Introduction
Hepatocellular carcinoma (HCC) is the third leading
cause of cancer mortality in the world, and the five-year survival
rate is lower than 5% (1). It has an
unfavorable prognosis due to its spread, metastases and high rate
of recurrence (2). Currently,
treatment with platinum-based chemotherapy is one the main means of
drug therapy for HCC. Cisplatin, the first generation of platinum
drugs, is one of the most active anticancer chemotherapeutic drugs
(3). As a cell cycle non-specific
drug, it can inhibit effectively the proliferation of HCC by the
cytotoxic effect (4,5). The main targets of cisplatin are DNA,
RNA and proteins with strong affinity to the nucleus (6). Cisplatin impairs DNA replication by
producing DNA intra-strand cross-links and cisplatin-DNA complexes,
or by binding to nuclear and cytoplasmic proteins (7,8). Thus, it
is primarily considered as a DNA-damaging anticancer drug (5,6). However,
the precise regulatory mechanisms by which cisplatin induces
apoptosis and inhibits proliferation are not completely clear.
Recently, accumulating evidences suggested that long
noncoding RNAs (lncRNAs) have many biological functions, such as
regulation of transcription, modulation of nuclear structure and
function, carcinogenesis and cancer progression (9–11). lncRNAs
are RNA molecules >200-bp long without protein coding functions
(7). lncRNAs, as epigenetic
regulators, are also associated with chemotherapy sensitivity in
cancers (8,12). The lnRNA H19 was found to induce
multidrug resistance 1 (MDR1)-associated drug resistance in liver
cancer cells through regulation of MDR1 promoter methylation
(13). In addition, the lncRNA H19
was markedly upregulated in liver cancer, while metastasis
associated lung adenocarcinoma transcript 1 was upregulated in
human and murine HCC, and the HOX transcript antisense RNA (HOTAIR)
levels increased in human HCC and a liver cancer cell line
(14). Therefore, lncRNAs may serve
as biomarkers for treatment response in cancers.
Furthermore, it was suggested that lncRNAs may play
an important role in regulating gene expression (10). The functions of lncRNAs are mainly
carried out by their secondary structure; however, this is
difficult to decipher (15). Due to
the considerable challenge of exploring the lncRNAs functions, a
co-expression-based method has been developed, in which lncRNA
functions are predicted based on the functions of their
co-expressed protein-coding genes (16). Genes that have similar expression
patterns under multiple conditions tend to be involved in the same
pathways. The co-expressed protein-coding genes are potentially
regulated by the corresponding lncRNAs.
Taken together, some lncRNAs would be involved in
antitumor effects and/or resistance to cisplatin by regulating gene
expression. In this study, we investigated the differential
expression of lncRNAs in HepG2 cells at different times of
cisplatin exposure. Four differentially expressed lncRNAs were
identified and their co-expressing genes were obtained. The aim of
the present study is to identify lncRNAs that may be valuable
biomarkers of cisplatin-based functions and chemoresponse, and also
candidates for therapy targets in HCC.
Materials and methods
Cell culture and main reagents
HepG2 (a human HCC cell line) was purchased from the
Institute of Biochemistry and Cell Biology, Chinese Academy of
Sciences (Shanghai, China). Cells were maintained in RPMI 1640
medium (Invitrogen; Thermo Fisher Scientific, Inc., Waltham, MA,
USA) with 10% fetal calf serum (Invitrogen; Thermo Fisher
Scientific, Inc.), 100 U/ml penicillin (Sigma-Aldrich; Merck
Milipore, Darmstadt, Germany) and 100 µg/ml streptomycin
(Sigma-Aldrich; Merck Milipore) at 37°C, 5% CO2. The
Cell Cycle Detection kit was purchased from Beyotime Institute of
Biotechnology (Suzhou, China). Anti-p21 [also known as
cyclin-dependent kinase inhibitor 1A (CDKN1A)] (bs-10129R),
anti-tumor protein p53 inducible protein 3 (TP53I3) (bs-6144R) and
anti-wild-type p53-induced phosphatase 1 (Wip1, also known as
PPM1D) (bs-2447R) antibodies were purchased from Beijing
Biosynthesis Biotechnology Co., Ltd. (Beijing, China). The RNeasy
Mini kit was purchased from Qiagen GmbH (Hilden, Germany), while
First-strand cDNA Synthesis kit was purchased from Tiangen Biotech
Co., Ltd. (Beijing, China). The LightCycler DNA Master SYBR Green I
kit was purchased from Roche Diagnostics GmbH (Mannheim,
Germany).
Microarray data set
Microarray data from the microarray data set
GSE38122 (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE38122)
was collected from the Gene Expression Omnibus database. This study
investigated differential gene expression in the messenger RNA
(mRNA) content of the HepG2 cell line upon 12, 24 and 48 h of
exposure to 7 µM cisplatin (Sigma-Aldrich; Merck Milipore) and its
solvent. In a total of 18 arrays, three biological replicates were
performed per compound/solvent at three time points. The samples
were examined with GeneChip® Human Genome U133 Plus 2.0
(HG-U133 Plus 2.0) Array from Affymetrix, Inc. (Santa Clara, CA,
USA).
Reannotation and identification of
differentially expressed lncRNAs
The probes on the HG-U133 Plus 2.0 array were
reannotated for human lncRNAs using noncoding RNA function
annotation server (ncFANs), as showed in its website (15). Differentially expressed lncRNAs were
identified by the fold-change method (17). The lncRNAs with a fold-change value of
>2.0 or <0.5 were considered as differentially expressed
lncRNAs.
Obtaining co-expressing genes for each
differentially expressed lncRNA
To obtain co-expressing genes for each
differentially expressed lncRNA, we calculated the Pearson's
correlation coefficient (PCC) between each differentially expressed
lncRNA and all the genes across all 18 samples. The genes with a
strict cut-off (PCC >0.9 or <-0.9) were identified as
co-expressing genes. Then, pathway enrichment was implemented to
identify the affected pathways of lncRNA co-expressing genes using
Database for Annotation, Visualization and Integrated Discovery
(DAVID) 6.7 (18).
Confirmation of differentially
expressed lncRNAs by reverse transcription-quantitative polymerase
chain reaction (qPCR)
HepG2 cells were harvested at 3, 6, 12 and 24 h
after cisplatin exposure (7 µM). Total RNA was extracted and
purified from HepG2 cells by RNeasy Mini kit (Qiagen GmbH).
Complementary DNA was obtained using First-strand cDNA Synthesis
kit (Tiangen Biotech Co., Ltd.). qPCR was performed on an ABI
StepOnePlus Real-Time PCR System (Applied Biosystems; Thermo Fisher
Scientific, Inc.) using the LightCycler DNA Master SYBR Green I kit
(Roche Diagnostics GmbH). The primer pairs were synthesized by
Sangon Biotech Co., Ltd. (Shanghai, China). The primers used for
qPCR had the following sequences: ENSG00000224818 sense,
5′-CTCTGGAGGGAGCAAGGA-3′ and antisense, 5′-TGGACTCTGAGGGACTGG-3′;
ENSG00000256185 sense, 5′-GGCACTTTTCAGAACATC-3′ and antisense,
5′-TGTCGTGTATCACAGCAT-3′; ENSG00000260912 sense,
5′-CGACCACCTATTCCACTT-3′ and antisense, 5′-GCCAGGAAGGCTCAAATC-3′;
and ENSG00000267194 sense, 5′-AAAACCCACCTCCAGCAC-3′ and antisense,
5′-GCGGCAATCCGTAAAGAA-3′. The PCR conditions were as follows: 95°C
for 5 min, followed by 30 cycles of 94°C for 30 sec, 55°C for 30
sec and 72°C for 30 sec, and then 72°C for 5 min. Fluorescence
values were collected, using the GADPH RNA expression level as
internal control. The relative content of mRNA was calculated
according to the following formula: Fold-change =
2−Δ(ΔCq), where Cq is the quantification cycle, ΔCq =
Cq(target)-Cq(GAPDH) and Δ(ΔCq) =
ΔCq(treated)-ΔCq(untreated).
Cell cycle analysis
Cells were seeded into 6-well plates at
1×106 cells/ml for 24 h and divided into two groups:
Control group (untreated HepG2 cells) and cisplatin treatment HepG2
group (7 µM). Cells were harvested, washed with PBS twice,
centrifuged at room temperature (RT) for 10 min at 600 × g,
resuspended in PBS and fixed in ice-cold 70% ethanol overnight at
4°C. Cells were washed and resuspended with PBS, stained with
propidium iodide for 30 min at 37°C in the dark, and then analyzed
on a flow cytometer with the Cell Cycle Detection kit (Beyotime
Institute of Biotechnology). A BD FACSAria flow cytometer (BD
Biosciences, Franklin Lakes, NJ, USA) was used, and the data were
analyzed using the QuantiCALC software version 1.0 (BD
Biosciences).
Immunofluorescence and microscopic
analysis
HepG2 cells were cultured on circular cover slips
and treated with cisplatin (7 µM) for 12 and 24 h. Then, cells were
washed with Hank's solution, fixed with 4% paraformaldehyde
solution for 30 min at RT, and permeabilized with 0.1% Triton X-100
(dissolved in PBS) for 10 min. Next, cells were washed with PBS
three times and blocked in 0.1% Triton X-100 [dissolved in PBS with
5% goat serum (Invitrogen; Thermo Fisher Scientific, Inc.)] for 1 h
at RT. After blocking, cells were incubated with 50 µl anti-TP53I3
antibody (1:100), anti-Wip1 antibody (1:200) or anti-p21 antibody
(1:100) at 4°C overnight. Then, cells were washed three times with
PBS, and incubated with Alexa 555-conjugated secondary antibodies
(1:200; A21428; Invitrogen; Thermo Fisher Scientific, Inc.) or with
Alexa 488-conjugated secondary antibodies (1:200; A11008;
Invitrogen; Thermo Fisher Scientific, Inc.) for 1 h at RT in the
dark. Immunofluorescent images were examined and analyzed using the
Olympus FV1000 confocal system (Olympus Corporation, Tokyo, Japan).
Protein expression levels were determined using the mean
fluorescence intensity values of the samples with the FV10 ASW 1.7
software (Olympus Corporation).
Statistical analysis
The values are expressed as the mean ± standard
deviation. Statistical analyses were performed using analysis of
variance and Student's t-test. P≤0.05 was considered to
indicate a statistically significant difference. All statistical
analyses were performed using the SPSS 17.0 software package (SPSS,
Inc., Chicago, IL, USA).
Results
Differentially expressed lncRNA
following exposure to cisplatin
Using ncFANs, several differentially expressed
lncRNAs were identified. The increasing numbers of differentially
expressed lncRNAs were showed in HepG2 cells as the exposure time
for cisplatin increased. There were 6, 26 and 86 differentially
expressed lncRNAs identified after treatment for 12, 24 and 48 h,
respectively. In all these three time points, four lncRNAs were
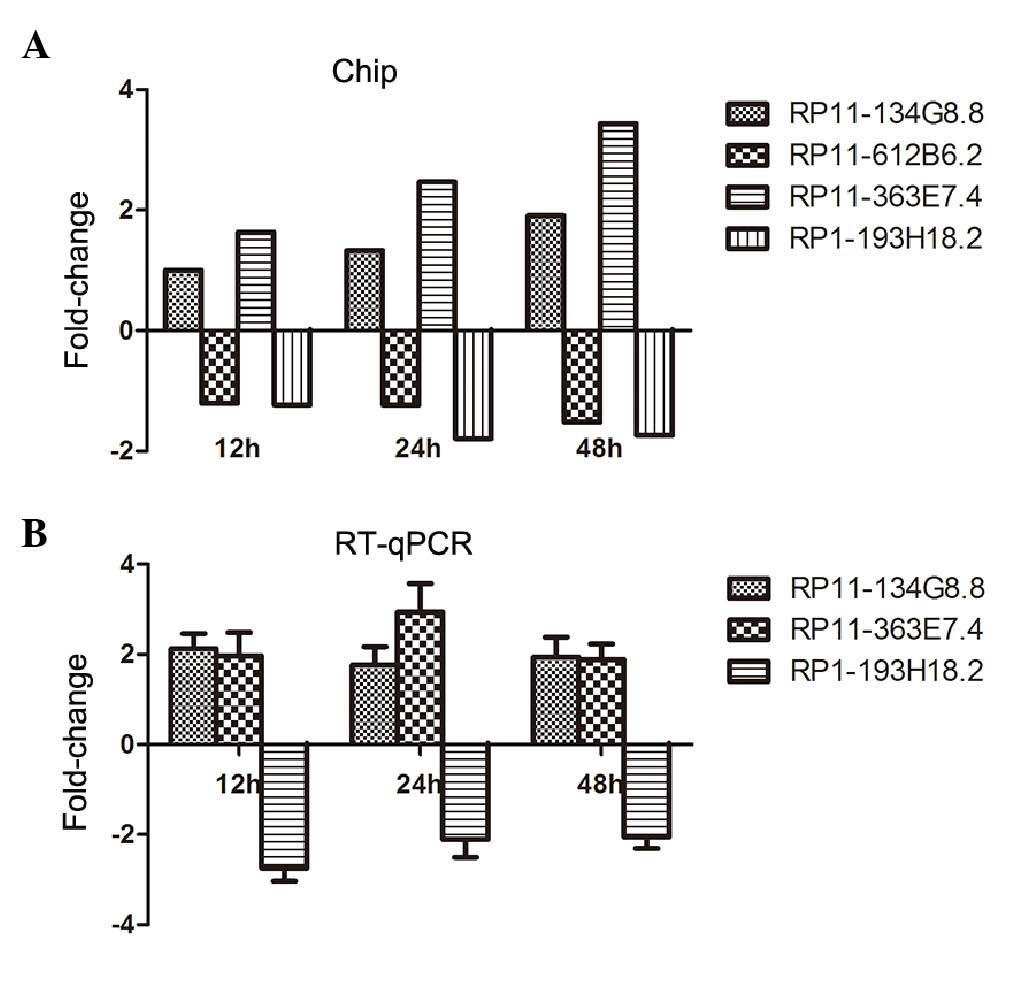
differentially expressed. They were RP11-134G8.8, RP11-612B6.2,
RP11-363E7.4 and RP1-193H18.2 (Fig.
1A). RP11-134G8.8 and RP11-363E7.4 were upregulated after
treatment with cisplatin, whereas RP11-612B6.2 and RP1-193H18.2
were downregulated.
lncRNA-gene co-expression network and
functional enrichment
After calculating the PCC between each continuous
differentially expressed lncRNAs and all genes, significant
co-expressed genes were obtained for these differentially expressed
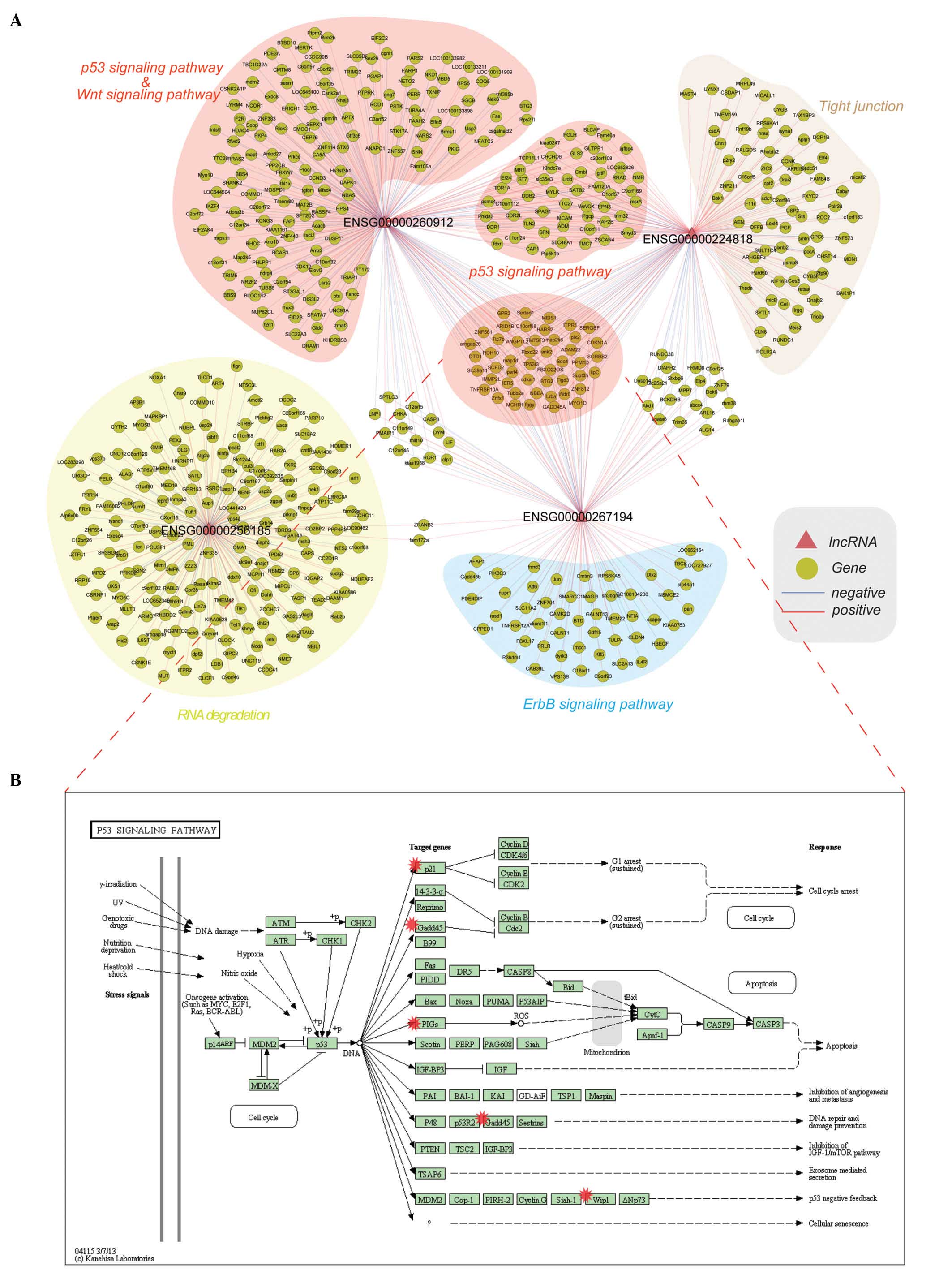
lncRNAs. Then, the lncRNA-gene co-expression network was
constructed, in which nodes were lncRNAs and genes. lncRNAs and
genes were connected if they were significantly co-expressed
(Fig. 2A). Some genes were found to
be co-expressed with only one lncRNA, whereas other genes were
co-expressed with more than one lncRNA. Functional annotation of
the corresponding co-expressed genes was implemented to explore the
potential function of these lncRNAs by using the DAVID 6.7 tool.
The results demonstrated that they were annotated in some
well-documented cancer-related pathways (for example, the p53
signaling pathway and the mitogen-activated protein kinase
signaling pathway) (Table I).
Particularly, some genes were co-expressed with three lncRNAs (red
background in the center of Fig. 2A)
and they were annotated into the p53 signaling pathway (Fig. 2B).
 | Table I.Annotated pathways of co-expressing
genes of the corresponding differentially expressed lncRNAs. |
Table I.
Annotated pathways of co-expressing
genes of the corresponding differentially expressed lncRNAs.
| lncRNA | Pathway | P-value | Genes |
|---|
| RP1-193H18.2 | hsa04115:p53
signaling pathway | <0.001 | CDKN1A, PPM1D,
CASP8, PMAIP1, GADD45B, GADD45A |
|
| hsa04912:GnRH
signaling pathway |
0.005 | JUN, CAMK2D, HBEGF,
ITPR1, MAP2K6 |
|
| hsa04012:ErbB
signaling pathway |
0.020 | CDKN1A, JUN,
CAMK2D, HBEGF |
|
| hsa04010:MAPK
signaling pathway |
0.040 | RPS6KA5, DUSP14,
JUN, GADD45B, GADD45A, MAP2K6 |
| RP11-134G8.8 | hsa04115:p53
signaling pathway | <0.001 | CDKN1A, PPM1D,
TP53I3, EI24, LRDD, DDB2, SFN, GADD45A |
|
| hsa05219:bladder
cancer |
0.080 | HRAS, CDKN1A,
PGF |
|
|
hsa00561:glycerolipid metabolism |
0.090 | CEL, AKR1B1,
LIPC |
| RP11-363E7.4 | hsa04115:p53
signaling pathway | <0.001 | LRDD, ZMAT3, RRM2B,
SFN, PMAIP1, SESN1, RFWD2, EI24, TP53I3, PPM1D, CDKN1A, CCND3,
CASP8, DDB2, MDM2, FAS, PERP, GADD45A |
|
|
hsa00970:aminoacyl-tRNA biosynthesis |
0.003 | PSTK, FARS2, NARS2,
HARS2, LARS2, DTD1 |
|
| hsa04110:cell
cycle |
0.038 | ANAPC1, CDKN1A,
CCND3, ANAPC1P1, MDM2, SFN, GADD45A, TUBB2A, TUBB6, |
|
| hsa04540:Gap
junction |
0.040 | TUBA4A, ITPR1,
MAP2K5 |
|
| hsa04120:ubiquitin
mediated proteolysis |
0.050 | RFWD2, ANAPC1,
FBXW7, ANAPC1P, TRIM32, DDB2, MDM2, NKD1, CSNK2A1, |
|
| hsa04310:Wnt
signaling pathway |
0.070 | CCND3, PPP2CB,
CSNK2A1P, NFATC2, TBL1X |
| RP11-612B6.2 | hsa03018:RNA
degradation |
0.090 | EXOSC4, CNOT2,
ZCCHC7 |
lncRNA validation
In order to confirm the four predicted lncRNAs, qPCR
was performed at three time points after cisplatin exposure (12, 24
and 48 h). GAPDH served as an internal control. The results in
Fig. 1B show the standardized
fold-changes in lncRNA expression in HepG2 cells incubated with
cisplatin for different periods of time. After 12, 24 and 48 h of
incubation, cisplatin significantly decreased the RP1-193H18.2
level by 2.75, 2.11 and 2.06-fold, respectively. RP11-134G8.8
expression increased by 1.77–2.13-fold, and the RP11-363E7.4 level
was also upregulated by 1.89–2.94-fold. These trends in lncRNA
expression are consistent with the microarray results. However,
RP11-612B6.2 lncRNA was not amplified (data not shown).
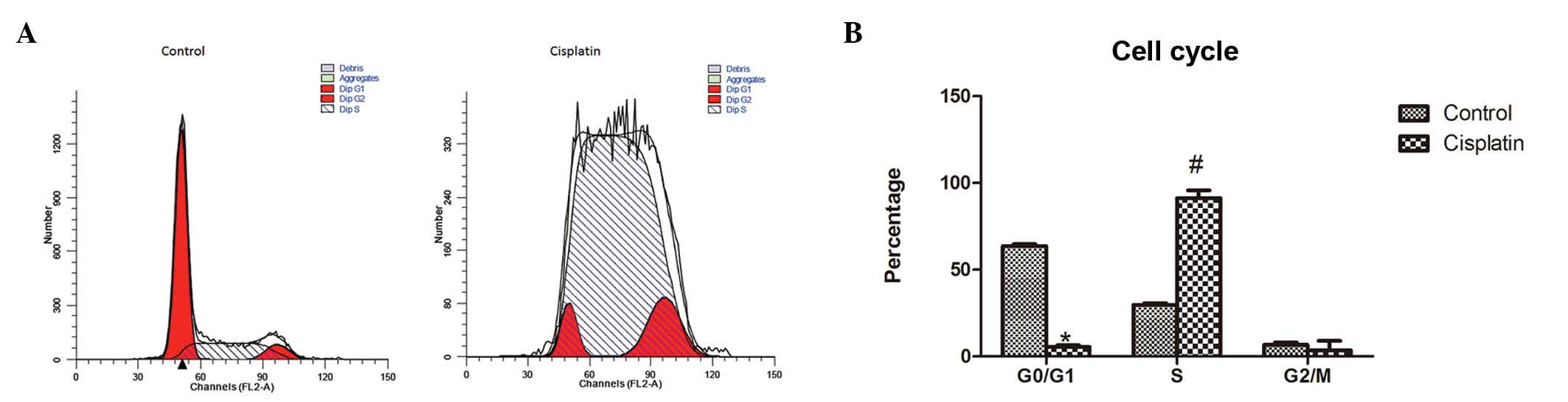
Cell cycle test
To evaluate HepG2 cell proliferation, cell cycle was
analyzed by flow cytometry. Flow cytometric analysis showed that
the percentage of cells in the G0/G1 phase was significantly lower
in HepG2 cells with cisplatin treatment for 24 h than in HepG2
cells under normal culture conditions (5.46±0.99 vs. 63.62±1.06,
P<0.01), whereas the percentage of cells in the S phase was
significantly higher in the cisplatin treatment group than in
untreated HepG2 cells (91.15±4.59 vs. 29.73±0.95, P<0.01). No
significant difference was noted in the percentage of cells in the
G2/M phase in the two groups of cells (3.39±5.48 vs. 6.65±1.42,
P>0.05). This result showed that cisplatin induced the arrest in
the S phase of the cell cycle (Fig.
3).
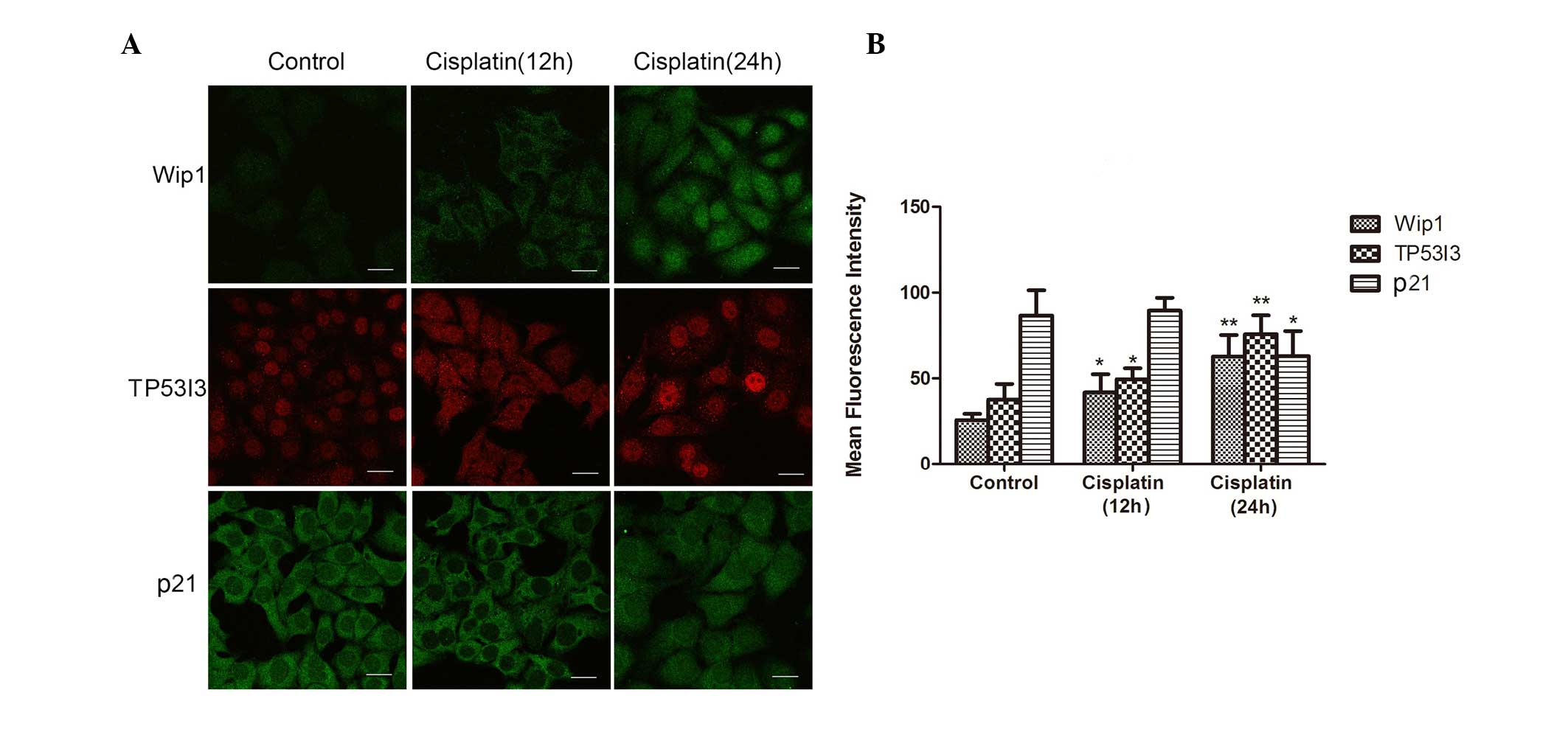
Immunofluorescence staining
To evaluate the co-expression genes of these lncRNAs
in HepG2 cells after cisplatin treatment, three genes (TP53I3,
CDKN1A and PPM1D) were chosen. Immunofluorescence was carried out
for their proteins: TP53I3, p21 (CDKN1A) and Wip1 (PPM1D). The
results showed that the expression level of TP53I3 and Wip1 was
upregulated in the cisplatin group compared with that of the
control group. Inversely, compared with the control group, the
expression level of p21 was downregulated in the cisplatin group
(Fig. 4).
Discussion
It has been reported that lncRNAs possess important
roles in gene regulation, though lncRNAs once were considered
incapable of encoding proteins (19).
Recently, the biological functions of lncRNAs have been received
increased attention, and abnormal expression of some lncRNAs was
found in HCC (1). However, only a few
lncRNAs have been elucidated as targets of cancer diagnosis and
therapy (19). Bioinformatics
analyses or microarray have been helpful in lncRNAs research. In
this study, four continuous differentially expressed lncRNAs
(RP11-134G8.8, RP11-612B6.2, RP11-363E7.4 and RP1-193H18.2) were
identified in cisplatin-treated HepG2 cells by analyzing microarray
data. Furthermore, RP11-134G8.8, RP11-363E7.4 and RP1-193H18.2 were
verified by qPCR. RP11-134G8.8 and RP11-363E7.4 were upregulated,
while RP1-193H18.2 was downregulated. To explore the functions of
the differentially expressed lncRNAs, we obtained and identified 57
significant co-expressing genes and pathways where the genes were
involved in, such as the p53, ErbB, Wnt and gonadotropin-releasing
hormone signaling pathways (Table I).
Obviously, many co-expressing genes of the three lncRNAs
(RP11-134G8.8, RP11-363E7.4 and RP1-193H18.2) were involved in the
p53 signaling pathway, which showed the most significant difference
among all pathways. Thus, after analyzing all data by
bioinformatics, cell cycle was examined, and three key genes
(CDKN1A, TP53I3 and PPM1D) in the p53 signaling pathway (20–23) were
verified, which may be useful for exploring the deeply regulated
mechanism of lncRNAs in HepG2 cells with cisplatin treatment.
CDKN1A/p21 is a cyclin-dependent kinase inhibitor
involved in carcinogenesis by regulating the cell cycle progression
at different phases (24,25). As a famous downstream target of p53,
changed expression of CDKN1A was found in various cancers and
therapeutic processes (26–29). Fluoroquinolones were reported to have
the ability to penetrate pancreatic tissue, and are usually
associated with loss or downregulation of the CDK inhibitors
p21/p27 as well as with the mutational inactivation of p53
(26). Significantly upregulated p21
at both the gene and protein levels was found in MCF-7 cells
treated with Dillenia suffruticosa root dichloromethane
extract, and the results suggested that the induction of
G0/G1-phase cell cycle arrest in MCF-7 cells was achieved via the
p53/p21 pathway (21). In human lung
adenocarcinoma cells, it was found that the upregulation of the
lncRNA HOTAIR contributes to cisplatin resistance, at least in
part, through the regulation of p21 expression (30). It seems that protein/gene expression
and cell cycle arrest are different in different cancers and
conditions (17,22–26). In
this study, CDKN1A was negatively regulated by RP1-193H18.2 and
positively regulated by RP11-134G8.8 and RP11-363E7.4 at the gene
level. The expression of p21 was downregulated after cisplatin
treatment for 24 h at the protein level, contrarily to the
upregulated p21 expression reported by Qu et al (27). However, it is easy to understand that
the loss of p21 expression could be the result of apoptosis, which
is induced by cisplatin. Additionally, there is a big interaction
network between p21 and other regulatory factors; thus, further
research on p21 is required.
TP53I3 is also induced by the tumor suppressor p53
and is thought to be involved in p53-mediated cell death (23,31).
p53-inducible gene 3 (PIG3) contributes to early cellular response
to DNA damage and is a precursor of the apoptosis pathway that
determines the fate of a cell in response to cellular stress
(32). Our results showed that TP53I3
was negatively regulated by RP1-193H18.2 but positively regulated
by RP11-134G8.8 and RP11-363E7.4 at the gene level, and PIG3 showed
upregulated expression at the protein level. This could be the cell
response to cisplatin at early times, trying to regulate the cell
homeostasis (33). Indeed, the
induction of p53 could also explain these results, since the p53
signaling pathway is involved in biological changes of HepG2 cells
under cisplatin treatment.
As a gene in the p53 signaling pathway, PPM1D
performs many physiological functions, including cell signaling,
apoptosis and cell cycle progression (22,34). The
protein PPM1D is a member of the protein phosphatase 2C (PP2C)
family of Ser/Thr protein phosphatases (32,34). PPM1D
is a stress-responsive PP2C phosphatase that plays a key role in
stress signaling (35). In addition,
it was suggested that PPM1D is associated with carcinogenesis
(36,37). It negatively regulates the DNA damage
response through the dephosphorylation and inactivation of p53,
ataxia telangiectasia mutated, p38 and checkpoint kinase 1/2
(38). In recent years, PPM1D was
considered as a prognostic marker and potential therapeutic target
in several cancers (39,40). In this study, RP1-193H18.2 played a
negative regulatory role for PPM1D at the gene level, while
RP11-134G8.8 and RP11-363E7.4 were positive factors. As a result,
upregulated protein levels of PPM1D were observed. This could be a
response of the cell to the stress induced by cisplatin.
Furthermore, it was found that PPM1D played an important role in
promoting cisplatin resistance, and as a novel downstream target of
Akt, PPM1D mediates its action of conferring cisplatin resistance
to gynecological cancer cells (41).
PPM1D could also be induced by p53 to maintain the homeostasis in
cells (42). In addition, Cao et
al reported that PPM1D plays a role in the cell cycle via p21
in dogs (43). Our research has
identified opposite expression tends for p21 and PPM1D within short
time of cisplatin treatment. However, more experiments must be
designed for confirming if there is an association between them in
humans.
Obvious cell cycle arrest at the S phase was induced
by cisplatin in this study. These results are different from those
of a previous report (27), despite
the fact that a similar dose of cisplatin was used. The only
difference is the exposure time to cisplatin. We hypothesize that
cisplatin may induce S phase arrest at only early times when cells
are in the state of stress. This could be one of direct reactions
of cells to the DNA duplicate damage induced by cisplatin. The
change of cell cycle should be explicated for all relative
factors.
In summary, the lncRNAs RP11-134G8.8, RP11-363E7.4
and RP1-193H18.2 were differentially expressed in HepG2 cells after
cisplatin treatment. These lncRNAs may play an important role by
regulating the expression of genes that are co-expressed with them.
In addition, cell cycle arrest could be induced at the S phase when
cells were treated with cisplatin for a short time. As a classic
pathway, the p53 signaling pathway contributes to the effect of
cisplatin and its induced resistance. Therefore, the lncRNAs
RP11-134G8.8, RP11-363E7.4 and RP1-193H18.2 and their co-expressed
genes, which annotated into the p53 signaling pathway, could be
potential biomarkers for cisplatin treatment. This study will help
to understand the lncRNAs functions in HepG2 cells under cisplatin
treatment.
Acknowledgements
The present study was supported by the National
Science Foundation of China (Beijing, China; grant no. 30873131)
and and by the Jilin Province Development and Reform Commission
(Jilin, China; grant no. 2016C023).
Glossary
Abbreviations
Abbreviations:
|
HCC
|
hepatocellular carcinoma
|
|
lncRNA
|
long noncoding RNA
|
References
|
1
|
Hou JK, Huang Y, He W, Yan ZW, Fan L, Liu
MH, Xiao WL, Sun HD and Chen GQ: Adenanthin targets peroxiredoxin
I/II to kill hepatocellular carcinoma cells. Cell Death Dis.
5:e14002014. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Yang X, Xie X, Xiao YF, Xie R, Hu CJ, Tang
B, Li BS and Yang SM: The emergence of long non-coding RNAs in the
tumorigenesis of hepatocellular carcinoma. Cancer Lett.
360:119–124. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Marullo R, Werner E, Degtyareva N, Moore
B, Altavilla G, Ramalingam SS and Doetsch PW: Cisplatin induces a
mitochondrial-ROS response that contributes to cytotoxicity
depending on mitochondrial redox status and bioenergetic functions.
PLoS One. 8:e811622013. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Wang Y, Liu Y, Liu Y, Zhou W, Wang H, Wan
G, Sun D, Zhang N and Wang Y: A polymeric prodrug of cisplatin
based on pullulan for the targeted therapy against hepatocellular
carcinoma. Int J Pharm. 483:89–100. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Ziko L, Riad S, Amer M, Zdero R,
Bougherara H and Amleh A: Mechanical stress promotes
cisplatin-induced hepatocellular carcinoma cell death. Biomed Res
Int. 2015:4305692015. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Bose RN: Biomolecular targets for platinum
antitumor drugs. Mini Rev Med Chem. 2:103–111. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Chu G: Cellular responses to cisplatin.
The roles of DNA-binding proteins and DNA repair. J Biol Chem.
269:787–790. 1994.PubMed/NCBI
|
|
8
|
Galluzzi L, Senovilla L, Vitale I, Michels
J, Martins I, Kepp O, Castedo M and Kroemer G: Molecular mechanisms
of cisplatin resistance. Oncogene. 31:1869–1883. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Bonasio R and Shiekhattar R: Regulation of
transcription by long noncoding RNAs. Annu Rev Genet. 48:433–455.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Kung JT, Colognori D and Lee JT: Long
noncoding RNAs: Past, present and future. Genetics. 193:651–669.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Shi D, Zheng H, Zhuo C, Peng J, Li D, Xu
Y, Li X, Cai G and Cai S: Low expression of novel lncRNA
RP11-462C24.1 suggests a biomarker of poor prognosis in colorectal
cancer. Med Oncol. 31:312014. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Sun M and Kraus WL: From discovery to
function: The expanding roles of long noncoding RNAs in physiology
and disease. Endocr Rev. 36:25–64. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Fan Y, Shen B, Tan M, Mu X, Qin Y, Zhang F
and Liu Y: Long non-coding RNA UCA1 increases chemoresistance of
bladder cancer cells by regulating Wnt signaling. FEBS J.
281:1750–1758. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
George J and Patel T: Noncoding RNA as
therapeutic targets for hepatocellular carcinoma. Semin Liver Dis.
35:63–74. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Liao Q, Xiao H, Bu D, Xie C, Miao R, Luo
H, Zhao G, Yu K, Zhao H, Skogerbø G, et al: ncFANs: A web server
for functional annotation of long non-coding RNAs. Nucleic Acids
Res. 39:W118–W124. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Hou Z, Xu C, Xie H, Xu H, Zhan P, Yu L and
Fang X: Long noncoding RNAs expression patterns associated with
chemo response to cisplatin based chemotherapy in lung squamous
cell carcinoma patients. PLoS One. 9:e1081332014. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Sui W, Yan Q, Li H, Liu J, Chen J, Li L
and Dai Y: Genome-wide analysis of long noncoding RNA expression in
peripheral blood mononuclear cells of uremia patients. J Nephrol.
26:731–738. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
da W Huang, Sherman BT and Lempicki RA:
Systematic and integrative analysis of large gene lists using DAVID
bioinformatics resources. Nat Protoc. 4:44–57. 2009.PubMed/NCBI
|
|
19
|
Ponting CP, Oliver PL and Reik W:
Evolution and functions of long noncoding RNAs. Cell. 136:629–641.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Hall JR, Messenger ZJ, Tam HW, Phillips
SL, Recio L and Smart RC: Long noncoding RNA lincRNA-p21 is the
major mediator of UVB-induced and p53-dependent apoptosis in
keratinocytes. Cell Death Dis. 6:e17002015. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Foo JB, Yazan LS, Tor YS, Wibowo A, Ismail
N, How CW, Armania N, Loh SP, Ismail IS, Cheah YK and Abdullah R:
Induction of cell cycle arrest and apoptosis by betulinic acid-rich
fraction from Dillenia suffruticosa root in MCF-7 cells involved
p53/p21 and mitochondrial signalling pathway. J Ethnopharmacol.
166:270–278. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Ma X, Han J, Wu Q, Liu H, Shi S, Wang C,
Wang Y, Xiao J, Zhao J, Jiang J and Wan C: Involvement of
dysregulated Wip1 in manganese-induced p53 signaling and neuronal
apoptosis. Toxicol Lett. 235:17–27. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Flatt PM, Polyak K, Tang LJ, Scatena CD,
Westfall MD, Rubinstein LA, Yu J, Kinzler KW, Vogelstein B, Hill DE
and Pietenpol JA: p53-dependent expression of PIG3 during
proliferation, genotoxic stress and reversible growth arrest.
Cancer Lett. 156:63–72. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Gartel AL and Radhakrishnan SK: Lost in
transcription: p21 repression, mechanisms and consequences. Cancer
Res. 65:3980–3985. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Ando T, Kawabe T, Ohara H, Ducommun B,
Itoh M and Okamoto T: Involvement of the interaction between p21
and proliferating cell nuclear antigen for the maintenance of G2/M
arrest after DNA damage. J Biol Chem. 276:42971–42977. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Yadav V, Sultana S, Yadav J and Saini N:
Gatifloxacin induces S and G2-phase cell cycle arrest in pancreatic
cancer cells via p21/p27/p53. PLoS One. 7:e477962012. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Qu K, Lin T, Wei J, Meng F, Wang Z, Huang
Z, Wan Y, Song S, Liu S, Chang H, et al: Cisplatin induces cell
cycle arrest and senescence via upregulating P53 and P21 expression
in HepG2 cells. Nan Fang Yi Ke Da Xue Xue Bao. 33:1253–1259.
2013.PubMed/NCBI
|
|
28
|
Wang C, Chen Z, Ge Q, Hu J, Li F, Hu J, Xu
H, Ye Z and Li LC: Up-regulation of p21 (WAF1/CIP1) by miRNAs and
its implications in bladder cancer cells. FEBS Lett. 588:4654–4664.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Zhang H, Zhang X, Ji S, Hao C, Mu Y, Sun J
and Hao J: Sohlh2 inhibits ovarian cancer cell proliferation by
upregulation of p21 and downregulation of cyclin D1.
Carcinogenesis. 35:1863–1871. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Liu Z, Sun M, Lu K, Liu J, Zhang M, Wu W,
De W, Wang Z and Wang R: The long noncoding RNA HOTAIR contributes
to cisplatin resistance of human lung adenocarcinoma cells via
downregualtion of p21 (WAF1/CIP1) expression. PLoS One.
8:e772932013. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Contente A, Dittmer A, Koch MC, Roth J and
Dobbelstein M: A polymorphic microsatellite that mediates induction
of PIG3 by p53. Nat Genet. 30:315–320. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Guan X, Liu Z, Wang L, Wang LE, Sturgis EM
and Wei Q: Functional repeats (TGYCC)n in the p53-inducible gene 3
(PIG3) promoter and susceptibility to squamous cell carcinoma of
the head and neck. Carcinogenesis. 34:812–817. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Li B, Shang ZF, Yin JJ, Xu QZ, Liu XD,
Wang Y, Zhang SM, Guan H and Zhou PK: PIG3 functions in DNA damage
response through regulating DNA-PKcs homeostasis. Int J Biol Sci.
9:425–434. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Shaltiel IA, Aprelia M, Saurin AT,
Chowdhury D, Kops GJ, Voest EE and Medema RH: Distinct phosphatases
antagonize the p53 response in different phases of the cell cycle.
Proc Natl Acad Sci USA. 111:7313–7318. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Lowe J, Cha H, Lee MO, Mazur SJ, Appella E
and Fornace AJ Jr: Regulation of the Wip1 phosphatase and its
effects on the stress response. Front Biosci (Landmark Ed).
17:1480–1498. 2012. View
Article : Google Scholar : PubMed/NCBI
|
|
36
|
Wang W, Zhu H, Zhang H, Zhang L, Ding Q
and Jiang H: Targeting PPM1D by lentivirus-mediated RNA
interference inhibits the tumorigenicity of bladder cancer cells.
Braz J Med Biol Res. 47:1044–1049. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Li GB, Zhang XL, Yuan L, Jiao QQ, Liu DJ
and Liu J: Protein phosphatase magnesium-dependent 1delta (PPM1D)
mRNA expression is a prognosis marker for hepatocellular carcinoma.
PLoS One. 8:e607752013. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Yagi H, Chuman Y, Kozakai Y, Imagawa T,
Takahashi Y, Yoshimura F, Tanino K and Sakaguchi K: A small
molecule inhibitor of p53-inducible protein phosphatase PPM1D.
Bioorg Med Chem Lett. 22:729–732. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Tan DS, Lambros MB, Rayter S, Natrajan R,
Vatcheva R, Gao Q, Marchio C, Geyer FC, Savage K, Parry S, et al:
PPM1D is a potential therapeutic target in ovarian clear cell
carcinomas. Clin Cancer Res. 15:2269–2280. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Peng TS, He YH, Nie T, Hu XD, Lu HY, Yi J,
Shuai YF and Luo M: PPM1D is a prognostic marker and therapeutic
target in colorectal cancer. Exp Ther Med. 8:430–434.
2014.PubMed/NCBI
|
|
41
|
Ali AY, Kim JY, Pelletier JF, Vanderhyden
BC, Bachvarov DR and Tsang BK: Akt confers cisplatin
chemoresistance in human gynecological carcinoma cells by
modulating PPM1D stability. Mol Carcinog. 54:1301–1314. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Park HK, Panneerselvam J, Dudimah FD, Dong
G, Sebastian S, Zhang J and Fei P: Wip1 contributes to cell
homeostasis maintained by the steady-state level of Wtp53. Cell
Cycle. 10:2574–2582. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Cao R, Zhang J, Zhang M and Chen X: PPM1D
regulates p21 expression via dephoshporylation at serine 123. Cell
Cycle. 14:641–647. 2015. View Article : Google Scholar : PubMed/NCBI
|