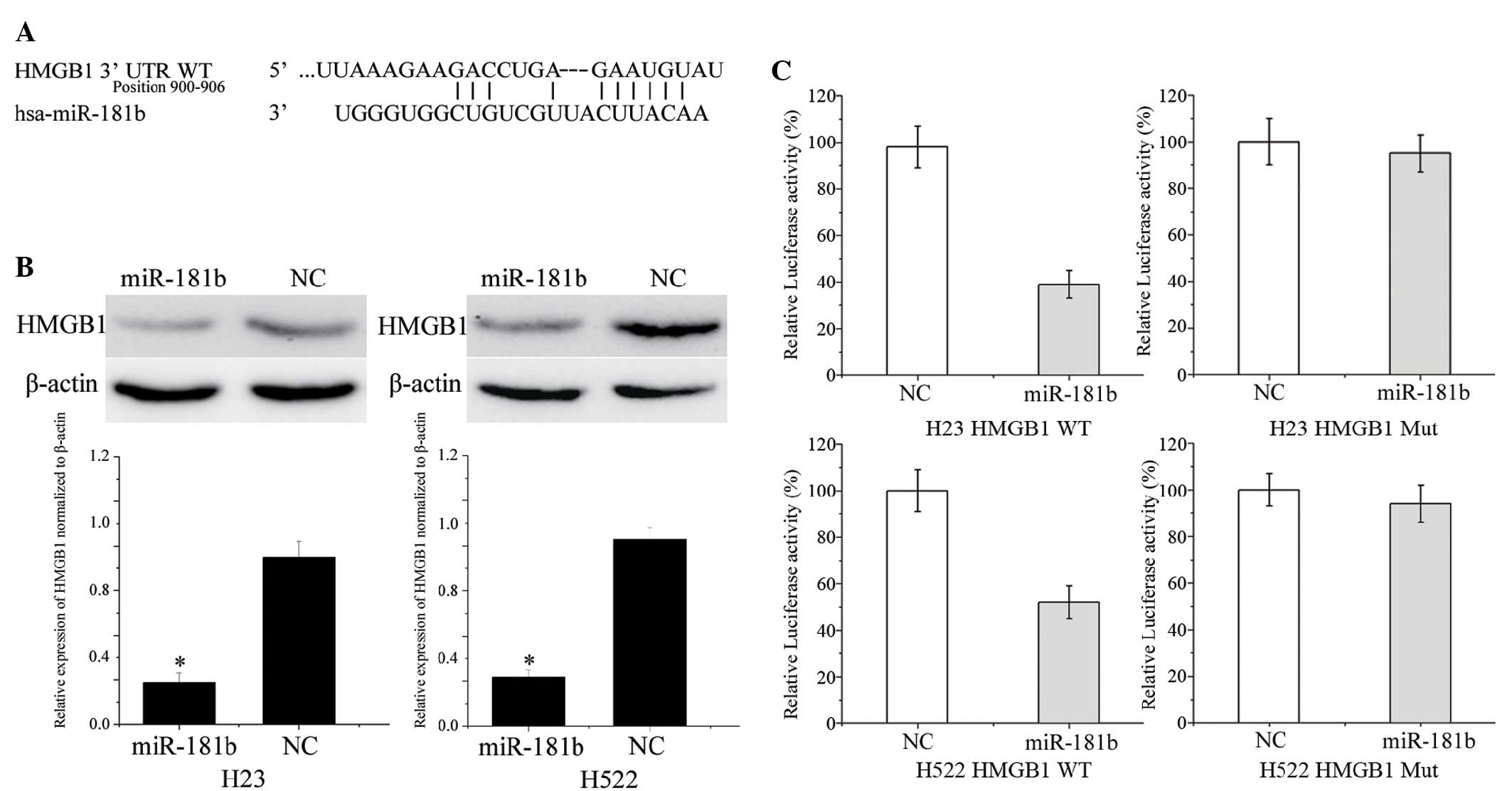
To identify the target of miR-181b in NSCLC, a
public database (TargetScan; http://www.targetscan.org) was used. HMGB1 was
predicted to be a target of miR-181b (Fig. 3A). To verify whether miR-181b directly
targets HMGB1, western blotting was performed to investigate
whether HMGB1 was decreased after transfection with miR-181b in
NSCLC H23 and H522 cells. As shown in Fig. 3B, HMGB1 was significantly
downregulated in the NSCLC cells following transfection with
miR-181b when compared with the NC (P=0.015 and 0.018 for H23 cells
and H522 cells, respectively).
Furthermore, luciferase reporter assays were
conducted. Luciferase reporter assays showed that miR-181b
inhibited the WT (P=0.026 for H23; P=0.350 for H522) but not the
MUT luciferase activity of HMGB1 in the NSCLC H23 and H522 cells
(Fig. 3C). Overall, HMGB1 was a
direct target gene of miR-181b in NSCLC.
Discussion
Lung cancer is considered to be the leading cause of
cancer-related mortality (24). The
poor prognosis of patients with early-stage lung cancer is
associated with lymph node metastasis and distant metastasis at the
time of presentation (25). A
previous study revealed that lymph node metastasis indicated high
tumor malignancy and a poor prognosis; the patients usually
succumbed to the associated complications caused by metastatic
loci, but did not succumb to the primary loci-induced symptoms
(26). Distant metastasis is a key
factor to determine a patient's prognosis, and is one of the most
common causes for the failure of treatment for NSCLC (27). The processes inducing and stimulating
metastasis are complex and remain unclear. Therefore, an urgent
requirement to search for novel sensitive, reliable biomarkers, and
novel therapeutic targets and approaches must be sought to block
NSCLC metastasis.
miR-181b belongs to the miR-181 family, which
comprises four members: miR-181a, miR-181b, miR-181c and miR-181d
(20). Previous studies have measured
the altered level of miR-181b in multiple tumors and in
leukemia/lymphoma (20). The miRNA
functions as a tumor suppressor or a tumor promoter in different
human malignancies. In one study, miR-181b was found to be
downregulated in aggressive B-cell chronic lymphocytic leukemia
compared with chronic lymphocytic leukemia (21). In human glioma, the expression of
miR-181b was downregulated and the transfection of miR-181b
inhibited cell growth, apoptosis and invasion (22). By contrast, Jiang et al
verified that miR-181b was significantly upregulated in gastric
tumor tissues compared with normal gastric tissues. Kaplan-Meier
survival analysis also found that the low expression of miR-181b
may be closely associated with better patient overall survival
(28). The present study expanded on
the data on the expression and function of miR-181b in cancer.
The present results provided evidence that miR-181b
was downregulated in NSCLC, which prompted the hypothesis that
miR-181b may have a tumor suppressive role in NSCLC carcinoma
development and progression. Through the overexpression of miR181b
in two NSCLC cell lines, the present study notably showed that
miR-181b reduced cell migration and invasion, also suggesting a
tumor suppressive role of miR-181b in metastasis. Thus, these
results may have clinical implications in the future.
The identification of miR-181b target genes is
critical for understanding the role of the miRNA in tumorigenesis,
and it is also important for searching for novel therapeutic
targets. Studies have verified that miR-181b regulates oncogenic
expression in human cells, including CYLD (29), NOVA1 (30), MDM2 (16), HMGB1 (31), MCL-1 (31), LATS2 (32) and Bcl-2 (33) cells. Therefore, upregulating miR-181b
or providing analogous pharmaceutical compounds exogenously should
be effective cancer therapies resulting from the upregulation of
these oncogenic transcripts. In the present study, the results
showed that miR-181b inhibited cell migration and invasion by
directly targeting HGMB1. This finding suggested that miR-181b
could be used for the development of novel therapeutic treatments
for NSCLC.
HMGB1, located on human chromosome 13q12, is a
member of the high mobility group protein super-family (34), and has been reported to be involved in
multiple biological processes, including cancer progression,
angiogenesis, invasion and metastatic development (35). The upregulation of HMGB1 is found in
almost all types of tumors (36).
HMGB1 has also been found to be downregulated in NSCLC at the mRNA
and protein levels, and is correlated with cancer progression, cell
proliferation and metastasis (36,37). The
findings suggest that HMGB1 possesses an oncogenic role and is a
potential novel therapeutic target. In the present study, it was
found that miR-181b controls HMGB1 expression to regulate NSCLC
cell migration and invasion. As a result, further investigation is
required into its predictive value for the early detection of tumor
recurrence and in target therapy drugs to block the invasive nature
of NSCLC.
In conclusion, the present study showed that
miR-181b was downregulated in NSCLC, and associated with the TNM
classification and metastasis. It was also verified that miR-181b
inhibits NSCLC cell metastasis by the downregulation of HMGB1.
Future studies are required to address whether the predictive and
therapeutic potential of miR-181b may be fully realized in cancer
treatment. If so, the use of miR-181b may be of future benefit for
the treatment of patients with NSCLC.
References
|
1
|
He D, Wang J, Zhang C, Shan B, Deng X, Li
B, Zhou Y, Chen W, Hong J, Gao Y, et al: Down-regulation of
miR-675-5p contributes to tumor progression and development by
targeting pro-tumorigenic GPR55 in non-small cell lung cancer. Mol
Cancer. 14:732015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
DeSantis CE, Lin CC, Mariotto AB, Siegel
RL, Stein KD, Kramer JL, Alteri R, Robbins AS and Jemal A: Cancer
treatment and survivorship statistics, 2014. CA Cancer J Clin.
64:252–271. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Yang Y, Liu L, Zhang Y, Guan H, Wu J, Zhu
X, Yuan J and Li M: MiR-503 targets PI3K p85 and IKK-β and
suppresses progression of non-small cell lung cancer. Int J Cancer.
135:1531–1542. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Zhang B, Liu T, Wu T, Wang Z, Rao Z and
Gao J: microRNA-137 functions as a tumor suppressor in human
non-small cell lung cancer by targeting SLC22A18. Int J Biol
Macromol. 74:111–118. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Crinò L, Weder W, van Meerbeeck J and
Felip E: ESMO Guidelines Working Group: Early stage and locally
advanced (non-metastatic) non-small-cell lung cancer: ESMO clinical
practice guidelines for diagnosis, treatment and follow-up. Ann
Oncol. 21(Suppl 5): v103–v115. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Gupta GP and Massagué J: Cancer
metastasis: Building a framework. Cell. 127:679–695. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Yu X, Wei F, Yu J, Zhao H, Jia L, Ye Y, Du
R, Ren X and Li H: Matrix metalloproteinase 13: A potential
intermediate between low expression of microRNA-125b and increasing
metastatic potential of non-small cell lung cancer. Cancer Genet.
208:76–84. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Zhu X, Gao G, Chu K, Yang X, Ren S, Li Y,
Wu H, Huang Y and Zhou C: Inhibition of RAC1-GEF DOCK3 by
miR-512-3p contributes to suppression of metastasis in non-small
cell lung cancer. Int J Biochem Cell Biol. 61:103–114. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Iorio MV and Croce CM: MicroRNA
dysregulation in cancer: Diagnostics, monitoring and therapeutics.
A comprehensive review. EMBO Mol Med. 4:143–159. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Engels BM and Hutvagner G: Principles and
effects of microRNA-mediated post-transcriptional gene regulation.
Oncogene. 25:6163–6169. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
van Rooij E, Purcell AL and Levin AA:
Developing microRNA therapeutics. Circ Res. 110:496–507. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Chen C, Zhao Z, Liu Y and Mu D:
microRNA-99a is downregulated and promotes proliferation, migration
and invasion in non-small cell lung cancer A549 and H1299 cells.
Oncol Lett. 9:1128–1134. 2015.PubMed/NCBI
|
|
13
|
Ambros V: The functions of animal
microRNAs. Nature. 431:350–355. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Bartel DP: MicroRNAs: Genomics,
biogenesis, mechanism, and function. Cell. 116:281–297. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Broderick JA and Zamore PD: MicroRNA
therapeutics. Gene Ther. 18:1104–1110. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Sun YC, Wang J, Guo CC, Sai K, Wang J,
Chen FR, Yang QY, Chen YS, Wang J, To TS, et al: MiR-181b
sensitizes glioma cells to teniposide by targeting MDM2. BMC
Cancer. 14:6112014. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Ventura A and Jacks T: MicroRNAs and
cancer: Short RNAs go a long way. Cell. 136:586–591. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Yang T, Thakur A, Chen T, Yang L, Lei G,
Liang Y, Zhang S, Ren H and Chen M: MicroRNA-15a induces cell
apoptosis and inhibits metastasis by targeting BCL2L2 in non-small
cell lung cancer. Tumour Biol. 36:4357–4365. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Lewis BP, Shih IH, Jones-Rhoades MW,
Bartel DP and Burge CB: Prediction of mammalian microRNA targets.
Cell. 115:787–798. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Yang J, Liu H, Wang H and Sun Y:
Down-regulation of microRNA-181b is a potential prognostic marker
of non-small cell lung cancer. Pathol Res Pract. 209:490–494. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Visone R, Veronese A, Balatti V and Croce
CM: MiR-181b: New perspective to evaluate disease progression in
chronic lymphocytic leukemia. Oncotarget. 3:195–202. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Shi ZM, Wang XF, Qian X, Tao T, Wang L,
Chen QD, Wang XR, Cao L, Wang YY, Zhang JX, et al: MiRNA-181b
suppresses IGF-1R and functions as a tumor suppressor gene in
gliomas. RNA. 19:552–560. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) Method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Wang RJ, Zheng YH, Wang P and Zhang JZ:
Serum miR-125a-5p, miR-145 and miR-146a as diagnostic biomarkers in
non-small cell lung cancer. Int J Clin Exp Pathol. 8:765–771.
2015.PubMed/NCBI
|
|
25
|
Reungwetwattana T, Weroha SJ and Molina
JR: Oncogenic pathways, molecularly targeted therapies, and
highlighted clinical trials in non-small-cell lung cancer (NSCLC).
Clin Lung Cancer. 13:252–266. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Mountain CF and Dresler CM: Regional lymph
node classification for lung cancer staging. Chest. 111:1718–1723.
1997. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Wang R, Chen XF and Shu YQ: Prediction of
non-small cell lung cancer metastasis-associated microRNAs using
bioinformatics. Am J Cancer Res. 5:32–51. 2014.PubMed/NCBI
|
|
28
|
Jiang J, Zheng X, Xu X, Zhou Q, Yan H,
Zhang X, Lu B, Wu C and Ju J: Prognostic significance of miR-181b
and miR-21 in gastric cancer patients treated with S-1/Oxaliplatin
or Doxifluridine/Oxaliplatin. PLoS One. 6:e232712011. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Li D, Jian W, Wei C, Song H, Gu Y, Luo Y
and Fang L: Down-regulation of miR-181b promotes apoptosis by
targeting CYLD in thyroid papillary cancer. Int J Clin Exp Pathol.
7:7672–7680. 2014.PubMed/NCBI
|
|
30
|
Zhi F, Wang Q, Deng D, Shao N, Wang R, Xue
L, Wang S, Xia X and Yang Y: MiR-181b-5p downregulates NOVA1 to
suppress proliferation, migration and invasion and promote
apoptosis in astrocytoma. PLoS One. 9:e1091242014. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Lu F, Zhang J, Ji M, Li P, Du Y, Wang H,
Zang S, Ma D, Sun X and Ji C: miR-181b increases drug sensitivity
in acute myeloid leukemia via targeting HMGB1 and Mcl-1. Int J
Oncol. 45:383–392. 2014.PubMed/NCBI
|
|
32
|
Xia Y and Gao Y: MicroRNA-181b promotes
ovarian cancer cell growth and invasion by targeting LATS2. Biochem
Biophys Res Commun. 447:446–451. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Cai B, An Y, Lv N, Chen J, Tu M, Sun J, Wu
P, Wei J, Jiang K and Miao Y: miRNA-181b increases the sensitivity
of pancreatic ductal adenocarcinoma cells to gemcitabine in vitro
and in nude mice by targeting BCL-2. Oncol Rep. 29:1769–1776.
2013.PubMed/NCBI
|
|
34
|
Ferrari S, Finelli P, Rocchi M and Bianchi
ME: The active gene that encodes human high mobility group 1
protein (HMG1) contains introns and maps to chromosome 13.
Genomics. 35:367–371. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Yao S, Zhao T and Jin H: Expression of
MicroRNA-325-3p and its potential functions by targeting HMGB1 in
non-small cell lung cancer. Biomed Pharmacother. 70:72–79. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Shen X, Hong L, Sun H, Shi M and Song Y:
The expression of high-mobility group protein box 1 correlates with
the progression of non-small cell lung cancer. Oncol Rep.
22:535–539. 2009.PubMed/NCBI
|
|
37
|
Sun KK, Ji C, Li X, Zhang L, Deng J, Zhong
N and Wu XY: Overexpression of high mobility group protein B1
correlates with the proliferation and metastasis of lung
adenocarcinoma cells. Mol Med Rep. 7:1678–1682. 2013.PubMed/NCBI
|