Introduction
Human desumoylating isopeptidase 2 (DESI-2), also
known as PPPDE peptidase domain-containing protein 1 (PPPDE-1) was
initially identified through large-scale genome sequencing
(1–3).
DESI-2 is ubiquitously distributed in the majority of organs and
tissues, and its altered expression has been detected in certain
cancer types (4). Bioinformatics have
indicated that DESI-2 contains a PPPDE domain and belongs to the
DESI protein family (5). As another
member of the DESI family, DESI-1 has been reported to be a
deSUMOylase that reverses protein SUMOylation (6). Therefore, DESI-2 is expected to possess
deSUMOylase activity to regulate protein SUMOylation, which may
serve important roles in diverse cellular processes (7,8).
Although previous studies have determined certain
functions of DESI-2 (9–11), its precise underlying molecular
mechanisms have yet to be elucidated. A previous study demonstrated
that the overexpression and depletion of DESI-2 in embryos was able
to induce developmental defects in Xenopus and zebrafish
animal models, indicating that DESI-2 is crucial for the
maintenance of basic cellular activities (9,10). In our
previous study, it was demonstrated that DESI-2 is located at the
Golgi apparatus in the cytoplasm and that its expression levels are
decreased in pancreatic cancer tissues (4). It has been identified that DESI-2
overexpression may induce apoptosis in A549 human lung
adenocarcinoma cells, HeLa cervical cancer cells and PANC-1
pancreatic cancer cells (11). In the
current study, the alterations in the cell transcriptional profile
following the overexpression of DESI-2 were evaluated using an mRNA
microarray; the key signaling pathways in pancreatic cancer tissues
were also examined. The aim of the present study was to elucidate
the biological functions of DESI-2 in cancer development and
progression.
Materials and methods
Materials
The anti-DESI-2 primary antibody (#20517-1-AP) was
purchased from Proteintech Group, Inc. (Rosemont, IL, USA). Primary
antibodies targeting BH3 interacting-domain death agonist (BID;
#sc-373939), retinoid X receptor (RXR; #sc-553), cytochrome
c (#sc-13156), caspase-3 (#sc-271028), Rho-associated
protein kinase (ROCK; #sc-17794) and β-actin (#sc-81178) were
purchased from Santa Cruz Biotechnology, Inc. (Dallas, TX, USA).
The anti-Ras homolog gene family member A (RhoA) primary antibody
(#ab54835) was purchased from Abcam (Cambridge, MA, USA).
Horseradish peroxidase (HRP)-conjugated secondary antibodies,
including goat anti-rabbit immunoglobulin (Ig)G (#ZB-2301) and goat
anti-mouse IgG (#ZB-2305), were purchased from ZSGB-Bio (Beijing,
China). The radioimmunoprecipitation assay (RIPA) lysis buffer was
from the Beyotime Institute of Biotechnology (Haimen, China). A
Histostain-Plus kit and an Immobilon Western Chemiluminescent HRP
Substrate were purchased from Invitrogen (Thermo Fisher Scientific,
Inc., Waltham, MA, USA) and EMD Millipore (Billerica, MA, USA),
respectively.
Cell culture and transfection
PANC-1 human epithelial pancreatic cancer cells
(ATCC, Manassas, VA, USA) were cultured in Gibco Dulbecco's
modified Eagle's medium supplemented with 10% fetal bovine serum
(both from Thermo Fisher Scientific, Inc.) at 37°C in a humidified
incubator with an atmosphere containing 5% CO2. The open
reading frames of DESI-2 (GenBank accession: NM_016076) were
subcloned into a plasmid pcDNA3.1(+) vector (Invitrogen; Thermo
Fisher Scientific, Inc.) between the BamHI and XhoI
sites according to the manufacturer's instructions, in order to
obtain the recombinant plasmid vector pcDNA3.1-DESI-2. The
pcDNA3.1-DESI-2 and empty control vector were transfected into
PANC-1 cells using Lipofectamine® 2000 (Invitrogen;
Thermo Fisher Scientific, Inc.) according to the manufacturer's
instructions. Following a 24-h transfection, the cells were
collected and prepared for subsequent experiments.
Microarray
The Human 12×135K Gene Expression Array was
manufactured by Roche NimbleGen, Inc. (Madison, WI, USA). Briefly,
total RNA was isolated from PANC-1 cells using Trizol reagent
(Invitrogen; Thermo Fisher Scientific, Inc.) and quantified using a
NanoDrop ND-1000 spectrophotometer (NanoDrop Technologies; Thermo
Fisher Scientific, Inc.). The integrity of the RNA was assessed by
electrophoresis on a 1% denaturing agarose gel. Double-stranded
cDNA (ds-cDNA) was then synthesized from 1-µg RNA samples using a
SuperScript ds-cDNA Synthesis kit (Invitrogen; Thermo Fisher
Scientific, Inc.) and labeled with Cy3 fluorescence (Roche
NimbleGen, Inc.). The microarrays were hybridized with Cy3-labeled
ds-cDNA (Hybridization System; Roche NimbleGen, Inc.) and imaged
using an Axon GenePix 4000B microarray scanner (Molecular Devices
LLC, Sunnyvale, CA, USA). The images were imported into NimbleScan
software version 2.5 (Roche NimbleGen, Inc.) for grid alignment and
expression data analysis. Expression data were normalized through
quantile normalization and the Robust Multichip Average algorithm
included in the NimbleScan software. The differentially expressed
genes (fold change ≥2.00) were identified through fold change
filtering by Agilent GeneSpring GX software version 11.5.1 (Agilent
Technologies, Inc., Santa Clara, CA, USA). Hierarchical clustering
analysis was also performed using the Agilent GeneSpring GX
software version 11.5.1. Gene Ontology (GO; www.geneontology.org) and Kyoto Encyclopedia of Genes
and Genomes (KEGG; http://www.genome.jp/kegg/) analyses were performed to
determine the respective roles of the differentially expressed
genes.
Western blot analysis
To extract the proteins, PANC-1 cells were collected
and lysed using RIPA lysis buffer on ice prior to being centrifuged
at 12,000 × g for 30 min at 4°C. Following the determination
of protein concentration using a Pierce bicinchoninic acid assay
kit (Thermo Fisher Scientific, Inc.), the protein samples (50
µg/lane, reduced with 50 mM dithiothreitol) were separated by 12%
SDS-PAGE under denaturing conditions. Subsequently, the separated
proteins were transferred onto polyvinylidene fluoride membranes
and blocked in phosphate-buffered saline with 0.05%
Tween® 20 (PBST) containing 5% skimmed milk for 2 h. The
membranes were washed three times with PBST and incubated with the
primary antibodies against DESI-2, RhoA, ROCK, RXR, BID, cytochrome
c, caspase-3 and β-actin (dilution, 1:800) for 2 h at room
temperature. The membranes were then washed three times with PBST
and probed with the HRP-conjugated secondary antibodies (dilution,
1:10,000) for 1 h at room temperature. Following three washes with
PBST, the target blots were visualized using Immobilon Western
Chemiluminescent HRP Substrate. In order to detect cytochrome
c in the cytoplasm, a Mitochondria Isolation kit (Beyotime
Institute of Biotechnology) was used to remove the mitochondria,
according to the manufacturer's instructions, and these were
subsequently subjected to western blotting. The western blot
experiments were each performed in triplicate to assess the
relative protein abundance.
Immunohistochemistry
Pancreatic ductal adenocarcinoma tissue specimens
were obtained from 96 patients with pancreatic cancer (age range,
31–74 years; 54 males and 42 females) who had undergone surgery at
the West China Hospital of Sichuan University (Chengdu, China). The
institutional ethics committee of Sichuan University approved the
present study and informed consent was obtained from all patients
enrolled in the study. The formalin-fixed and paraffin-embedded
tissue samples were cut into 5-µm-thick sections using a microtome.
The sections were dewaxed in xylene and rehydrated in graded
alcohol. Following heat-induced epitope retrieval, the sections
were probed for DESI-2, RXR and RhoA with the corresponding primary
antibodies (dilution, 1:200) at 4°C overnight. After washing for
3×5 min in PBS, the sections were stained using an
avidin-biotin-peroxidase complex method and visualized with
diaminobenzidine (Beyotime Institute of Biotechnology) according to
the manufacturer's instructions. The sections were imaged with a
Leica DM2500 microscope and analyzed using Leica Application Suite
software version 3.8 (Leica Microsystems, Inc., Buffalo Grove, IL,
USA). DESI-2 expression level was scored as a staining intensity
score (0, negative; 1, mild; 2, moderate; and 3, strong) multiplied
by a score representing the percentage of positively staining cells
(0, none; 1, 1–25%; 2, 26–50%; 3, 51–75%; and 4, 76–100%),
resulting in an overall scoring scale of 0–12 (12). According to the median expression
level of DESI-2, the cancer samples were divided into DESI-2-higher
and DESI-2-lower tissue samples.
Statistical analysis
GO analysis was used to analyze differentially
expressed genes with respect to GO categories. The P-value denotes
the significance of the GO term enrichment in the differentially
expressed gene list (threshold for significance, P<0.05).
Biological pathways that were significantly enriched among the
differentially expressed genes were determined by pathway analysis
based on the KEGG database (P<0.05).
Results
Quality control and analysis of mRNA
microarray
The recombinant vector pc-DNA3.1(+) containing
DESI-2 was transiently overexpressed in PANC-1 cells, with an empty
vector as the control. The total RNA was isolated from the cells
and subjected to quality control prior to mRNA microarray. Agarose
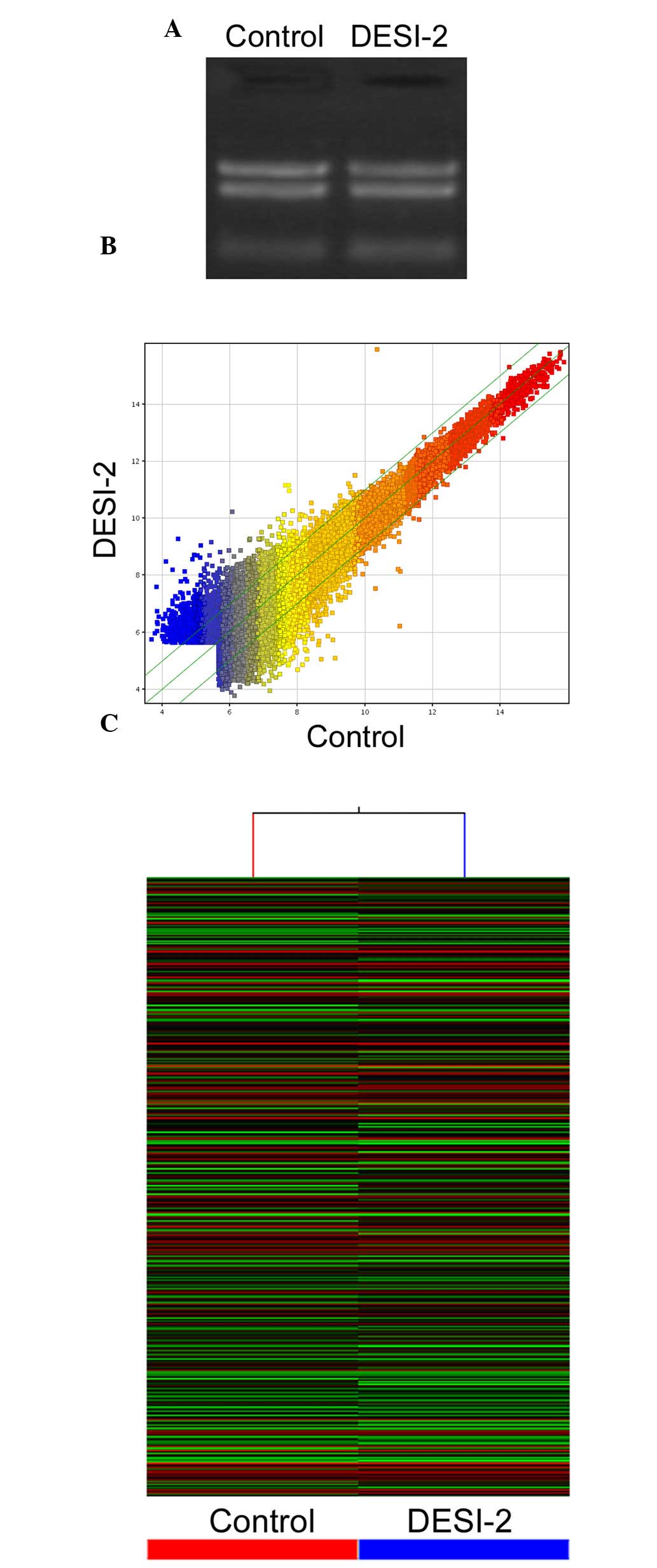
gel electrophoresis demonstrated that the isolated RNA samples were
able to be separated into three distinct bands of 28s RNA, 18s RNA
and 5s RNA (Fig. 1A). The gene
expression variations were assessed and visualized using the
scatter-plot method to identify differential genes (Fig. 1B). Hierarchical cluster analysis was
performed based on their expression levels and presented in the
dendrogram. The results revealed distinguishable gene expression
profiles between DESI-2 overexpressing cells and the control cells
(Fig. 1C).
Alterations in cell signaling pathways
following the overexpression of DESI-2
A total of 45,033 genes were examined by NimbleGen
Human Gene Expression Microarrays. The genes were considered to be
differentially expressed when the mRNA abundance exceeded the fold
change threshold (fold change ≥2.0). The altered expression
included a total of 1,766 upregulated and 1,643 downregulated
genes. These differential genes were subsequently subjected to
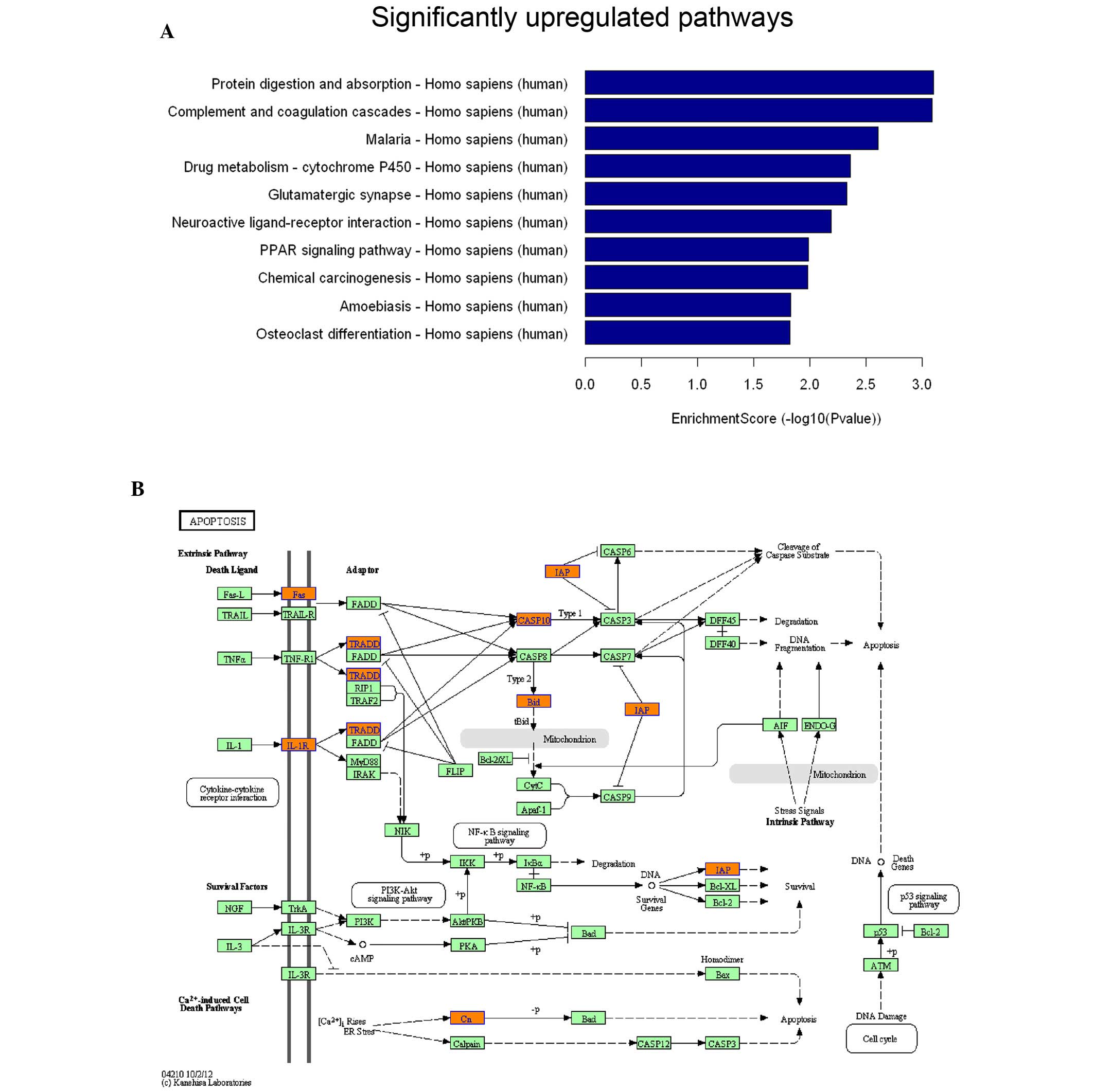
pathway analysis using the KEGG database. The upregulated genes
were primarily distributed in protein degradation, cell death and
differentiation (Fig. 2A; only the
top 10 altered pathways are presented). It is noteworthy that a
number of genes associated with apoptosis signaling, including Fas,
caspase-10, BID and TRADD, exhibited increased expression levels
(Fig. 2B). The downregulated genes
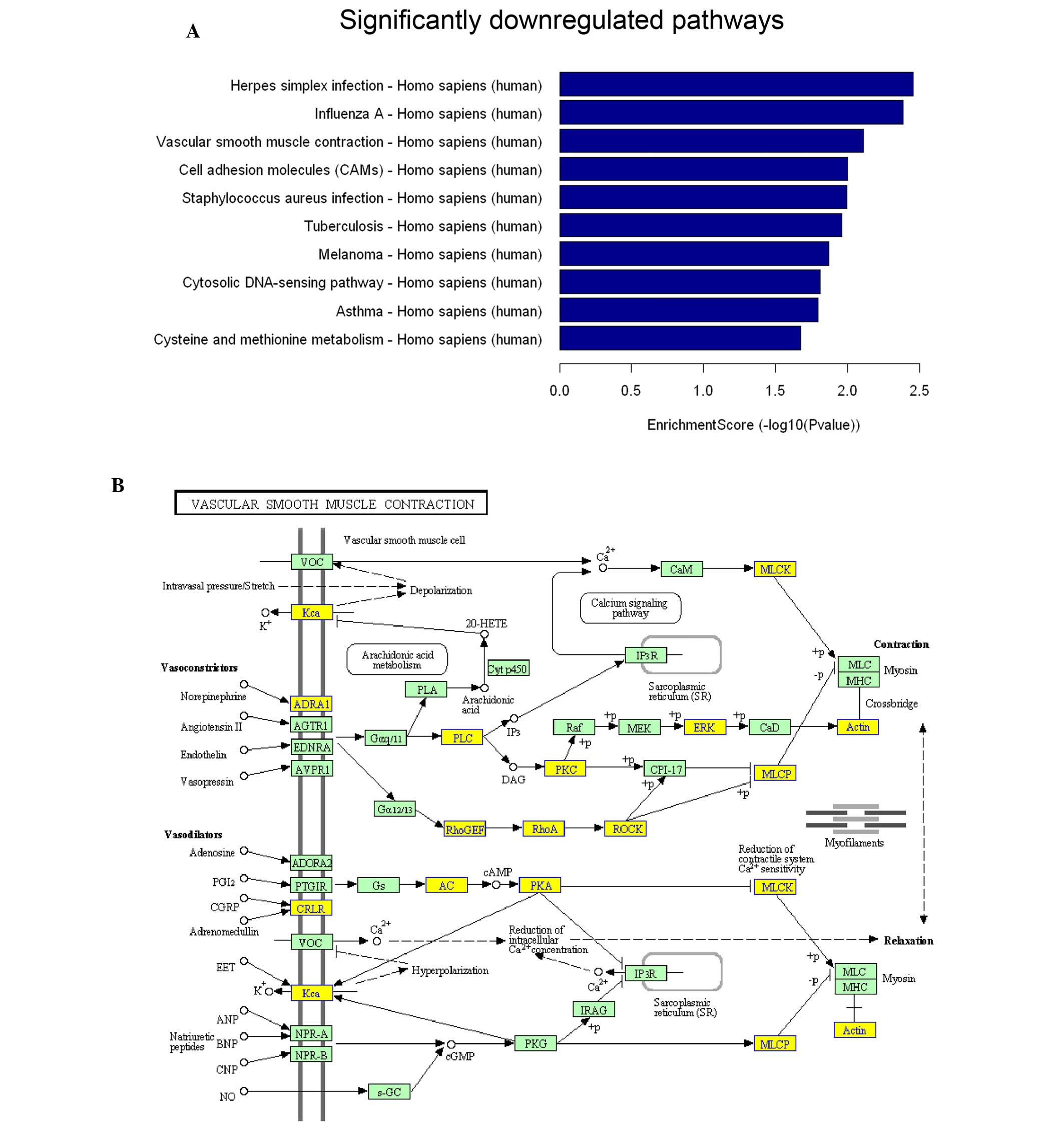
were principally distributed in metabolism, cell movement and
adhesion (Fig. 3A; only the top 10
altered pathways are presented). The critical genes Rho-guanine
nucleotide exchange factor (GEF), RhoA and ROCK in the Rho
signaling pathway exhibited decreased expression levels (Fig. 3B).
Western blotting to determine the
differential expression of key genes
As changes to the mRNA must be evaluated at the
protein level, the key signaling pathways that were altered in
DESI-2 overexpressing cells were further examined. Western blotting
demonstrated that RXR and BID are upregulated in PANC-1 cells that
overexpress DESI-2 (Fig. 4). BID has
an essential role in the apoptosis pathway and, therefore, its
downstream signaling pathways, including the release of cytochrome
c and the activation of caspase-3, were further
investigated. As hypothesized, cleaved caspase-3 and the emergence
of cytochrome c in cytoplasm were detected in the cells
overexpressing DESI-2, but not in the control PANC-1 cells
(Fig. 4). These results suggest that
endogenous apoptosis may be induced by the overexpression of DESI-2
in PANC-1 cells.
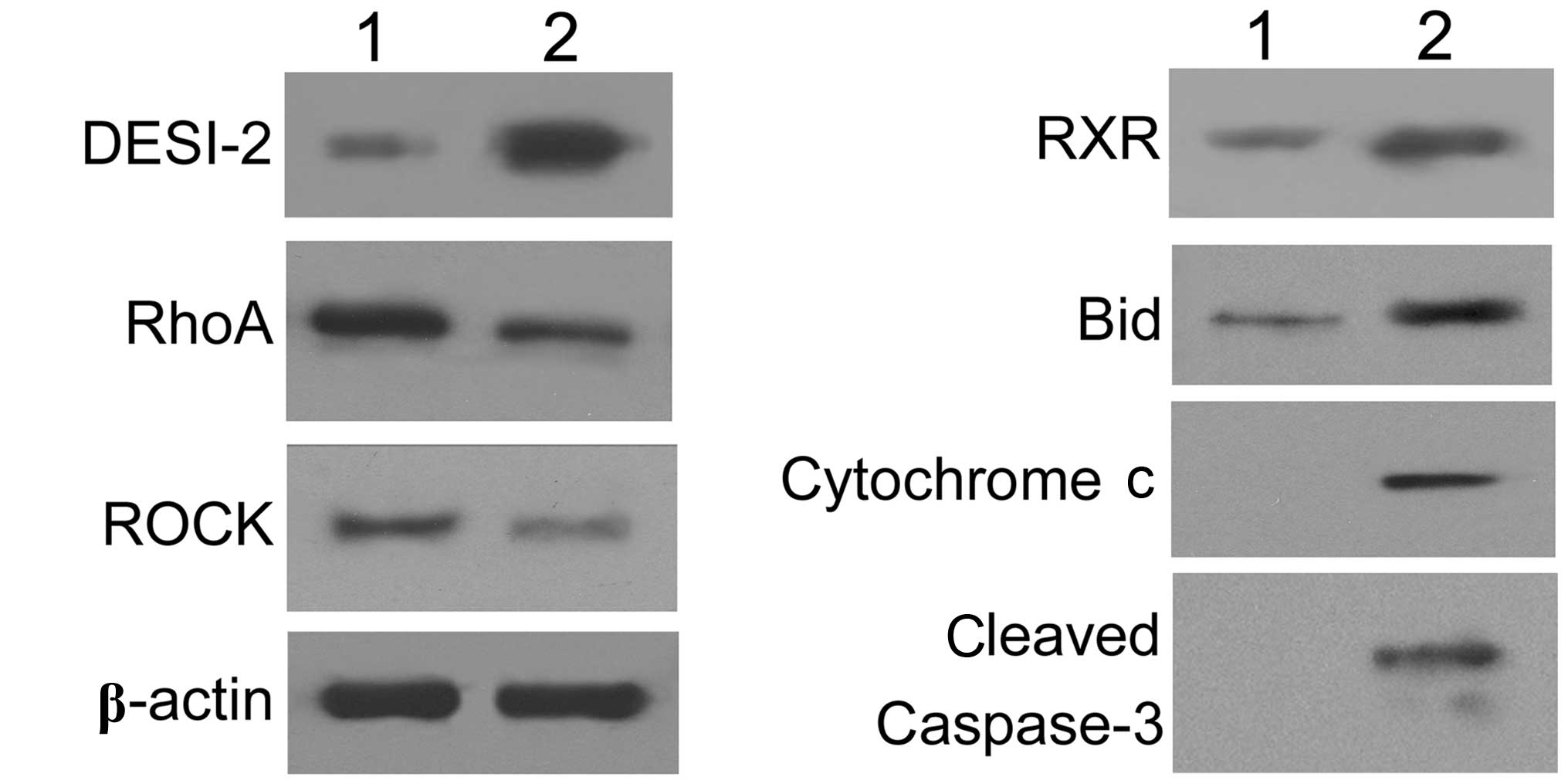 | Figure 4.The essential factors involved in cell
movement and apoptosis were detected by western blotting (1,
untreated cells; 2, DESI-2-overexpressing cells). Following the
overexpression of DESI-2 in PANC-1 cells, the key factors
identified as significantly altered in the mRNA microarray were
selected for examination at the protein level. Western blot
analysis revealed that RhoA and ROCK were downregulated, whereas
DESI-2, RXR and BID were upregulated in DESI-2-overexpressing
cells. Additionally, cytoplasmic cytochrome c and cleaved
caspase-3 were detected. β-actin was used as the internal control.
DESI-2, desumoylating isopeptidase 2; RhoA, Ras homolog gene family
member A; ROCK, Rho-associated protein kinase; RXR, retinoid X
receptor; BID, BH3 interacting-domain death agonist. |
With regard to the downregulated mRNA, western
blotting demonstrated that RhoA and ROCK exhibited decreased
expression levels in DESI-2 overexpressing PANC-1 cells,
corresponding with the changes in their mRNA (Fig. 4). These results indicated that the
RhoA signaling pathway was downregulated following the
overexpression of DESI-2 in PANC-1 cells.
Expression features of RhoA and RXR in
pancreatic cancer tissues with various DESI-2 levels
RhoA and RXR exhibit marked changes in their
expression profiles in DESI-2 overexpressing cells; thus, the
expression features of RhoA and RXR in pancreatic cancer tissues
were further investigated. A total of 96 pancreatic ductal
adenocarcinoma tissue specimens were collected and subjected to
immunohistochemistry to analyze the expression levels of DESI-2,
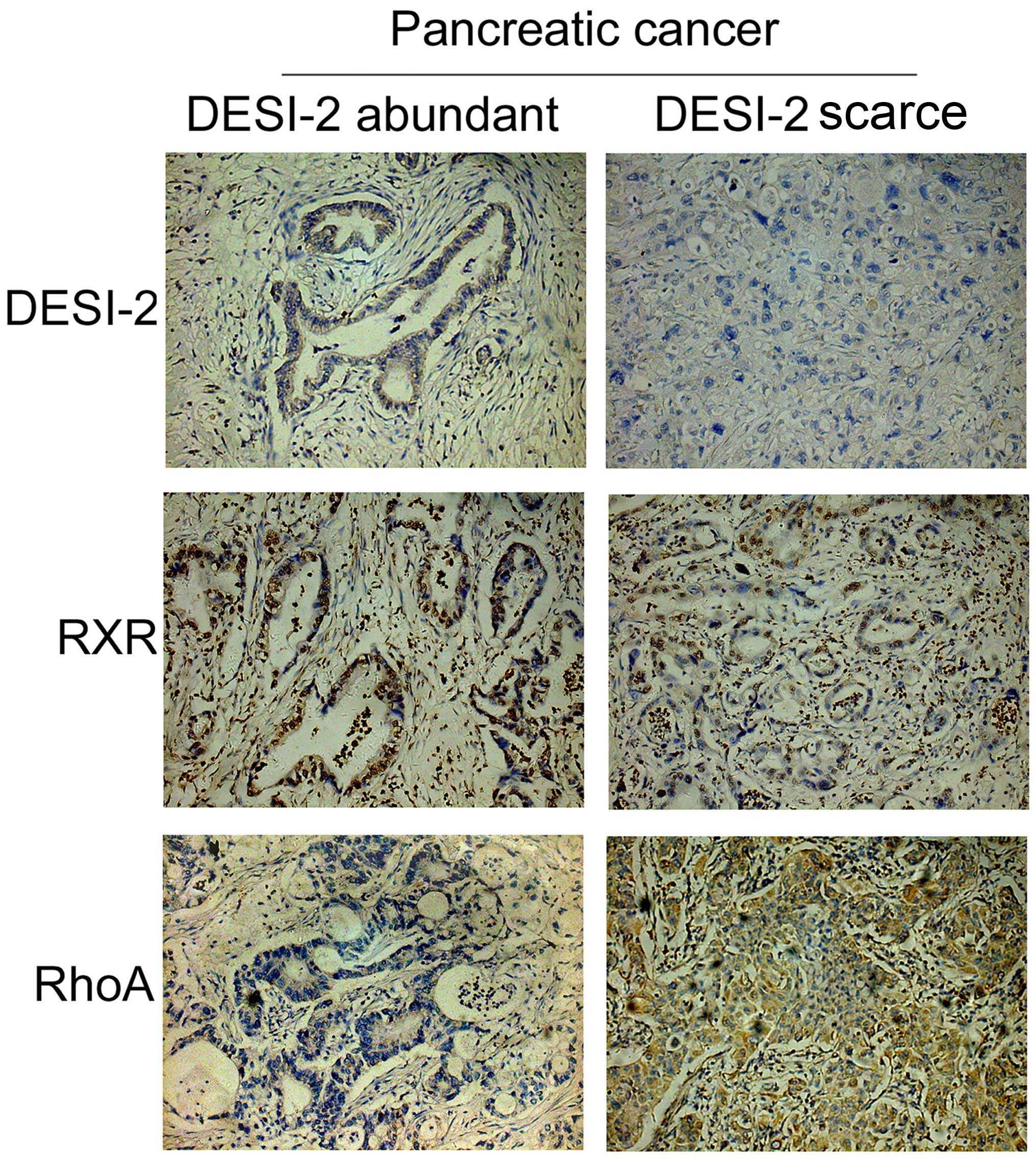
RhoA and RXR. The tissue samples were divided into two groups
according to the expression levels of DESI-2, which included DESI-2
abundant and DESI-2 scarce pancreatic tumor tissues (Fig. 5). Immunohistochemistry demonstrated
that RXR exhibited similar changes in expression levels to DESI-2,
with higher expression levels in DESI-2 abundant pancreatic tumor
tissues (Fig. 5). However, RhoA
expression exhibited lower expression levels in DESI-2-abundant
pancreatic tumor tissues, contrary to the expression profile of
DESI-2 (Fig. 5).
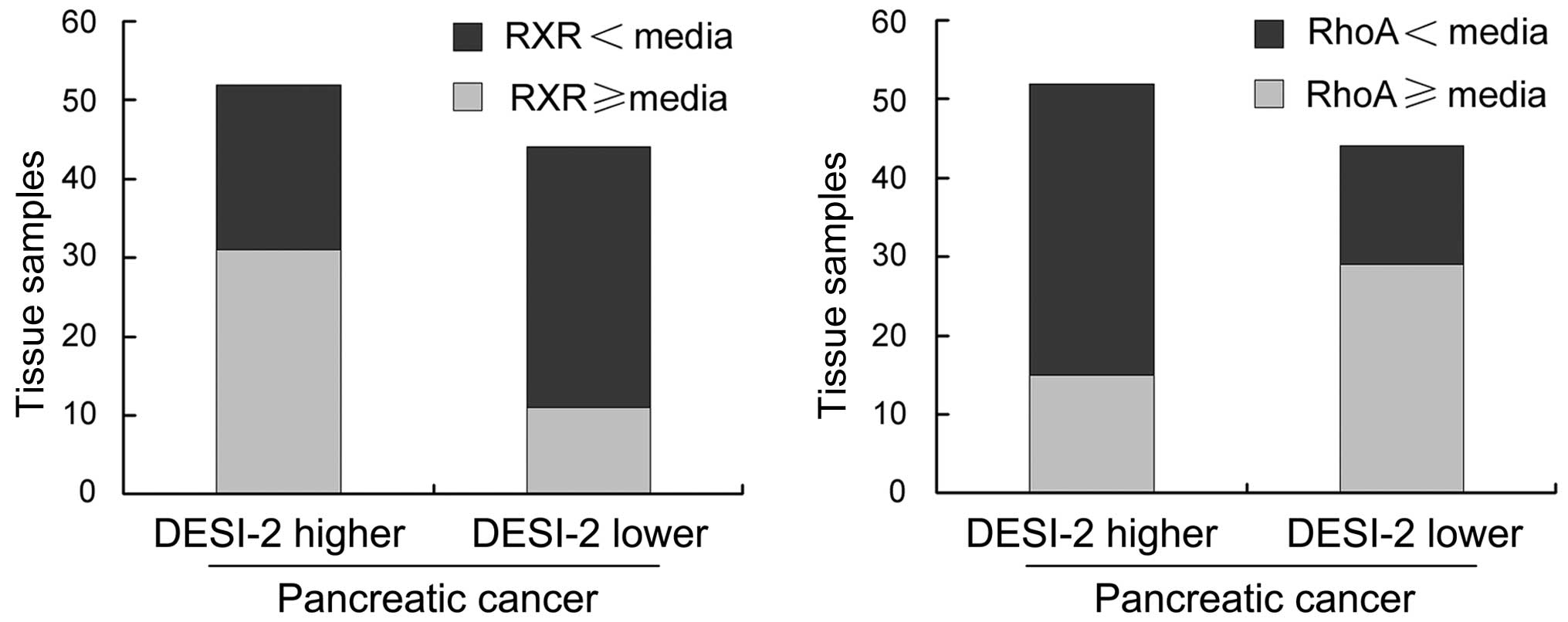
These results were established by further analysis.
The 96 tissue samples were divided into the DESI-2-higher (52
tissue samples) and DESI-2-lower (44 tissue samples) cancer groups,
according to the median expression level of DESI-2 present. The
tissue samples with RXR or RhoA expression levels higher or lower
than the median expression level are presented in Fig. 6. The proportion of tissues with RXR
expression levels exceeding the median is markedly increased in
DESI-2-higher cancer (31/52), compared with DESI-2-lower cancer
(11/44). However, the proportion of tissues with RhoA expression
levels that exceed the median is markedly decreased in
DESI-2-higher cancer (15/52), compared with DESI-2-lower cancer
(29/44).
Discussion
In the present study, the transcription profiles of
a set of specific genes in DESI-2-overexpressing PANC-1 cells were
investigated using analysis of mRNA microarrays. To the best of our
knowledge, this is the first study to comprehensively analyze the
associated signaling pathways following the induced overexpression
of DESI-2 in pancreatic cancer. Our previous study (12) investigated the altered protein
expression profiles in A549 human lung adenocarcinoma cells with
DESI-2 overexpression, using a proteomic strategy consisting of
two-dimensional electrophoresis (2-DE) coupled with mass
spectrometry (13). Although several
proteins were identified with the aforementioned proteomic
strategy, it was challenging to elucidate the comprehensive
pathways due to the limitations of 2-DE, including protein
solubility, throughput and abundance. Therefore, mRNA microarray
data may facilitate further investigations into the functions of
DESI-2 from alternative perspectives.
As mRNA microarrays reflect the transcription
profile of the genome, the protein expression levels must be
further examined to determine the actual changes in the expression
levels of specific genes. In our previous study, it was identified
that the overexpression of DESI-2 was able to induce cell apoptosis
through a mitochondrial apoptosis pathway; decreased B-cell
lymphoma-2 (Bcl-2) expression levels, increased of Bcl-2-associated
x-protein (Bax) expression levels, the release of cytochrome
c and the activation of caspase-3 were observed (11). In the current study, microarray
analysis identified that upstream BID expression levels were
increased, which was corroborated at the protein level. As an
essential apoptosis-associated protein, BID may be cleaved into
tBID, which then activates Bax and Bcl-2 antagonist/killer,
triggering the mitochondrial apoptosis pathway (14–16). The
results suggest a mechanism that may underlie the DESI-2-induced
apoptosis observed in our previous study (11).
RXR was another identified upregulated gene in
DESI-2 overexpressing PANC-1 cells. The changes in RXR expression
are important due to its essential role in the peroxisome
proliferator-activated receptor (PPAR) signaling pathway. As a
transcription factor, RXR binds to PPAR-α, -β and -γ to form
RXR-PPAR complexes, which may subsequently induce tumor protein p53
transcription and nitric oxide production to further activate the
mitochondrial apoptosis pathway (17–19). The
results in this study suggest that a significant decrease in RXR
expression levels in DESI-2-lower pancreatic cancer tissues
indicates a potential association between RXR and DESI-2 expression
levels in the development of pancreatic cancer.
With regard to the downregulated signaling pathways
revealed by microarray analysis, it was identified that RhoGEF,
RhoA and ROCK all exhibited decreased protein expression in
DESI-2-overexpressing PANC-1 cells. RhoA has an important role in
cell adhesion and proliferation (20–22).
Numerous previous studies have reported associations between RhoA
expression and development of pancreatic cancer (23,24). The
relatively low RhoA expression levels in DESI-2-higher pancreatic
cancer tissues, which were identified in this study, suggested that
high DESI-2 expression levels may be a marker for a lower degree of
malignancy in pancreatic cancer.
In conclusion, the results of the present study
revealed a comprehensive gene expression and transcription profile
in DESI-2-overexpressing cells, and also evaluated the essential
signaling factors at the protein level. The altered signaling
pathways may be considered as potential molecular mechanisms
underlying the functions of DESI-2 in the regulation of certain
biological processes. Additionally, the gene expression analysis of
pancreatic cancer tissues demonstrated that DESI-2 may be an
indicator of malignancy in pancreatic cancer.
Acknowledgements
This study was supported by the National Natural
Science Foundation of China (grant no. 81101530).
References
|
1
|
Gerhard DS, Wagner L, Feingold EA, Shenmen
CM, Grouse LH, Schuler G, Klein SL, Old S, Rasooly R, Good P, et
al: The status, quality, and expansion of the NIH full-length cDNA
project: The mammalian gene collection (MGC). Genome Res.
14:2121–2127. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Gregory SG, Barlow KF, McLay KE, Kaul R,
Swarbreck D, Dunham A, Scott CE, Howe KL, Woodfine K, Spencer CC,
et al: The DNA sequence and biological annotation of human
chromosome 1. Nature. 441:315–321. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Burkard TR, Planyavsky M, Kaupe I,
Breitwieser FP, Bürckstümmer T, Bennett KL, Superti-Furga G and
Colinge J: Initial characterization of the human central proteome.
BMC Syst Biol. 5:172011. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
He Y, Wang J, Gou L, Shen C, Chen L, Yi C,
Wei X and Yang J: Comprehensive analysis of expression profile
reveals the ubiquitous distribution of PPPDE peptidase domain 1, a
Golgi apparatus component and its implications in clinical cancer.
Biochimie. 95:1466–1475. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Lai CH, Chou CY, Ch'ang LY, Liu CS and Lin
W: Identification of novel human genes evolutionarily conserved in
Caenorhabditis elegans by comparative proteomics. Genome Res.
10:703–713. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Shin EJ, Shin HM, Nam E, Kim WS, Kim JH,
Oh BH and Yun Y: DeSUMOylating isopeptidase: A second class of SUMO
protease. EMBO Rep. 13:339–346. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Geiss-Friedlander R and Melchior F:
Concepts in sumoylation: A decade on. Nat Rev Mol Cell Biol.
8:947–956. 2007. View
Article : Google Scholar : PubMed/NCBI
|
|
8
|
Kim KI and Baek SH: SUMOylation code in
cancer development and metastasis. Mol Cells. 22:247–253.
2006.PubMed/NCBI
|
|
9
|
Yan F, Ruan XZ, Yang HS, Yao SH, Zhao XY,
Gou LT, Ma FX, Yuan Z, Deng HX and Wei YQ: Identification,
characterization, and effects of Xenopus laevis PNAS-4 gene on
embryonic development. J Biomed Biotechnol. 2010:1347642010.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Yao S, Xie L, Qian M, Yang H, Zhou L, Zhou
Q, Yan F, Gou L, Wei Y, Zhao X and Mo X: Pnas4 is a novel regulator
for convergence and extension during vertebrate gastrulation. FEBS
Lett. 582:2325–2332. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Yan F, Gou L, Yang J, Chen L, Tong A, Tang
M, Yuan Z, Yao S, Zhang P and Wei Y: A novel pro-apoptosis gene
PNAS4 that induces apoptosis in A549 human lung adenocarcinoma
cells and inhibits tumor growth in mice. Biochimie. 91:502–507.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Gou L, Wang W, Tong A, Yao Y, Zhou Y, Yi C
and Yang J: Proteomic identification of RhoA as a potential
biomarker for proliferation and metastasis in hepatocellular
carcinoma. J Mol Med (Berl). 89:817–827. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Gou LT, Tong AP, Yan F, Yuan Z, He F, Wang
W, Zhou Y, Chen LJ, Tang MH and Yang JL: Altered protein-expressing
profile in hPNAS4-induced apoptosis in A549 human lung
adenocarcinoma cells. J Cell Biochem. 108:1211–1219. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Sarosiek KA, Chi X, Bachman JA, Sims JJ,
Montero J, Patel L, Flanagan A, Andrews DW, Sorger P and Letai A:
BID preferentially activates BAK while BIM preferentially activates
BAX, affecting chemotherapy response. Mol Cell. 51:751–765. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Ren D, Tu HC, Kim H, Wang GX, Bean GR,
Takeuchi O, Jeffers JR, Zambetti GP, Hsieh JJ and Cheng EH: BID,
BIM and PUMA are essential for activation of the BAX- and
BAK-dependent cell death program. Science. 330:1390–1393. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Moldoveanu T, Grace CR, Llambi F, Nourse
A, Fitzgerald P, Gehring K, Kriwacki RW and Green DR: BID-induced
structural changes in BAK promote apoptosis. Nat Struct Mol Biol.
20:589–597. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Yamazaki K, Shimizu M, Okuno M,
Matsushima-Nishiwaki R, Kanemura N, Araki H, Tsurumi H, Kojima S,
Weinstein IB and Moriwaki H: Synergistic effects of RXR alpha and
PPAR gamma ligands to inhibit growth in human colon cancer
cells-phosphorylated RXR alpha is a critical target for colon
cancer management. Gut. 56:1557–1563. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Ziouzenkova O and Plutzky J: Retinoid
metabolism and nuclear receptor responses: New insights into
coordinated regulation of the PPAR-RXR complex. FEBS Lett.
582:32–38. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
IJpenberg A, Tan NS, Gelman L, Kersten S,
Seydoux J, Xu J, Metzger D, Canaple L, Chambon P, Wahli W and
Desvergne B: In vivo activation of PPAR target genes by RXR
homodimers. EMBO J. 23:2083–2091. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Zhang W, Huang Y and Gunst SJ: The small
GTPase RhoA regulates the contraction of smooth muscle tissues by
catalyzing the assembly of cytoskeletal signaling complexes at
membrane adhesion sites. J Biol Chem. 287:33996–34008. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Bhadriraju K, Yang M, Ruiz S Alom, Pirone
D, Tan J and Chen CS: Activation of ROCK by RhoA is regulated by
cell adhesion, shape, and cytoskeletal tension. Exp Cell Res.
313:3616–3623. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Hu T, Guo H, Wang W, Yu S, Han L, Jiang L,
Ma J, Yang C, Guo Q and Nan K: Loss of p57 expression and RhoA
overexpression are associated with poor survival of patients with
hepatocellular carcinoma. Oncol Rep. 30:1707–1714. 2013.PubMed/NCBI
|
|
23
|
Peruta M Della, Giagulli C, Laudanna C,
Scarpa A and Sorio C: RHOA and PRKCZ control different aspects of
cell motility in pancreatic cancer metastatic clones. Mol Cancer.
9:612010. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Dittert DD, Kielisch C, Alldinger I, Zietz
C, Meyer W, Dobrowolski F, Saeger HD and Baretton GB: Prognostic
significance of immunohistochemical RhoA expression on survival in
pancreatic ductal adenocarcinoma: A high-throughput analysis. Hum
Pathol. 39:1002–1010. 2008. View Article : Google Scholar : PubMed/NCBI
|