Introduction
Esophageal cancer (EC) has been well studied in the
literature, being classified as sixth highest in terms of new
cancer cases per year and the seventh leading cause of
cancer-related mortality, worldwide (1–3). The
incidence rate of EC is four-fold higher among males when compared
with that among women (1,2). A total of 450,000 new cases of EC are
reported annually worldwide, among which 380,000 cases are expected
to succumb to the disease within 12 months following diagnosis
(1). In developed countries, EC
accounts for ~6% of malignant gastrointestinal cancer cases, with
an extremely high mortality rate; <10% of patients are expected
to survive >5 years after diagnosis (1–3). There are
two main types of EC, namely squamous cell carcinoma and
adenocarcinoma. Certain genes, for example, oncogenes, tumor
suppressor genes, metastasis genes and apoptosis genes, can have a
giant impact on the development of EC (4). Risk factors including, alcohol
consumption, smoking, chronic inflammation of the esophageal
mucosa, achalasia, Plummer-Vinson syndrome, Barrett's esophagus and
dietary factors (micronutrient and vitamin deficiencies,
consumption of hot or highly processed products or food
contaminated with spores or nitrogenous compounds) are considered
increase the probability of EC development (1,3). Notably,
EC exhibits a significantly higher occurrence among smokers when
compared with non-smokers. Both tobacco and alcohol consumption
increase the risk of EC (1).
Nowadays, a variety of methods exist to examine EC and more is
known about the disease, however, further research is necessary in
order to take effective steps to prevent and treat it (5). EC treatment choice is dependent on the
developmental stage of the disease. For example, in cases of early
cancer diagnosis, esophagectomy is possible (6). However, at later developmental stages,
surgical anastomosis is performed using Kirschne-Akiyama gastric
tube or fundus rotation gastroplasty techniques (7). In cases of unresectable EC, patients are
treated with a combination of chemo- and radiotherapy (8). Laser therapy may also be used as an
effective method for improving the patient's ability to swallow
(9).
microRNAs (miRNAs/miRs) are non-coding small RNA
molecules, well-conserved across diverse groups, for example,
animals, plants and viruses (10).
miRNAs can regulate the translation of mRNA and integrate different
regulatory pathways for abnormal and normal cell development
(11). It has been demonstrated that
certain miRNAs have promising therapeutic potential (12). A number of miRNAs have been found to
exhibit potential as prognostic factors in various cancer types,
including miR155 and let-7a-2 (lung adenocarcinoma), miR21, miR-9-3
and miR145 (breast carcinoma) and miR18 and miR20 (hepatocellular
carcinoma) (13–15).
For the present study, four genes connected with
cancer-related pathways (PDGFA, MET, MMP9 and
SERPINE1), as well as four miRNA genes (miR-21, miR-134,
miR-205 and miR-495) were chosen. Angiogenesis describes the
formation of new, thin-walled blood vessels from pre-existing
vasculature. It is involved in numerous physiological and
pathological processes, including chronic inflammation, ischemic
diseases and solid tumor formation (16). Metastatic cascades are initiated by
the invasion of tissues surrounding the tumor, followed by tumor
cell infiltration of the surrounding blood vessels. Tumor cells are
then transported further into the organism leading to the formation
of metastatic colonies (17). These
processes are prevalent in EC and are considered to be the primary
cause of mortality (18).
PDGFA, MET, MMP9 and SERPINE1 have been
identified as the most promising genes involved in those processes
based on miRNA gene expression analysis and literature data
(19). PDGFA is involved in
angiogenesis, while MET, MMP9 and SERPINE1 are
involved in the invasion and metastasis of cancer (20,21).
Materials and methods
EC cell lines and culture
conditions
For the experiments, four different cell lines were
used: Three EC lines (KYSE-30, KYSE-150 and KYSE-270), along with a
Het-1A immortalized human esophageal epithelial cell line as a
negative control. The three EC lines were obtained from the
European Collection of Cell Cultures (Salisbury, UK), and the
control cell line was ordered from the American Type Culture
Collection (Manassas, VA, USA). The cancer cell lines were grown in
a medium which consisted of Ham F12 and RPMI 1640 in a 1:1 ratio,
with 2 mM L-glutamine and 2% fetal bovine serum (Sigma-Aldrich, St.
Louis, MO, USA). Quantum 286 medium (Sigma-Aldrich) was used to
grow the Het-1A cell line. Standard conditions of 37°C, 5% (v/v)
CO2 and a relative humidity of 100% were used for the
cultures, without antibiotics.
Contamination-free conditions in the
cell culture room
Each experiment with cell lines was performed by a
researcher with a number of years of experience with this type of
work in a range of high-quality laboratories. In these
institutions, antibiotics are not recommended for use in the cell
culture room. A high-standard culture cell room was used for these
experiments. Microscopic observations were performed every working
day (Monday to Saturday) for the control of cell conditions. All
liquids (i.e., media and buffers) and plastic equipment were
sterile.
RNA isolation from cell lines
Small RNAs from the cell lines were recovered using
the mirVana™ miRNA Isolation kit (Life Technologies, Carlsbad, CA,
USA) according to the manufacturer's instructions. The quality and
concentration of RNA was analyzed by the Picodrop Microliter UV/Vis
Spectrophotometer (Picodrop Ltd., Hinxton, UK). RNA was used for
the reverse transcription (RT) process directly after
isolation.
miRNA genes analysis in cell
lines
RT reactions for miRNA were performed with Megaplex™
Pools for microRNA Expression Analysis (Life Technologies). RT-qPCR
was performed according to the manufacturer's instructions. The
concentration of RNA used for conversion to cDNA was 700 ng/µl, and
the total volume of the RT polymerase chain reaction (PCR) reagents
was 4.5 µl. The reactions were performed under the following
conditions: 40 cycles of three steps: 16°C for 120 sec, 42°C for 60
sec and 50°C for 1 sec. Following completion of the cycles, a final
step was performed at 85°C for 5 min. TaqMan® Array
Human MicroRNA A (Life Technologies) was used to analyze 377 human
microRNAs in the sample according to the manufacturer's
instructions (22). Following
analysis of the results and the associated literature, four miRNA
genes were picked: miR-21, miR-134, miR-205 and miR-495.
Significant differences in miR-134, miR-205 and miR-495 gene
expression between cancer cell lines and controls have been
identified previously (2), and miR21
expression has often been analyzed in the literature (23).
Analysis of cancer pathway genes in
cell lines: Quantitative PCR (RT-qPCR)
The RT2 Profiler™ PCR Array (Qiagen,
Venlo, Netherlands) was used to perform RT reactions for the genes
correlated with the cancer-related pathways. A total of 84 genes
representative of six biological pathways associated with
transformation and tumorigenesis were checked with the Cancer
PathwayFinder PCR Array (Qiagen) to choose four genes. The
reactions were performed according to the manufacturer's
instructions (19). The concentration
of RNA used for conversion to cDNA was 1 µg/µl, and the total
volume of the RT PCR reagents was 20 µl. The reactions were
performed under the following conditions: 25°C for 5 min, 42°C for
5 min, 55°C for 15 min and 95°C for 5 min. Following completion of
the cycles, a final step was performed at 85°C for 5 min. After
analysis of the results and knowledge from literature, four miRNA
genes were picked: MET, MMP9, PDGFA and
SERPINE1.
Tissue samples from patients
Esophageal tumors and adjacent non-cancerous
specimens were collected from a total of 126 cancerous and
non-cancerous samples from 63 patients undergoing surgical
resection, primarily for diagnosis. Non-cancerous samples were
taken from tumor-free tissue at least 5 cm from the tumor-like
lesions. Samples were collected between 2005 and 2013 at the M.
Kopernik Provincial Specialist Hospital in Lodz (Lodz, Poland).
Each diagnosis was confirmed through microscopic examination by
licensed pathologists and written informed consent for use of the
tissues in research studies was provided by each patient. Tissues
were put through a paraffin cell block preparation procedure.
Samples were kept at 4°C in dry storage conditions. The Bioethics
Committee for Research on Humans at the Medical University of Lodz
(Lodz, Poland) approved this project as one following the
Declaration of Helsinki. All patient information, including gender,
age, alcohol consumption, smoking status, history of cancer, other
cancers and histopathology is recorded in Table I. All patients belonged to the
Caucasian population.
 | Table I.Clinical and pathological
characteristics for patients (n=63) with EC and the association
between MMP9 and SERPINE1 expression and patient
clinicopathological factors. |
Table I.
Clinical and pathological
characteristics for patients (n=63) with EC and the association
between MMP9 and SERPINE1 expression and patient
clinicopathological factors.
|
Characteristics | Patients, n
(%) | P-value (MMP9;
SERPINE1) |
|---|
| Gender |
| P=0.505;
P=0.817 |
|
Female | 16 (25.4) |
|
|
Male | 47 (74.6) |
|
| Age,
yearsa |
|
P=0.011b;
P=0.044b |
|
<45 | 5 (7.9) |
|
|
45–55 | 12 (19.0) |
|
|
56–65 | 34 (54.0) |
|
|
>65 | 12 (19.0) |
|
| Type of EC |
| P=0.241;
P=0.235 |
|
SCC | 38 (60.3) |
|
| AC | 22 (34.9) |
|
|
BSCC | 1 (1.6) |
|
| CS | 1 (1.6) |
|
|
ASC | 1 (1.6) |
|
| AJCC system |
| P=0.504;
P=0.543 |
|
I+II | 22 (34.9) |
|
|
III+IV | 28 (44.4) |
|
| No
data | 13 (20.6) |
|
| Lymph node
metastasis |
| P=0.309;
P=0.934 |
|
Negative | 33 (52.4) |
|
|
Positive | 17 (27.0) |
|
| No
data | 13 (20.6) |
|
| Family history of
cancer |
| P=0.465;
P=0.639 |
|
Yes | 5 (7.9) |
|
| No | 48 (92.1) |
|
| Other cancers |
| P=0.534;
P=0.526 |
|
Yes | 15 (23.8) |
|
| No | 48 (76.2) |
|
| Alcohol
consumption |
| P=0.743;
P=0.366 |
|
Rarely | 39 (61.9) |
|
|
Often | 24 (38.1) |
|
| Smoking status |
| P=0.826;
P=0.008b |
|
Never | 14 (22.2) |
|
|
Ever | 49 (77.8) |
|
| Packs of
cigarettes/day |
| P=0.057;
P=0.942 |
|
0 | 14 (22.2) |
|
| ≤2 | 34 (54.0) |
|
|
>2 | 15 (23.8) |
|
| Years of
smoking |
| P=0.081;
P=0.032b |
|
<20 | 11 (23.4) |
|
|
21–30 | 15 (31.9) |
|
|
>31 | 21 (44.7) |
|
RNA isolation from formalin-fixed,
paraffin-embedded (FFPE) samples
Total RNA of the EC and non-cancerous specimens was
extracted from FFPE tissues using the High Pure FFPE RNA Micro kit
(Roche, Basel, Switzerland) according to the manufacturer's
instructions. The quality and concentration of RNA was analyzed by
the Picodrop Microfilter UV/Vis Spectrophotometer (Picodrop Ltd.).
Directly after isolation, RNA was used in the reverse transcription
process.
FFPE sample miRNA gene analysis:
RT-qPCR
The TaqMan Reverse Transcription kit (Life
Technologies) was used according to the manufacturer's instructions
in a UNO-Thermoblock from Biometra (Goettingen, Germany).
Commercial primers were used in each reaction with TaqMan Universal
Master Mix II without UNG (Life Technologies) according to the
manufacturer's instructions. RNU24 was used as a control (Table II). Three samples were amplified
simultaneously. The 7900HT Fast Real Time OCR System (Life
Technologies) was used.
 | Table II.TaqMan® MicroRNA Assay
primers used to amplify miRNA genes in quantitative polymerase
chain reaction. |
Table II.
TaqMan® MicroRNA Assay
primers used to amplify miRNA genes in quantitative polymerase
chain reaction.
| Gene | Assay ID |
|---|
| RNU24 | 001001 |
| miR-21 | 002438 |
| miR-134 | 001186 |
| miR-205 | 000509 |
| miR-495 | 001663 |
FFPE sample analysis for genes
correlated with cancer pathways: RT-qPCR
PCR primers (Table
III) were used with the addition of Brilliant II
SYBR® Master Mix (Agilent Technologies Inc., Santa
Clara, CA, USA). GAPDH was used as an endogenous control.
For MET, SERPINE1 and PDGFA, the reactions
were performed under the following conditions: 95°C for 10 min,
followed by 40 cycles of 95°C for 30 sec and 60°C for 25 sec. For
MMP9, the reactions were performed using the following
conditions: 95°C for 10 min, followed by 40 cycles of 95°C for 30
sec and 63°C for 30 sec. Three reactions were performed
simultaneously. Nuclease-free water was used as a control for each
experiment to ensure that there was no contamination. qPCR was
performed on the Agilent Technology Mx3005P instrument (Agilent
Technologies Inc.).
 | Table III.PCR primers used to amplify genes
correlated with cancer pathways in qPCR reaction. |
Table III.
PCR primers used to amplify genes
correlated with cancer pathways in qPCR reaction.
| Genes | Primers sequence
(5′-3′) | Product size,
bp | Annealing
temperature, °C |
|---|
| GAPDH | F:
AGCCACATCGCTCAGACA | 66 | 60.0 |
|
| R:
GCCCAATACGACCAAATCC |
| 63.0 |
| MET | F:
TTCTGACCGAGGGAATCAT | 82 | 60.0 |
|
| R:
CCTTCACTTCGCAGGCAG |
|
|
| MMP9 | F:
GAACCAATCTCACCGACAGG | 67 | 63.0 |
|
| R:
GCCACCCGAGTGTAACCATA |
|
|
| PDGFA | F:
CCCCTGCCCATTCGGAGGAAGAG | 227 | 60.0 |
|
| R:
TTGGCCACCTTGACGCTGCGGTG |
|
|
|
SERPINE1 | F:
CTCTCTCTGCCCTCACCAA | 183 | 60.0 |
|
| R:
GAAACTGTCTGAACATGTCG |
|
|
Statistical analysis
The Wilcoxon rank-sum test was used to appraise the
correlation between genes in the EC tumors and adjacent
non-cancerous specimens. Correlations between genes and
clinicopathological factors were tested using the Kruskal-Wallis
and Mann-Whitney U tests. Spearman's correlation coefficient was
also calculated. P<0.05 was used to indicate a statistically
significant difference. Survival was estimated using Kaplan-Meier
methods and the long-rank test. Calculations were prepared by
STATISTICA version 10 (StatSoft, Inc., Tulsa, OK, USA) and IBM SPSS
version 19 (IBM, Armonk, NY, USA).
Results
Expression of genes
Expression levels of miR-21, miR-134 and miR-205
were significantly higher in all the EC samples than in the
non-cancerous controls. Only miR-495 was not expressed in the FFPE
samples. All the cancer pathway-associated genes that has been
selected for the study on FFPE (MET, MMP9,
PDGFA and SERPINE1) exhibited significantly higher
expression levels in the 63 EC samples than in the adjacent
non-cancerous specimens (P<0.05) (Fig.
1).
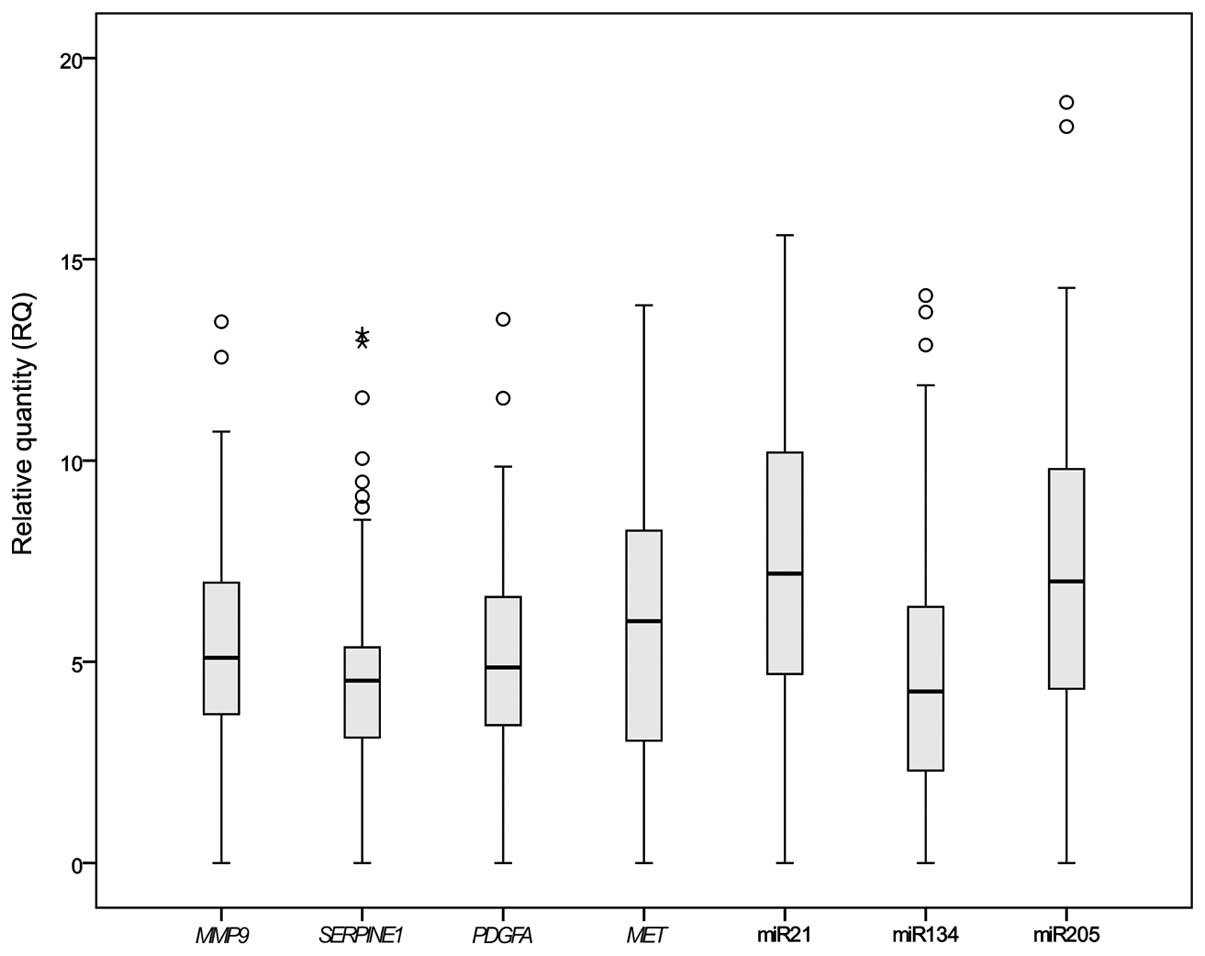 | Figure 1.Gene expression in esophageal cancer
samples. Boxplot showing the relative expression of genes. Ct
values from quantitative polymerase chain reaction were normalized
against RNU24, and the relative expression was calculated by the
2−∆∆Ct method. Outliers are represented by circles and
extreme outliers are represented by stars. The P-values for
MMP9, SERPINE1, PDGFA, MET, miR-21,
miR-134 and miR-205 were P=0.046, P=0.001, P=0.041, P=0.019,
P=0.045, P=0.002 and P=0.009, respectively, as determined by the
Mann-Witney U test. miR, microRNA. |
Association between genes and
clinicopathological factors
One significant correlation was found between the
miRNA genes and clinicopathological factors: Age was positively
correlated with miR-134 expression (P<0.039), as determined by
Spearman's rank correlation coefficient. The expression of two of
the cancer pathway-associated genes, MMP9 and
SERPINE1, was significantly correlated with the
clinicopathological factors. MMP9 was positively correlated
with age (P<0.002), while SERPINE1 was positively
correlated with smoking status (P<0.0015) and with the number of
years of smoking (P<0.029). All these results were acquired by
calculating Spearman's rank correlation coefficient. The
Kruskal-Wallis test and the Mann-Whitney U test confirmed this
data. MMP9 and SERPINE1 exhibited a significant
correlation with age (P=0.011 and P=0.044, respectively). In
addition, SERPINE1 was significantly correlated with smoking
and the number of years of smoking (P=0.008 and P=0.032,
respectively).
Correlation between genes
One positive correlation between an miRNA gene and
the expression of a cancer pathway-associated gene was proven for
miR-134 and MMP9, with statistical significance
(P<0.019), as determined using Spearman's rank correlation
coefficient.
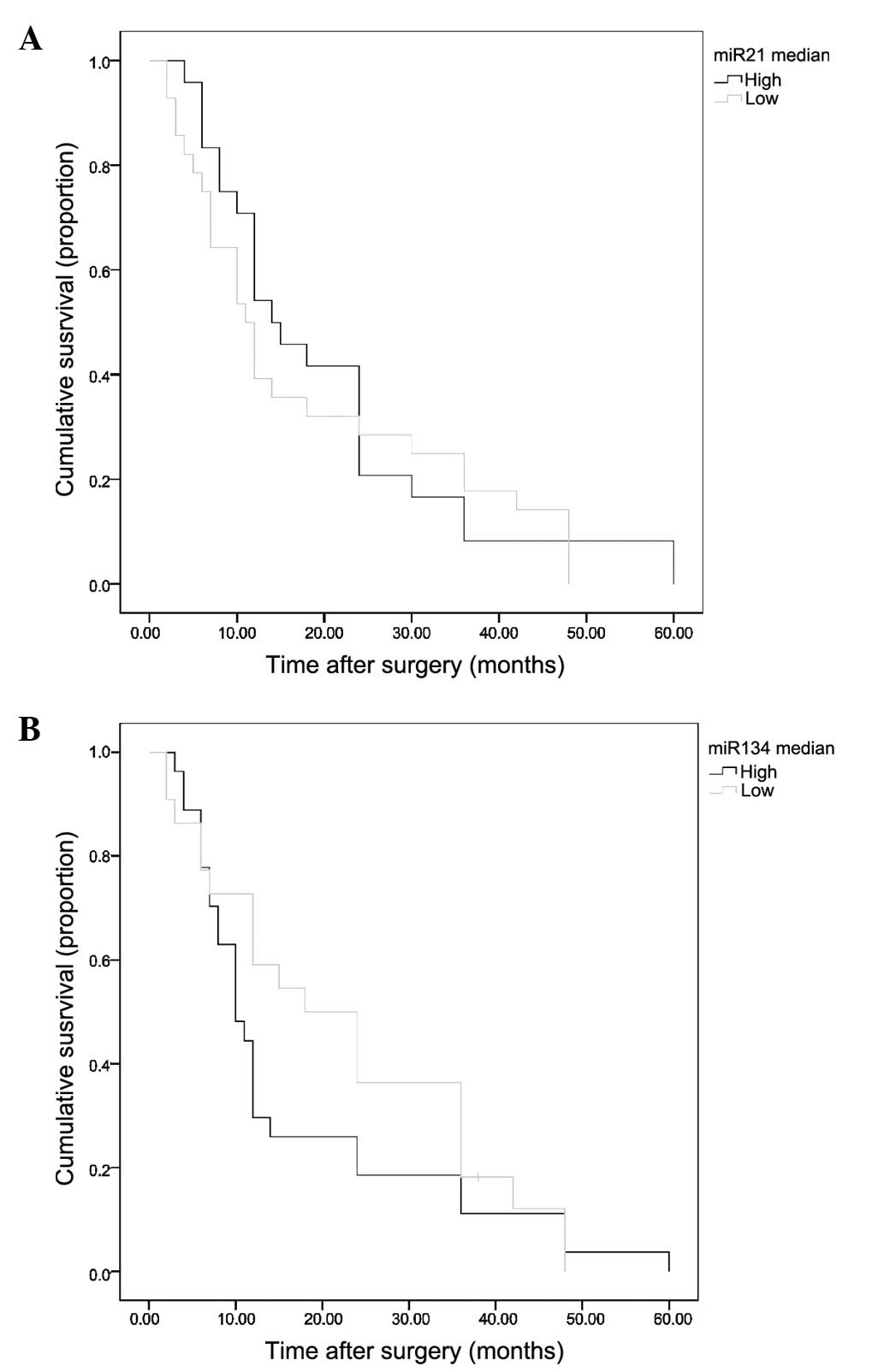
Survival analysis
The available survival data for 56 of the 63
patients (88.9%) analyzed against miR-21 and miR-134 expression is
shown (Fig. 2). Survival plots for
miR-21 and miR-134 showed differences between high and low
expression, but these differences were not significant (P=0.278 and
P=0.183, respectively). Gender, American Joint Committee on Cancer
system classification and type of EC were identified as invariable
upon analysis (data not shown).
Discussion
In the present study of EC, the expression of seven
genes was detected (miR-495 was not expressed in the FFPE samples).
For each of the genes, MET, MMP9, PDGFA,
SERPINE1, miR-21, miR-134 and miR-205, the results were
significant in comparison to adjacent non-cancerous specimens. The
study investigated whether the expression of these genes in EC was
correlated with clinicopathological factors and prognosis of EC
patients. For MMP9, SERPINE1 and miR-134, the results
were significant, as determined by the Kruskal-Wallis and
Mann-Whitney U tests, and Spearman's correlation coefficient. It is
noteworthy that for the first time in the literature a correlation
was detected between MMP9 and miR-134.
Different miRNA databases were used to analyze the
genes for the present study: miR2Disease (http://www.mir2disease.org/), miRecords (http://c1.accurascience.com/miRecords/),
miRSel (https://services.bio.ifi.lmu.de/mirsel/), miRTarBase
(http://mirtarbase.mbc.nctu.edu.tw/)
and miRWalk (http://www.umm.uni-heidelberg.de/apps/zmf/mirwalk/),
starBase v2.0 (http://starbase.sysu.edu.cn/) and DIANA Lab's TarBase
(http://diana.imis.athena-innovation.gr/DianaTools/index.php?r=tarbase/index).
In miR2Disease only EC-related data could be found on miR-21 and
miR-205 (24). There were records for
miR-134 and miR-495, but with regard to other diseases.
Only miR-21 and miR-205 were described in the
miRecords target miRNA search, but no information with regard to
the correlation of the genes and the EC pathways was mentioned.
MET was included in the target gene search results, but was
not associated with the miRNA genes of the present study. However,
in miRSel there was information on a correlation between miR-21 and
MMP9. miR-134 was correlated with PDGFRA, a receptor
for PDGFA, which was analyzed in the present study. None of
the 98 available records for miR-205 were connected with genes
correlated with cancer pathways. There was no record for miR-495.
For the miRTarBase, only one correlation between miR-21 and
MMP9 was found (25).
A notable correlation was shown by the miRWalk
database, according to which PDGFA is situated on the
miR-495 stem-loop. SEPRINE1 is located on the stem-loop of
miR-134 in the UTR. In starBase v2.0 no records were found. In
DIANA Lab's TarBase, only a correlation between miR-21 and
MMP9 was found. The aforementioned analyses are current as
of September 2014, and the list of miRNA databases is based on
those used by Vergoulis et al (26).
Houldsworth et al (27) identified an association between
MET and EC, while Gu et al (28) showed that MMP9 affected the
survival of patients with this disease. PDGFA was detected
in EC by Wong et al (29) and
Aoki et al (30) suggested
that SERPINE1 may be a potentially important index for the
prediction of post-operative illness.
A correlation between miR-134 and the duration of
survival with this disease has also been found (31). Guo et al demonstrated a
correlation between miR-495 and the gross pathological
classification of EC (32). There is
no information with regard to miR-134 and the genes analyzed for
this study in EC or even for other cancers in the case of
MMP9. There are no studies in the literature of miR-205 and
miR-495 and genes from this study regarding EC.
As MMP9 and SERPINE1 have shown
correlation with clinicopathological factors, information on their
proteins was obtained. Search Tool for the Retrieval of Interacting
Genes/Proteins has been used to study the interactions between
genes and their protein products (33). It was found that SERPINE1 and MMP9 are
separated in interactions by PLAU. It is known that there is a
strong correlation between SERPINE1 and MMP9 in
pancreatic cancer (34). There is
only one abstract with regard to this interaction in EC (35). Kajiwara et al measured the
expression of these two genes in EC, but did not find a significant
interaction between them (20).
In conclusion, the present study identified genes
with expression that was correlated with the clinicopathological
factors of EC. MMP9, SERPINE1 and miR-134 are the
most prognostic genes analyzed in this population-based study of
EC. Moreover, a positive correlation was established between
MMP9 and miR-134, which has not previously been reported
with regards to esophageal cancer.
Acknowledgements
This study was supported by a research project grant
from the Medical University of Lodz (no. 502-64-002).
References
|
1
|
Boyle P and Levin B: World Cancer Report
2008. IARC; Lyon, France: 2008
|
|
2
|
Jemal A, Bray F, Center MM, Ferlay J and
Ward:
|
|
3
|
Klimczak A, Bitner J and Szemraj J: Genes
responsible for etiopathogenesis in esophageal squamous cell
carcinoma. Postepy Biochem. 57:33–40. 2011.(In Polish). PubMed/NCBI
|
|
4
|
Lam AK: Molecular biology of esophageal
squamous cell carcinoma. Crit Rev Oncol Hematol. 33:71–90. 2000.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Klimczak A, Kuropatwa A, Lewkowski J and
Szemraj J: Synthesis of new N-arylamino (2-furyl) methylphosphonic
acid diesters and in vitro evaluation of their cytotoxicity against
esophageal cancer cells. Med Chem Res. 22:852–860. 2013. View Article : Google Scholar
|
|
6
|
Law S and Wong J: Therapeutic options for
esophageal cancer. J Gastroenterol Hepatol. 19:4–12. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Hartwig W, Strobel O, Schneider L, Hackert
T, Hesse C, Büchler MW and Werner J: Fundus rotation gastroplasty
vs. Kirschner-Akiyama gastric tube in esophageal resection:
Comparison of perioperative and long-term results. World J Surg.
32:1695–1702. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Ordu AD, Nieder C, Geinitz H, Kup PG,
Deymann LF, Scherer V, Combs SE and Fakhrian K: Radio(chemo)therapy
for locally advanced squamous cell carcinoma of the esophagus:
Long-term outcome. Strahlenther Onkol. 191:153–160. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Rabenstein T: Palliative endoscopic
therapy of esophageal cancer. Viszeralmedizin. 31:354–359. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Carrington JC and Ambros V: Role of
microRNAs in plant and animal development. Science. 301:336–338.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Kim VN, Han J and Siomi MC: Biogenesis of
small RNAs in animals. Nat Rev Mol Cell Biol. 10:126–139. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Thorsen SB, Obad S, Jensen NF, Stenvang J
and Kauppinen S: The therapeutic potential of microRNAs in cancer.
Cancer J. 18:275–284. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Yanaihara N, Caplen N, Bowman E, Seike M,
Kumamoto K, Yi M, Stephens RM, Okamoto A, Yokota J, Tanaka T, et
al: Unique microRNA molecular profiles in lung cancer diagnosis and
prognosis. Cancer cell. 9:189–198. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Iorio MV, Ferracin M, Liu CG, Veronese A,
Spizzo R, Sabbioni S, Magri E, Pedriali M, Fabbri M, Campiglio M,
et al: MicroRNA gene expression deregulation in human breast
cancer. Cancer Res. 65:7065–7070. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Murakami Y, Yasuda T, Saigo K, Urashima T,
Toyoda H, Okanoue T and Shimotohno K: Comprehensive analysis of
microRNA expression patterns in hepatocellular carcinoma and
non-tumorous tissues. Oncogene. 25:2537–2545. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Johnson KE and Wilgus TA: Vascular
Endothelial Growth Factor and Angiogenesis in the Regulation of
Cutaneous Wound Repair. Adv Wound Care (New Rochelle). 3:647–661.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Valastyan S and Weinberg RA: Tumor
metastasis: Molecular insights and evolving paradigms. Cell.
47:275–292. 2011. View Article : Google Scholar
|
|
18
|
Bal J: Biologia Molekularna W Medycynie.
Wydawnictwo Naukowe PWN; Warszawa: 2011, (In Polish).
|
|
19
|
Qiagen, . RT2 Profiler PCR Array Handbook.
Qiagen, USA: pp. 27–48. 2011
|
|
20
|
Kajiwara T, Hiasa Y, Nishina T, Matsumoto
T, Hori S, Nadano S, Iguchi H, Takeji S, Tsubouchi E, Ikeda Y, et
al: Maximum standardized uptake value in 18F-fluoro-2-deoxyglucose
positron emission tomography is associated with advanced tumor
factors in esophageal cancer. Mol Clin Oncol. 2:313–321.
2014.PubMed/NCBI
|
|
21
|
Kameyama H, Udagawa O, Hoshi T, Toukairin
Y, Arai T and Nogami M: The mRNA expressions and
immunohistochemistry of factors involved in angiogenesis and
lymphangiogenesis in the early stage of rat skin incision wounds.
Leg Med (Tokyo). 17:255–260. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Biosystems A: Megaplex™ Pools For microRNA
Expression Analysis. Biosystems A; USA: pp. 15–22. 2010
|
|
23
|
Akagi I, Miyashita M, Ishibashi O, Mishima
T, Kikuchi K, Makino H, Nomura T, Hagiwara N, Uchida E and Takizawa
T: Relationship between altered expression levels of MIR21, MIR143,
MIR145, and MIR205 and clinicopathologic features of esophageal
squamous cell carcinoma. Dis Esophagus. 24:523–530. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Feber A, Xi L, Luketich JD, Pennathur A,
Landreneau RJ, Wu M, Swanson SJ, Godfrey TE and Litle VR: MicroRNA
expression profiles of esophageal cancer. J Thorac Cardiovasc Surg.
135:255–260; discussion 260. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Giovannetti E, Funel N, Peters GJ, Del
Chiaro M, Erozenci LA, Vasile E, Leon LG, Pollina LE, Groen A and
Falcone A: MicroRNA-21 in pancreatic cancer: Correlation with
clinical outcome and pharmacologic aspects underlying its role in
the modulation of gemcitabine activity. Cancer Res. 70:4528–4538.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Vergoulis T, Vlachos IS, Alexiou P,
Georgakilas G, Maragkakis M, Reczko M, Gerangelos S, Koziris N,
Dalamagas T and Hatzigeorgiou AG: TarBase 6.0: Capturing the
exponential growth of miRNA targets with experimental support.
Nucleic Acids Res. 40:D222–D229. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Houldsworth J, Cordon-Cardo C, Ladanyi M,
Kelsen DP and Chaganti RS: Gene amplification in gastric and
esophageal adenocarcinomas. Cancer Res. 50:6417–6422.
1990.PubMed/NCBI
|
|
28
|
Gu ZD, Li JY, Li M, Gu J, Shi XT, Ke Y and
Chen KN: Matrix metalloproteinases expression correlates with
survival in patients with esophageal squamous cell carcinoma. Am J
Gastroenterol. 100:1835–1843. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Wong FH, Hu CP, Chen SC, Yu YT and Chang
C: Absence of genomes of DNA tumor viruses and expression of
oncogenes and growth factors in two esophageal carcinoma cell lines
of Chinese origin. Zhonghua Min Guo Wei Sheng Wu Ji Mian Yi Xue Za
Zhi. 25:59–68. 1992.PubMed/NCBI
|
|
30
|
Aoki K, Nishino N, Baba S, Urano T and
Takada A: Postoperative changes in plasma tissue-type plasminogen
activator and type 1 plasminogen activator inhibitor. Surg Today.
24:1039–1043. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Hamano R, Miyata H, Yamasaki M, Kurokawa
Y, Hara J, Moon JH, Nakajima K, Takiguchi S, Fujiwara Y, Mori M and
Doki Y: Overexpression of miR-200c induces chemoresistance in
esophageal cancers mediated through activation of the Akt signaling
pathway. Clin Cancer Res. 17:3029–3038. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Guo Y, Chen Z, Zhang L, Zhou F, Shi S,
Feng X, Li B, Meng X, Ma X, Luo M, et al: Distinctive microRNA
profiles relating to patient survival in esophageal squamous cell
carcinoma. Cancer Res. 68:26–33. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Snel B, Lehmann G, Bork P and Huynen MA:
STRING: A web-server to retrieve and display the repeatedly
occurring neighbourhood of a gene. Nucleic Acids Res. 28:3442–3444.
2000. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Wagner M, Kleeff J, Friess H, Buchler MW
and Korc M: Enhanced expression of the type II transforming growth
factor-beta receptor is associated with decreased survival in human
pancreatic cancer. Pancreas. 19:370–376. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Pidgeon GP, Allott E, Donohoe C, Cathcart
M, McGarrigle S, Cummins R, Kay E and Reynolds J: Adipose tissue
upregulates MMP9 expression in oesophageal cancer promoting
invasion and migration in vitro and correlates with obesity status
and poor differentiation in vivo. Cancer Res. 71:(Suppl): abstract
1556. 2011. View Article : Google Scholar
|