Introduction
Activating transcription factor (ATF) 2 is a member
of the ATF/cyclic AMP-responsive element binding protein family of
transcription factors, which harbors leucine zipper and DNA binding
domains (1), and exhibits both
oncogenic and tumor-suppressor functions (2–4). ATF2 is
important for normal cellular development and survival, and is also
involved in the response to stress and DNA damage in the context of
normal and tumorigenesis processes (5). The transactivating potential of ATF2 is
stimulated to a higher level by a broad group of agents causing DNA
damage and other types of cellular stress, including
short-wavelength ultraviolet (UV) light, osmotic stress and
inflammatory cytokines (6,7). ATF2 is expressed ubiquitously, and
elevated expression of ATF2 was detected in skin carcinomas
(8) and melanoma (9). It was reported that the ATF2 expression
levels were significantly induced under oxidative stress in
immortalized human urothelial cells treated with arsenic (10).
Translational control is important in the control of
gene expression, by which, cells can respond to rapid changes in
physiological conditions, including DNA damage and cellular stress
(11,12). One of the common mechanisms of
translational control is the induction of changes in the
translation initiation site on the messenger RNA (mRNA) molecule,
which may occur via two distinct mechanisms, cap-dependent scanning
and internal ribosome entry (IRE) (13). The latter mechanism requires the
formation of a complex RNA structural element termed IRE segment
(IRES), which is located in the 5′-untranslated region (UTR) of the
mRNA molecule (14). To date, IRES
elements have been mainly identified in mRNAs involved in
regulating gene expression during development, differentiation,
cell cycle progression, cell growth, apoptosis and stress,
including nuclear factor-κB repressing factor, X-chromosome linked
inhibitor of apoptosis protein (XIAP), vascular endothelial growth
factor, c-Myc, fibroblast growth factor-2 and Smad mRNAs (15–19). The
most notable difference in cellular IRES sequence from the UTR data
sets is that IRES-containing UTRs are generally long (>150
bases) and GC rich (20).
Multiple cellular stresses lead to the inhibition of
translation; however, there are certain cellular mRNAs that are
selectively translated under these conditions to protect cells from
stress, the majority of which contain an IRES element (21). Yang et al noticed that tumor
necrosis factor receptor-associated factor 1 protein expression can
be stimulated under cellular stress conditions, and its translation
is regulated by IRES (22). In
addition, it was reported that ATF4 splice variant, whose protein
expression was induced by thapsigargin and tunicamycin in THP-1
cells, contains an IRES structure (23).
Adriamycin (ADM) and paclitaxel (PTX) are two
effective antitumor agents that are widely used in hepatocellular
carcinoma therapy (24,25). Both can induce cell apoptosis and
cause cellular stress (26). In the
present study, ATF2 protein expression in the human hepatoma cell
line Bel7402 was observed to be induced following treatment with
ADM or PTX, and it was demonstrated that the 5′-UTR of ATF harbors
a bona fide IRES structure localized between nt −299 to nt
−269 upstream of the start codon. The IRES activity of ATF2 was low
in the mouse embryo fibroblast cell line National Institutes of
Health (NIH)-3T3 and in the human colon carcinoma cell line HCT-8,
while it was high in the human embryonic kidney (HEK) 293 cell line
and in the human hepatoma cell line Bel7402. Furthermore, the
present study was demonstrated that ATF2 IRES activity was also
elevated following treatment with cellular stress-inducing
chemotherapeutic drugs such as PTX and ADM.
Materials and methods
Plasmid construction
The bicistronic vector pRF was constructed by
inserting the firefly luciferase (FL or FLUC) gene from the plasmid
pGL-6 (Beyotime Institute of Biotechnology, Haimen, China) into the
linker region of the plasmid pRL-SV4 (Promega Corporation, Madison,
WI, USA) downstream of the Renilla luciferase (RL or RLUC)
gene. To insert the FLUC fragment, a polymerase chain reaction
(PCR) was performed on the pGL-6 plasmid with primers FLUCF
5′-GCTCTAGACTCGAGCATATGGAATTCGGCGCGCCCATGGAAGATGCCAAAAACATTAAGAAGGGCCC-3′
(forward) and FLUCR 5′-AAATATGCGGCCGCTTACACGGCGATCTTGCCGCCCTT-3′
(reverse). The PCR was conducted in a 50-µl reaction mixture
containing 2X PrimeSTAR Max Premix (Takara Biotechnology Co., Ltd.,
Dalian, China), 0.2 µM forward and reverse primers, and 200 ng
genomic DNA, which was purchased from the Beyotime Institute of
Biotechnology. The PCR was conducted at 98°C for 3 min, followed by
32 cycles of 98°C for 1 min, 55°C for 10 sec and 72°C for 1 min,
and 72°C for 5 min. The amplified product was digested with
Bsp119 and XbaI, and subsequently cloned into
pRL-SV40, thus generating pRF. In this vector, the two cistrons
were separated by ~20 base pairs (bp) of the intercistronic linker
region containing several unique endonuclease restriction sites.
The expression of bicistronic mRNA is driven by a simian virus (SV)
40 promoter. The full length 5′-UTR complementary DNA (cDNA) of
ATF2, nuclear respiratory factor (NRF) and XIAP were obtained by
gene synthesis (Sangon Biotech Co., Ltd., Shanghai, China), while
serial truncations of ATF2 were produced by PCR amplification with
forward and reverse primers (Table I)
containing EcoRI and NdeI endonuclease restriction
sites, respectively. The PCR was conducted in a 50-µl reaction
mixture containing 2X PrimeSTAR Max Premix, 0.2 µM forward and
reverse primers, and 200 ng genomic DNA. The PCR was conducted at
98°C for 3 min, followed by 32 cycles of 98°C for 1 min, varying
annealing temperatures (as indicated in Table I) and 72°C for 10 sec, and 72°C for 5
min. All these 5′-UTR cDNAs were each inserted immediately upstream
of the translation start codon of the FLUC gene in the dual
luciferase vector pRF at the EcoRI and NdeI
sites.
 | Table I.Oligonucleotide primers used in
polymerase chain reaction for plasmids construction. |
Table I.
Oligonucleotide primers used in
polymerase chain reaction for plasmids construction.
| Vector (nt) | Primer sequence,
5′-3′ | Temperature,
°C |
|---|
| pR-ATF2-F
(31–299) | F1:
GGAATTCCATATGGAAGCCTGTGGGAGCCC | 50.0 |
|
| R1:
CGGAATTCAAGTTGAATAACTTATCACATTC |
|
| pR-ATF2-F
(51–299) |
F2:GGAATTCCATATGGCCTTTAAAGTGCCGTTCAG | 50.0 |
|
| R2:
CGGAATTCAAGTTGAATAACTTATCACATTC |
|
| pR-ATF2-F
(76–299) | F3:
GGAATTCCATATGTCCTCCAGGGGTGCTTTGTA | 50.0 |
|
| R3:
CGGAATTCAAGTTGAATAACTTATCACATTC |
|
| pR-ATF2-F
(141–299) | F4:
GGAATTCCATATGACTAGTGACCTGGAAAGGGT | 50.0 |
|
| R4:
CGGAATTCAAGTTGAATAACTTATCACATTC |
|
| pR-ATF2-F
(1–229) | F5:
GGAATTCCATATGGTTGTCAGTCCGATCTCGC | 52.0 |
|
| R5:
CGGAATTCATTTACTCTGCTTCCAGGAATACC |
|
| pR-ATF2-F
(1–159) | F6:
GGAATTCCATATGGTTGTCAGTCCGATCTCGC | 52.8 |
|
| R6:
CGGAATTCCCCTTTCCAGGTCACTAGTTC |
|
| pR-ATF2-F
(1–89) | F7:
GGAATTCCATATGGTTGTCAGTCCGATCTCGCG | 54.5 |
|
| R7:
CGGAATTCGCACCCCTGGAGGAAAAGG |
|
Cell culture and DNA transfection
The HEK293 cell line (Type Culture Collection of the
Chinese Academy of Sciences, Shanghai, China) was cultured in
Dulbecco's modified Eagle medium (Gibco; Thermo Fisher Scientific,
Inc., Waltham, MA, USA), while the Bel7402, HCT-8 and NIH-3T3 cell
lines (Type Culture Collection of the Chinese Academy of Sciences)
were cultured in RPMI 1640 medium (Gibco; Thermo Fisher Scientific,
Inc.) with 10% (vol/vol) fetal bovine serum (Gibco; Thermo Fisher
Scientific, Inc.), penicillin (100 U/ml) and streptomycin (100
µg/ml). All cells were kept at 37°C in a humidified atmosphere with
5% CO2. Transient transfection was performed using
Lipofectamine 2000 (Invitrogen; Thermo Fisher Scientific,
Inc.)according to the manufacturer's protocol. The cell media were
replaced with complete culture media at 6 h post-transfection, and
24–48 h later, the cells were rinsed twice with phosphate-buffered
saline, and cell extracts were then prepared using 5X Passive Lysis
Buffer (Promega Corporation).
Dual luciferase assay
FLUC and RLUC activities were measured using the
Dual-Luciferase Reporter Assay System (Promega Corporation)
according to the manufacturer's protocol, with the exception that
only 100 µl of each reagent was used. The signals were measured
with a luminometer.
RNA isolation and reverse
transcription (RT)-PCR
Total RNA was isolated from the cells using TRIzol
reagent (Beyotime Institute of Biotechnology), and was reverse
transcribed into cDNA using oligo(dT)18 primer (Takara
Biotechnology Co., Ltd.) and PrimeScript Reverse Transcriptase
(Takara Biotechnology Co., Ltd.). PCR was next performed using
specific primers, which were designed according to the
corresponding sequences of the ATF2 5′-UTR mRNA: ATF2F
5′-GCACGTAATGACAGTGTCATTGT-3′ (forward) and ATF2R
5′-ATGTGGGCTGTGCAGTTTGTG-3′ (reverse). The PCR was conducted in a
50-µl reaction mixture containing 2X PrimeSTAR Max Premix, 0.2 µM
forward and reverse primers, and 250 ng total RNA. The PCR was
conducted at 98°C for 3 min, followed by 32 cycles of 98°C for 1
min, 51.6°C for 15 sec and 72°C for 18 sec, and 72°C for 5 min.
Upon electrophoresis on a 1.5% agarose gel, the PCR products were
visualized with ethidium bromide staining under UV light. DNA bands
were analyzed using ImageJ software (NIH, Bethesda, MD, USA).
Western blotting
Whole-cell proteins were obtained using
radioimmunoprecipitation assay buffer containing 1 mM phenylmethane
sulfonyl fluoride. The same quantity of total protein was loaded
onto each lane of the gel, and the proteins were electrophoresed on
8% polyacrylamide gels containing 0.1% sodium dodecyl sulfate. The
resolved proteins were semi-dry transferred to a polyvinylidene
fluoride membrane. The membrane was blocked with 0.5% bovine serum
albumin (Beyotime Institute of Biotechnology) at room temperature
for ~3 h, prior to be incubated with the corresponding primary
antibodies [anti-ATF2 (1:5,000; ab32061; Abcam, Cambridge, MA, USA)
or anti-β-actin (1:1,000; AA128; Beyotime Institute of
Biotechnology), which was used as an internal reference] at room
temperature for 2 h. Next, the membrane was incubated with the
secondary antibody Goat Anti-Rabbit HRP (IgG H&L) (1:3,000;
ab6721; Abcam) at room temperature for 1.5–2.0 h. Finally, the
antigen-antibody complexes were visualized by enhanced
chemiluminescence (Thermo Fisher Scientific, Inc.).
Statistical analysis
Each experiment was performed ≥3 times. All
experimental data were analyzed using GraphPad Prism 5.0 (GraphPad
Software, Inc., La Jolla, CA, USA) and presented as mean values ±
standard error of the mean. Statistical analysis was performed
using the Student's t-test. P<0.05 was considered to indicate a
statistically significant difference.
Results
Protein expression of ATF2 is induced
at the translational level following treatment with
chemotherapeutic drugs in Bel7402 cells
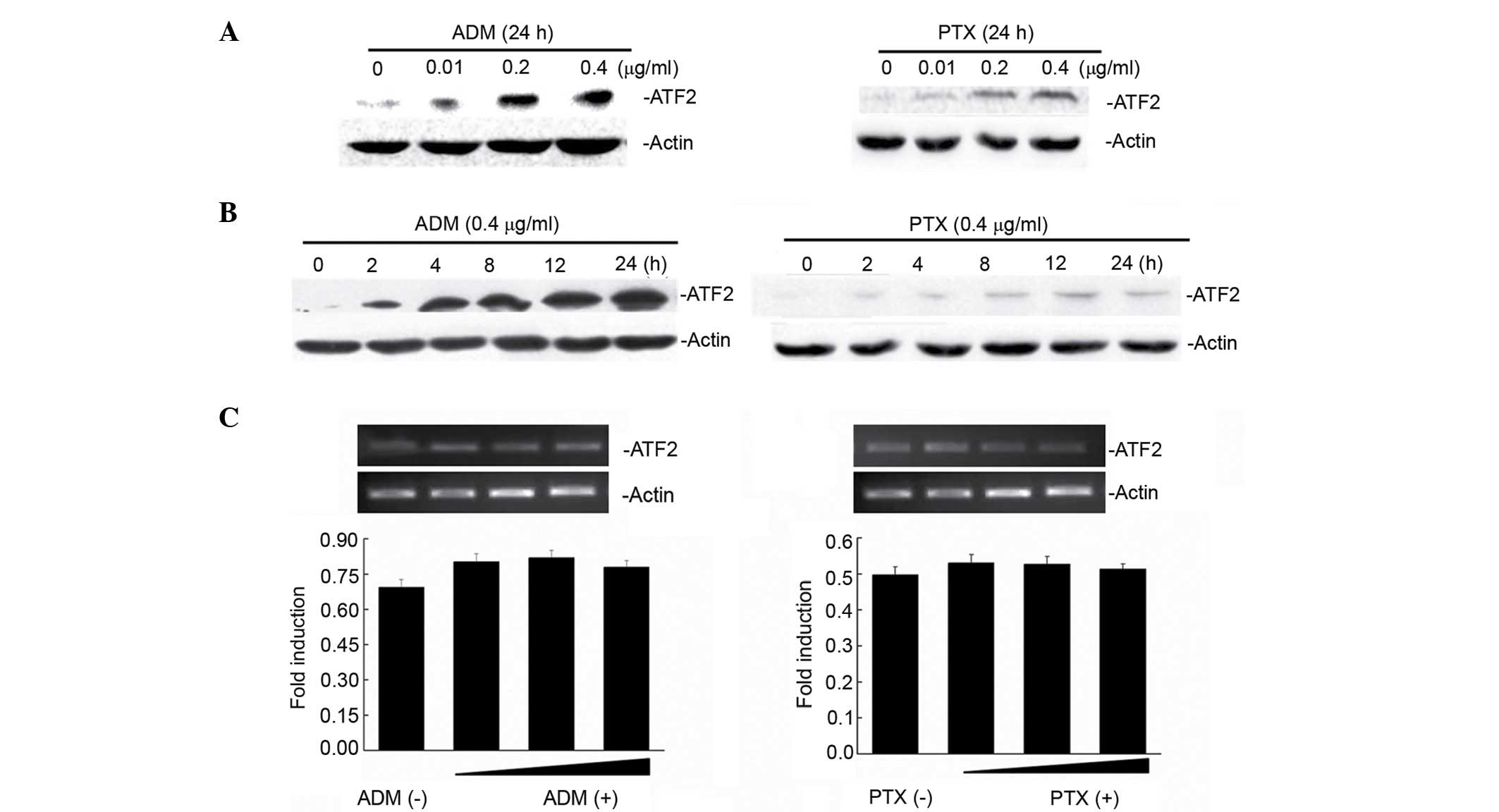
ATF2 exhibited very low expression in Bel7402 cells
(Fig. 1A). Notably, when Bel7402
cells were treated with increasing concentrations (0.01, 0.2 and
0.4 µg/ml) of two chemotherapeutic drugs (ADM and PTX), ATF2
protein expression was induced following treatment with ADM and PTX
(Fig. 1A) in a dose-dependent manner.
To demonstrate whether the ATF2 induction changed with the exposure
time to the drugs, ATF2 protein expression was detected at various
times following 0.4-µg/ml ADM and PTX treatment for 2, 4, 8, 12 and
24 h. Fig. 1B reveals that ATF2
protein expression is increased in a time-dependent manner
following treatment with ADM and PTX. Next, the mechanism by which
these drugs increased the levels of ATF2 in Bel7402 cells was
examined. Treatment of Bel7402 cells with either ADM or PTX did not
result in any significant change (P=0.552) in ATF2 mRNA levels
(Fig. 1C), indicating that
ADM/PTX-induced ATF2 expression occurs at the translational
level.
ATF2 5′-UTR-mediated IRE in mammalian
cultured cell lines
Since by searching the ATF2 gene in the human genome
database (http://www.ncbi.nlm.nih.gov/nuccore/NM_001880.3) it
was observed that the ATF2 5′-UTR is generally long (ATF2, 299 nt)
and rich in GC, the possibility that the ATF2 5′-UTR contains an
IRES that could regulate ATF2 protein synthesis following treatment
with chemotherapeutic drugs was examined. To evaluate whether ATF2
translation was initiated from an IRE, the ATF2 5′-UTR was inserted
between the two cistrons of a bicistronic vector, in which the
first cistron encodes the RLUC gene and the second cistron encodes
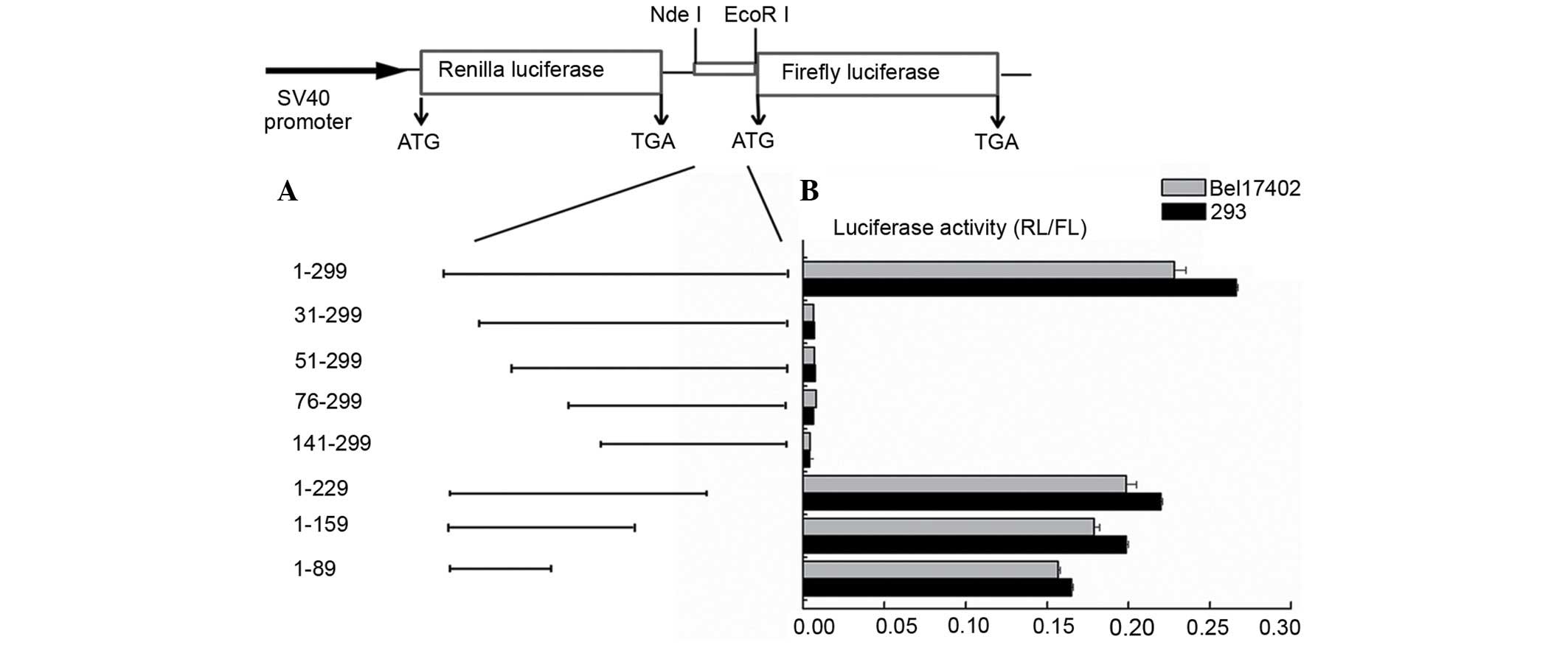
the FLUC gene (Fig. 2A). The first
cistron is proximal to the mRNA's cap structure, and therefore, is
expected to be translated by the conventional cap-dependent
translation mode. The second cistron is distal from the cap site;
therefore, it is expected to undergo translation only upon
insertion of an IRES element between the two cistrons (27). The bicistronic vectors pR-ATF2-F were
transfected into HEK293 Bel7402 and HCT-8 human cells, and into
murine NIH-3T3 cells. The pR-XIAP-F and pR-NRF-F vectors, which
contain the well-characterized cellular XIAP and NRF IRES, were
transfected as positive controls, while pRF was used as a negative
control. The RL and FL activities were determined as aforementioned
described. The IRES activity was calculated as the ratio of FLUC to
RLUC normalized to that of pRF-transfected cells. As indicated in
Fig. 2B, the ATF2 IRES was observed
to be active in the HEK293, Bel7402 and HCT-8 cell lines, but not
in the NIH-3T3 cell line. The ATF2 IRES activity was different in
each cell line, being high in Bel7402 and HEK293 cells (~5-fold
higher than the XIAP IRES activity). These results suggest that a
putative IRES element was present in the 5′-UTR of the ATF2 mRNA.
The difference in the levels of IRES activity observed in these
cell lines is probably due to intrinsic differences between these
cell types.
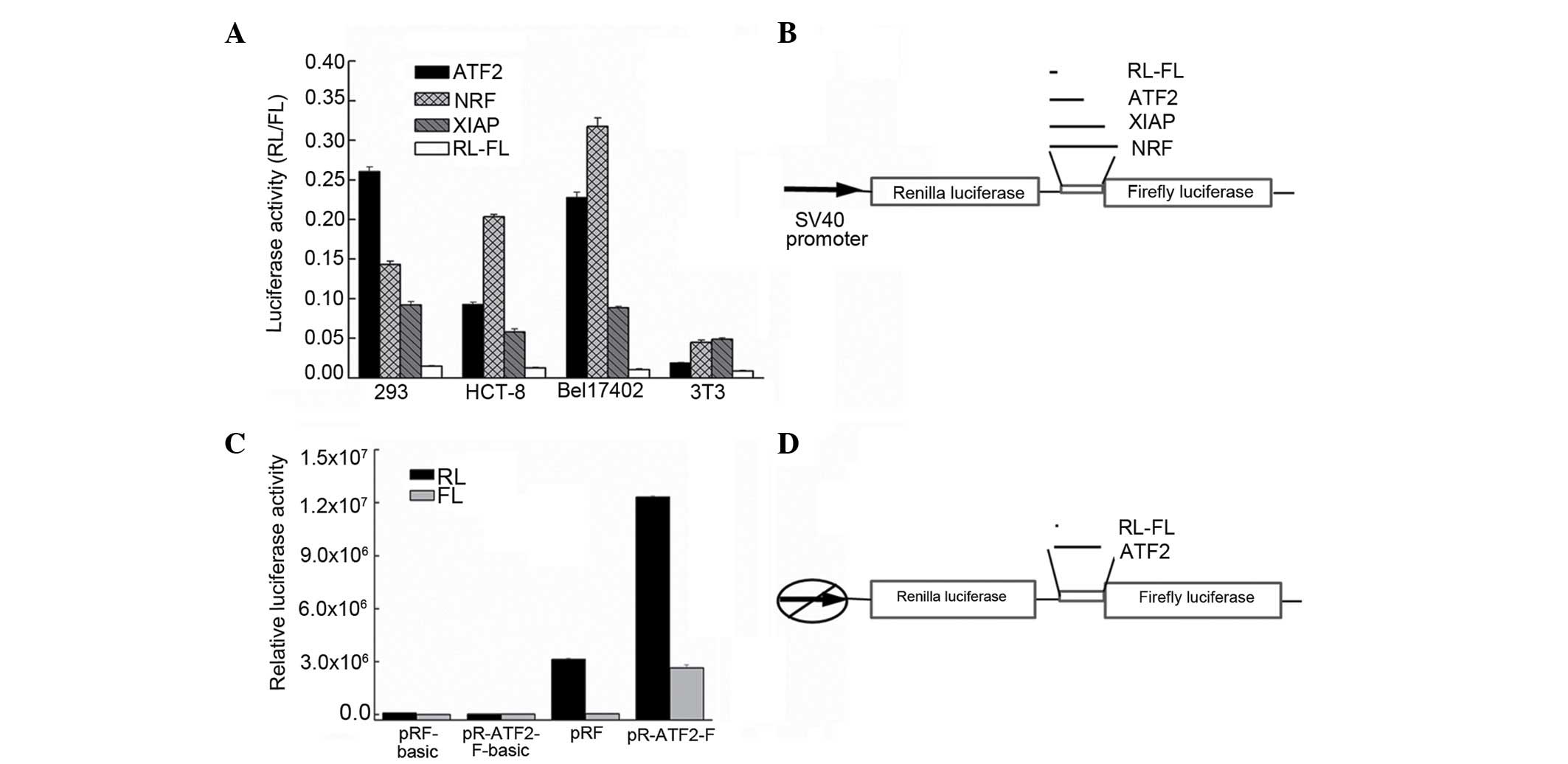 | Figure 2.Identification of the IRES activity in
the ATF2 5′-UTR. (A) The plasmids pRAF, pRXF, pRNF and pRF were
transfected into the cell lines HEK293, Bel7402, HCT-8 and NIH-3T3.
The IRES activity was expressed as the ratio of downstream cistron
expression to upstream cistron expression (FL/RL). The error bars
indicate the SD determined from ≥3 independent experiments
performed in triplicate. (B) Schematic representation of the
expression cassette of the dual-luciferase bicistronic constructs
pRF, pRAF, pRXF and pRNF. (C) The translation of the second cistron
is not due to reinitiation or aberrant messenger RNA species. To
test the cryptic promoter activity in the ATF2 5′-UTR, the sequence
was cloned upstream of the FL reporter, and the SV40 promoter was
removed. The empty vector was used as a negative control. The
plasmids were transfected into Bel7402 cells. The RL and FL
activities were measured after 24 h. The error bars indicate the SD
determined from ≥3 independent experiments performed in triplicate.
(D) Schematic representation of the expression cassette of the
dual-luciferase bicistronic constructs pRF and pRAF in which the
promoter was deleted. RL, Renilla luciferase; FL, firefly
luciferase; SV, simian virus; ATF2, activating transcription factor
2; NRF, nuclear respiratory factor; XIAP, X-linked inhibitor of
apoptosis protein; IRES, internal ribosome entry segment; UTR,
untranslated region; SD, standard deviation. |
Internal initiation of translation
mediated by the ATF2 5′-UTR is not due to cryptic promoter
activity
To exclude the possibility that the expression of
the second cistron in these two bicistronic mRNAs could be derived
from monocistronic RNAs due to the presence of a cryptic promoter
in the 5′-UTR, pR-ATF2-F-basic plasmids were generated, in which
the ATF2 5′-UTR was inserted in the promoterless pRF-basic vector
upstream of the RLUC coding sequence. The HEK293 and Bel7402 cell
lines were each transfected with the pRF plasmid (a vector
containing the SV40 early promoter and serving as a positive
control), the promoterless pRF-basic plasmid and the
pR-ATF2-F-basic plasmid. Luciferase activities were measured as
aforementioned described. Fig. 2C
indicates that the FLUC activity measured in
pR-ATF2-F-basic-transfected cells is comparable to that measured in
cells transfected with the pRF-basic plasmid, and is ~350-fold
lower than the activity produced by pR-ATF2-F. This result clearly
illustrated that the ATF2 5′-UTR was devoid of promoter
activity.
Mapping the ATF2 IRES
To further identify the core regions that promote
the internal initiation of translation, serial truncations of the
ATF2 5′ leader sequence were created (Fig. 3A). Subsequently, the activities of the
inserted sequences were assayed as aforementioned described. The
ability of these truncated sequences to promote the translation of
the mRNA was also compared with that of pRF. Deletions of 30 nt of
the 5′ end of ATF2 resulted in a marked decrease in IRES activity,
and further deletion of 52, 75 and 142 nt of the 5′ end of ATF2 led
to a similar reduction. Notably, at the 3′ end of ATF2, deletion of
70, 140 and 210 nt did not produce a significant decrease
(P=0.148), since the IRES activity remained almost the same as that
of the full-length ATF2 5′-UTR (Fig.
3B). These data indicated that the 5′ end of ATF2 is important
for positioning the IRES in context with the translational start
site, and the core IRES region of ATF2 contains 30 bases and
resided between nt −299 and −269. The results were the same when
different cells were transfected. These results demonstrated that
the full length of the ATF2 5′-UTR contributes to its maximal IRES
activity, and the 5′ 30 nt of the ATF2 5′ leader region are
essential for its IRES activity.
Induction of ATF2 IRES activity under
cellular stress conditions
It has been reported that, for certain mRNAs,
IRES-mediated initiation of protein translation is activated during
conditions such as cellular stress and apoptosis (28). In the present study, ATF2 protein
expression in Bel7402 cells was induced under treatment with ADM
and PTX (Fig. 1). To investigate
whether the ATF2 IRES activity is also affected by cellular stress,
the ATF2 activity was examined in Bel7402 cells treated with two
chemotherapeutic drugs (ADM and PTX), which induce a stress
condition in cells. Gene transfections and reporter assays were
performed to determine if ADM and PTX induced ATF2 IRES activity.
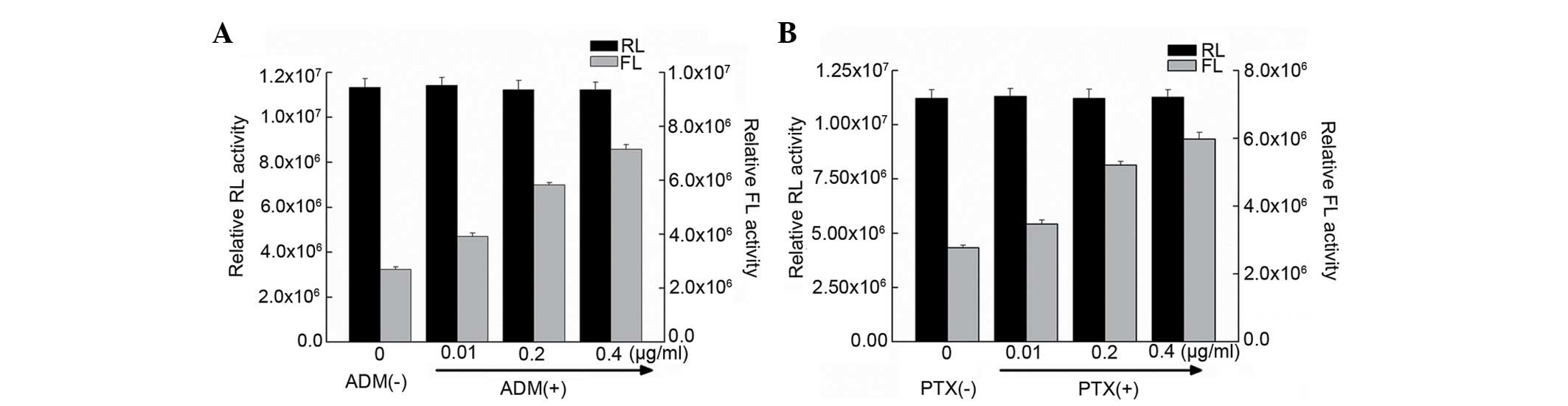
Bel7402 cells were transfected with the pRF plasmid, containing the
full-length fragment of the ATF2 5′-UTR sequence, and then treated
with ADM and PTX. It was observed that the FLUC activity in
transfected cells treated with ADM and PTX was higher than that of
untreated cells (Fig. 4).
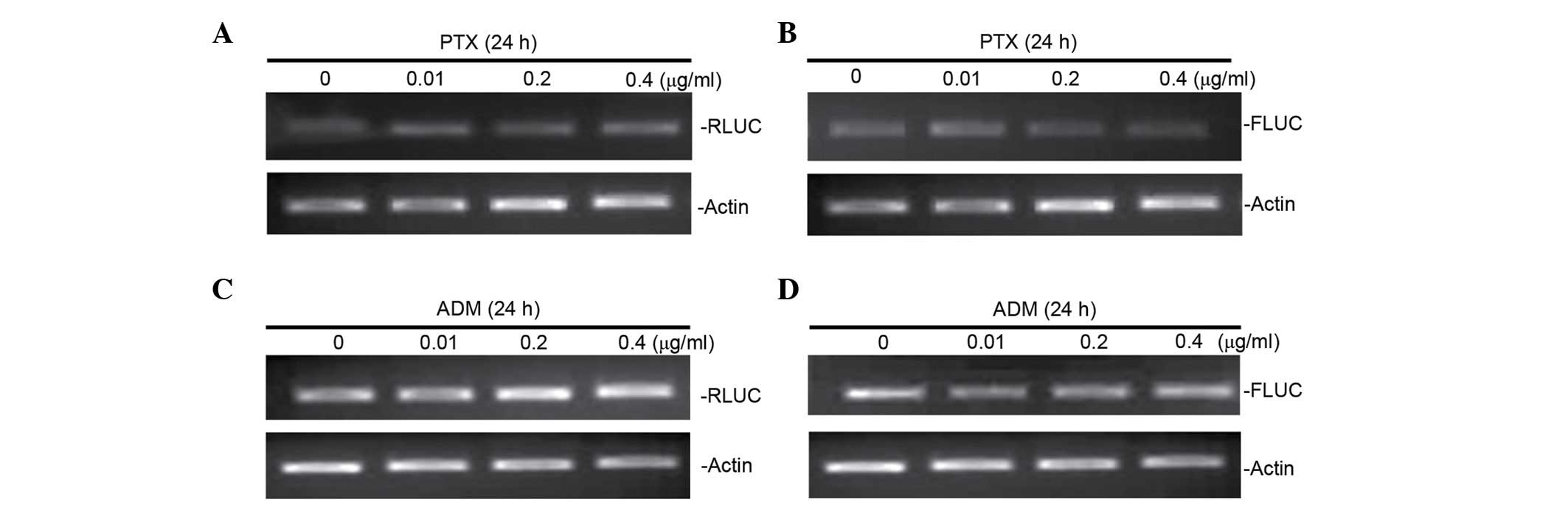
Furthermore, RT-PCR analyses of mRNA expression of FLUC were
performed in cells transfected with pRL-ATF2-FL following treatment
with ADM and PTX. It was observed that FLUC mRNA expression was not
altered by drugs-induced cellular stress (Fig. 5), suggesting that ADM and PTX induced
ATF2 IRES activity.
Discussion
The transcription factor ATF2 is a nuclear target of
stress-activated protein kinases such as p38, which are activated
by various extracellular stresses (29,30).
Previous studies demonstrated that the stimulation of ATF2 mainly
occurs by post-translational modification of protein subunits and
changes in binding efficiency (31,32).
However, the regulation at the translational level of ATF2 is
poorly understood. To the best of our knowledge, the present study
provided the first evidence of IRES-mediated translation of the
ATF2 5′-UTR by using bicistronic Renilla firefly luciferase
assays. The present study has revealed that the ATF2 IRES has
various IRES activities in different cell types. The differences in
ATF2 IRES activity between the different cells lines examined in
the current study may be due to the levels of expression of
IRES-transactivating factors (ITAFs) required for its function,
which are probably different in these cells (33).
By performing deletion mapping, the core IRES region
of the ATF2 mRNA was identified to be located between nt −299 and
−269 of the ATF2 5′-UTR. In certain cases, a specific and stable
RNA structure is necessary for IRES activity (34). For example, a module with a 9-nt motif
in the Gtx mRNA, a novel murine homeobox gene which in adult
animals is specifically expressed within glial cells of the central
nervous system, including the forebrain, and in germ cells of the
testis (35), recruits the 40S
ribosomal subunit by base pairing to the 18S ribosomal RNA (rRNA)
and initiates the internal translation (36), and in c-Myc mRNA, there are two 14-nt
segments that can recruit the 43S preinitiation complex and
activate the internal translation initiation (37). In line with these observations, our
data lead to the postulation that the 30-nt short fragment of the
ATF2 IRES is crucial for recruiting a ribosome and enhancing the
initiation of translation, and this core IRES region of ATF2 may
affect the translation efficiency through mRNA-rRNA interactions of
these complementary regions. To gain more insight into the
mechanism, further investigation is warranted to study the
differences in the ability of recruiting a ribosome by the
full-length ATF2 5′-UTR or the minimal ATF2 IRES region.
An important finding of the present study is that
IRES-regulated ATF2 translation is activated upon the treatment of
Bel7402 cells with two chemotherapeutic drugs, ADM and PTX. The
difference in the ability to stimulate the expression of ATF2
protein may due to the treatment capability of these two drugs;
perhaps the Bel7402 cells used in the present study have a greater
ability to evade death during ADM therapy compared with PTX
therpay. The expression and tissue distribution of ITAFs can change
during cellular stress, and thus regulate IRES activity (38,39). To
understand the mechanism of initiation of translation of the ATF2
IRES, it will be necessary to perform further experiments in order
to identify the ITAFs that regulate the IRES activity and to
clarify the functional association between each of these
proteins.
It has been reported that c-Jun/ATF2 heterodimers
can mediate a response to certain stress signals (31). Transcriptional activation domains and
the target genes of c-Jun/ATF2 heterodimers, which are implicated
in growth control, include c-Jun itself, cyclin D1 and cyclin A
(3). Notably, an IRES has been
demonstrated to be involved in the translation of the mRNA of
several ATF2 targets, including c-Jun and cyclin D1 (40,41). This
implies that IRES are very important in reposes to stress, and it
would be relevant to investigate the ITAFs required for these
IRES.
In the present study, it has been demonstrated that
ATF2 5′-UTR harbors an IRES element localized between nt −299 and
nt −269 upstream of the start codon, which would provide insights
into the structure-function association of ATF2 in IRES-mediated
translation. In addition, both ATF2 protein expression and IRES
activity were increased in Bel7402 cells following treatment with
the cellular stress-inducing chemotherapeutic drugs PTX and ADM,
indicating the significant role of ATF2 IRES response to cellular
stress in human hepatoma cells.
Acknowledgements
The present study was supported by the National
Natural Science Foundation of China (Beijing, China; grant no.
81101667) and the Natural Science Foundation of Jiangsu Province
(Nanjing, China; grant no. BK2009071).
References
|
1
|
Maekawa T, Sakura H, Kanei-Ishii C, Sudo
T, Yoshimura T, Fujisawa J, Yoshida M and Ishii S: Leucine zipper
structure of the protein CRE-BP1 binding to the cyclic AMP response
element in brain. Embo J. 8:2023–2028. 1989.PubMed/NCBI
|
|
2
|
van Dam H and Castellazzi M: Distinct
roles of Jun: Fos and Jun: ATF dimers in oncogenesis. Oncogene.
20:2453–2464. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Maekawa T, Shinagawa T, Sano Y, Sakuma T,
Nomura S, Nagasaki K, Miki Y, Saito-Ohara F, Inazawa J, Kohno T, et
al: Reduced levels of ATF-2 predispose mice to mammary tumors. Mol
Cell Biol. 27:1730–1744. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Bhoumik A and Ronai Z: ATF2: A
transcription factor that elicits oncogenic or tumor suppressor
activities. Cell Cycle. 7:2341–2345. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Yu T, Li YJ, Bian AH, Zuo HB, Zhu TW, Ji
SX, Kong F, Yin de Q, Wang CB, Wang ZF, et al: The regulatory role
of activating transcription factor 2 in inflammation. Mediators
Inflamm. 2014:9504722014. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Morrison DK and Davis RJ: Regulation of
MAP kinase signaling modules by scaffold proteins in mammals. Annu
Rev Cell Dev Biol. 19:91–118. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
van Dam H, Wilhelm D, Herr I, Steffen A,
Herrlich P and Angel P: ATF-2 is preferentially activated by
stress-activated protein kinases to mediate c-jun induction in
response to genotoxic agents. Embo J. 14:1798–1811. 1995.PubMed/NCBI
|
|
8
|
Papassava P, Gorgoulis VG, Papaevangeliou
D, Vlahopoulos S, van Dam H and Zoumpourlis V: Overexpression of
activating transcription factor-2 is required for tumor growth and
progression in mouse skin tumors. Cancer Res. 64:8573–8584. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Bhoumik A, Huang TG, Ivanov V, Gangi L,
Qiao RF, Woo SL, Chen SH and Ronai Z: An ATF2-derived peptide
sensitizes melanomas to apoptosis and inhibits their growth and
metastasis. J Clin Invest. 110:643–650. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Liu S, Wang F, Yan L, Zhang L, Song Y, Xi
S, Jia J and Sun G: Oxidative stress and MAPK involved into ATF2
expression in immortalized human urothelial cells treated by
arsenic. Arch Toxicol. 87:981–989. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Sheikh MS and Fornace AJ Jr: Regulation of
translation initiation following stress. Oncogene. 18:6121–6128.
1999. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Lackner DH and Bähler J: Translational
control of gene expression from transcripts to transcriptomes. Int
Rev Cell Mol Biol. 271:199–251. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Shatsky IN, Dmitriev SE, Terenin IM and
Andreev DE: Cap- and IRES-independent scanning mechanism of
translation initiation as an alternative to the concept of cellular
IRESs. Mol Cells. 30:285–293. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Stoneley M and Willis AE: Cellular
internal ribosome entry segments: Structures, trans-acting factors
and regulation of gene expression. Oncogene. 23:3200–3207. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Hellen CU and Sarnow P: Internal ribosome
entry sites in eukaryotic mRNA molecules. Genes Dev. 15:1593–1612.
2001. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Komar AA and Hatzoglou M: Internal
ribosome entry sites in cellular mRNAs: Mystery of their existence.
J Biol Chem. 280:23425–23428. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Oumard A, Hennecke M, Hauser H and
Nourbakhsh M: Translation of NRF mRNA is mediated by highly
efficient internal ribosome entry. Mol Cell Biol. 20:2755–2759.
2000. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Riley A, Jordan LE and Holcik M: Distinct
5′ UTRs regulate XIAP expression under normal growth conditions and
during cellular stress. Nucleic Acids Res. 38:4665–4674. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Shiroki K, Ohsawa C, Sugi N, Wakiyama M,
Miura K, Watanabe M, Suzuki Y and Sugano S: Internal ribosome entry
site-mediated translation of Smad5 in vivo: Requirement for a
nuclear event. Nucleic Acids Res. 30:2851–2861. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Baird SD, Turcotte M, Korneluk RG and
Holcik M: Searching for IRES. RNA. 12:1755–1785. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Nevins TA, Harder ZM, Korneluk RG and
Holcik M: Distinct regulation of internal ribosome entry
site-mediated translation following cellular stress is mediated by
apoptotic fragments of eIF4G translation initiation factor family
members eIF4GI and p97/DAP5/NAT1. J Biol Chem. 278:3572–3579. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Yang L, Gu L, Li Z and Zhou M: Translation
of TRAF1 is regulated by IRES-dependent mechanism and stimulated by
vincristine. Nucleic Acids Res. 38:4503–4513. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Chan CP, Kok KH, Tang HM, Wong CM and Jin
DY: Internal ribosome entry site-mediated translational regulation
of ATF4 splice variant in mammalian unfolded protein response.
Biochim Biophys Acta. 1833:2165–2175. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Fabrizio C, Stefano M, Laura G, Marco N,
Lorenzo M and Antonio O: Re: Baseline and early MR apparent
diffusion coefficient quantification as a predictor of response of
unresectable hepatocellular carcinoma to doxorubicin drug-eluting
bead chemoembolization. J Vasc Interv Radiol. 27:1456–1458. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Chen L, Liu Y, Wang W and Liu K: Effect of
integrin receptor-targeted liposomal paclitaxel for hepatocellular
carcinoma targeting and therapy. Oncol Lett. 10:77–84.
2015.PubMed/NCBI
|
|
26
|
Xi G, Hu X, Wu B, Jiang H, Young CY, Pang
Y and Yuan H: Autophagy inhibition promotes paclitaxel-induced
apoptosis in cancer cells. Cancer Lett. 307:141–148. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Pelletier J and Sonenberg N: Internal
initiation of translation of eukaryotic mRNA directed by a sequence
derived from poliovirus RNA. Nature. 334:320–325. 1988. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Komar AA and Hatzoglou M: Cellular
IRES-mediated translation: The war of ITAFs in pathophysiological
states. Cell Cycle. 10:229–240. 2014. View Article : Google Scholar
|
|
29
|
Gozdecka M, Lyons S, Kondo S, Taylor J, Li
Y, Walczynski J, Thiel G, Breitwieser W and Jones N: JNK suppresses
tumor formation via a gene-expression program mediated by ATF2.
Cell Rep. 9:1361–1374. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
De Graeve F, Bahr A, Sabapathy KT, Hauss
C, Wagner EF, Kedinger C and Chatton B: Role of the ATFa/JNK2
complex in Jun activation. Oncogene. 18:3491–3500. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Livingstone C, Patel G and Jones N: ATF-2
contains a phosphorylation-dependent transcriptional activation
domain. EMBO J. 14:1785–1797. 1995.PubMed/NCBI
|
|
32
|
Zeke A, Bastys T, Alexa A, Garai Á,
Mészáros B, Kirsch K, Dosztányi Z, Kalinina OV and Reményi A:
Systematic discovery of linear binding motifs targeting an ancient
protein interaction surface on MAP kinases. Mol Syst Biol.
11:8372015. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
King HA, Cobbold LC and Willis AE: The
role of IRES trans-acting factors in regulating translation
initiation. Biochem Soc Trans. 38:1581–1586. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Filbin ME and Kieft JS: Toward a
structural understanding of IRES RNA function. Curr Opin Struct
Biol. 19:267–276. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Komuro I, Schalling M, Jahn L, Bodmer R,
Jenkins NA, Copeland NG and Izumo S: Gtx: A novel murine
homeobox-containing gene, expressed specifically in glial cells of
the brain and germ cells of testis, has a transcriptional repressor
activity in vitro for a serum-inducible promoter. EMBO J.
12:1387–1401. 1993.PubMed/NCBI
|
|
36
|
Hu MC, Tranque P, Edelman GM and Mauro VP:
rRNA-complementarity in the 5′ untranslated region of mRNA
specifying the Gtx homeodomain protein: Evidence that base-pairing
to 18S rRNA affects translational efficiency. Proc Natl Acad Sci
USA. 96:1339–1344. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Cencig S, Nanbru C, Le SY, Gueydan C, Huez
G and Kruys V: Mapping and characterization of the minimal internal
ribosome entry segment in the human c-myc mRNA 5′ untranslated
region. Oncogene. 23:267–277. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Schepens B, Tinton SA, Bruynooghe Y,
Beyaert R and Cornelis S: The polypyrimidine tract-binding protein
stimulates HIF-1alpha IRES-mediated translation during hypoxia.
Nucleic Acids Res. 33:6884–6894. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Petz M, Them N, Huber H, Beug H and
Mikulits W: La enhances IRES-mediated translation of laminin B1
during malignant epithelial to mesenchymal transition. Nucleic
Acids Res. 40:290–302. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Blau L, Knirsh R, Ben-Dror I, Oren S,
Kuphal S, Hau P, Proescholdt M, Bosserhoff AK and Vardimon L:
Aberrant expression of c-Jun in glioblastoma by internal ribosome
entry site (IRES)-mediated translational activation. Proc Natl Acad
Sci USA. 109:E2875–E2884. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Shi Y, Sharma A, Wu H, Lichtenstein A and
Gera J: Cyclin D1 and c-myc internal ribosome entry site
(IRES)-dependent translation is regulated by AKT activity and
enhanced by rapamycin through a p38 MAPK- and ERK-dependent
pathway. J Biol Chem. 280:10964–10973. 2005. View Article : Google Scholar : PubMed/NCBI
|