Introduction
Liver cancer is one of the most common causes of
cancer-associated mortality worldwide (1). Its five-year survival rate is
significantly lower than that of other types of cancer (2), mainly because the majority of liver
cancer patients are at late stage when they are diagnosed (3). Therefore, it is necessary to identify
new biomarkers for early diagnosis of liver cancer (4).
MicroRNAs (miRNAs or miRs) are short non-coding RNAs
with an average length of 20–25 nt (5). By binding to the 3′ untranslated region
(UTR) of their target messenger (m)RNA molecules, miRNAs inhibit
the translation of their target mRNA molecules (6). However, in limited cases, the binding
sites are located in the 5′ UTR or in the coding region of the
target mRNA (6). miRNAs are important
in regulating various biological processes, including cell
development, cell proliferation, cell differentiation and cell
apoptosis. Several previous studies have reported that miRNAs
participate in the initiation and carcinogenesis of cancer
(7,8).
The carcinogenesis of liver cancer and its associated miRNAs have
become a research hotspot area (9).
The results of previous studies have revealed that miRNAs could be
candidate prognostic and diagnostic biomarkers for liver cancer
(9).
The present study, with miRNA expression microarray
data obtained from the Gene Expression Omnibus (GEO) database,
aimed to identify the key miRNAs involved in the carcinogenesis of
liver cancer. In order to obtain confident results, differentially
expressed miRNAs in liver cancer were identified by combining the
results of three independent methods: Fisher's exact test,
t-test and Wilcoxon test. Target genes of the selected
miRNAs were predicted also by three independent methods: DIANA
(10), miRanda (11) and TargetScan (12). Ingenuity Pathway Analysis (IPA) was
conducted using the targets of the identified miRNAs to explore the
underlying mechanisms of carcinogenesis of liver cancer.
Materials and methods
Microarray data of miRNA
From the GEO database (https://www.ncbi.nlm.nih.gov/geo/), the microarray
data of miRNA GSE6857 and GSE30297, which represent miRNA
expression profile data from 496 liver cancer patients and 35
normal controls, respectively, were obtained. The data sets were
based on the platforms GPL4700 and GPL8786, respectively. All raw
data, including the original CEL, GPR and SOFT files, were obtained
for further analysis.
Detection of differentially expressed
miRNAs in liver cancer
Normalization of the raw miRNA data was performed in
R platform (version 3.1.3; https://www.r-project.org/) using the Robust
Multi-array Analysis (RMA) method (13). The final log2-transformed RMA
expression values were then stored for further analysis. Three
independent tests, including Fisher's exact test, t-test and
Wilcoxon test, were used to identify significantly differentially
expressed miRNAs between liver cancer and normal control samples.
The miRNAs that were supported by the above three tests were
considered to be confidently involved in the carcinogenesis of
liver cancer.
Prediction of miRNAs target genes
Prediction of target genes for the differentially
expressed miRNAs was performed using current available methods,
including DIANA (10), miRanda
(11) and TargetScan (12). In order to reduce the false-positive
rate of target prediction, the predicted targets supported by at
least two independent methods were selected as reliable target
genes of miRNAs. In addition, miRNAs target genes with wet
experimental support in the TarBase 6.0 database (14) were also included in the final pathway
analysis.
IPA
The most significantly differentially expressed
miRNAs identified in the previous steps were selected for IPA. The
Ingenuity Knowledge database (http://www.ingenuity.com/products/ipa) and IPA tools
were used to identify the enriched roles of miRNAs and their target
genes in cellular functions, pathways and diseases.
Cell culture and transfection
The human liver cancer cell line HepG2 was obtained
from the Chinese Center for Type Culture Collection (Beijing,
China). The HepG2 cell line was first cultured in RPMI-1640 medium
(Thermo Fisher Scientific, Inc., Waltham, MA, USA) supplemented
with 10% fetal bovine serum (Thermo Fisher Scientific, Inc.). Cells
were maintained in a humidified atmosphere with 5% CO2
at 37°C. HepG2 cells were seeded in 24-well plates at
6×105 cells/well and incubated overnight. Transfection
of the miR-1297 mimics, anti-miR-1297, inactive control cel-mir-67
or pMIR-REPORT vector (all Thermo Fisher Scientific, Inc.) was
performed using Lipofectamine 2000 transfection reagent
(Invitrogen; Thermo Fisher Scientific, Inc.) with 300 nmol miRNA or
1 µg/ml DNA plasmid, respectively. Total proteins of HepG2 cells
were isolated at 48 h post-transfection using M-PER Reagent (Thermo
Fisher Scientific, Inc.).
Western blotting
Proteins were separated by 12% SDS-PAGE and then
transferred onto nitrocellulose membranes (Bio-Rad Laboratories,
Inc., Hercules, CA, USA). Membranes were blocked with 5% non-fat
milk and incubated with anti-retinoblastoma (RB)1 antibody (cat.
no. ab181616; Abcam, Shanghai, China) or anti-β-actin antibody
(cat. no. ab8227; Abcam) at 4°C overnight. Following extensive
washes with TBST, a secondary antibody (cat. no. ab150077; Abcam)
was added to the system. Finally, enhanced chemiluminescence
(Abcam) was used to detect the immunoreactive protein bands.
Cell proliferation assay
Cell proliferation assay was performed with Cell
Counting Kit-8 (CCK-8; Dojindo Molecular Technologies, Inc.,
Kumamoto, Japan). HepG2 liver cancer cells were plated at
6×105 cells/well in 24-well plates. Then, cells were
incubated in 10% CCK-8 at 37°C for color conversion. Proliferation
rates were detected at 24, 48 and 72 h post-transfection.
Luciferase assay
HepG2 cells were seeded at 6×105
cells/well in 24-well plates and incubated for 24 h. Subsequently,
the cells were co-transfected with 0.8 µg pGL3-RB1-3′UTR or
pGL3-RB1-3′UTR Mut plasmid or with 0.08 ng phRL-SV40 control vector
(all Promega Corporation, Madison, WI, USA), and with 100 nM
miR-1297 or inactive control RNA, using Lipofectamine 2000
(Invitrogen; Thermo Fisher Scientific, Inc.). The Renilla
luciferase and firefly luciferase activities were detected using a
dual luciferase assay (Promega Corporation) at 24 h
post-transfection.
Statistical analysis
Statistical analyses were performed using R (version
3.1.3; https://www.r-project.org/). Values were
expressed as means ± standard deviation. Differences between groups
were estimated with the Student's t-test. P<0.05 was
considered to indicate a statistically significant difference.
Results
Differentially expressed miRNAs in
liver cancer
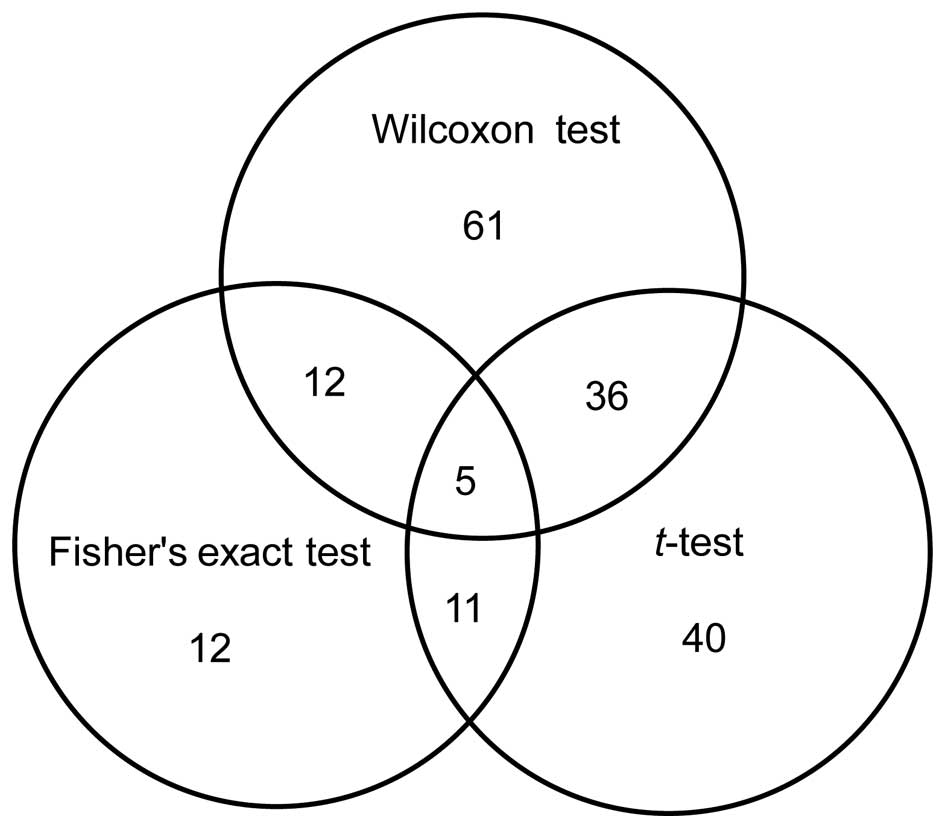
Based on the combined results of the three
independent tests, five miRNAs were identified to be significantly
differentially expressed in liver cancer (Fig. 1 and Table
I), including three upregulated and two downregulated miRNAs.
Among these miRNAs, miR-1297 had the most significant
deregulation.
 | Table I.Differentially expressed miRNAs
identified from three tests. |
Table I.
Differentially expressed miRNAs
identified from three tests.
| miRNA | t-test | Wilcoxon test | Fisher's exact
test | Log
(fold-change) |
|---|
| hsa-miR-1297 |
1.30×10−4 |
4.49×10−3 |
2.21×10−3 | 1.69 |
| hsa-miR-18a* |
7.15×10−4 |
1.23×10−3 |
1.46×10−2 | 1.40 |
| hsa-miR-183 |
1.43×10−3 |
1.70×10−3 |
2.48×10−2 | 1.28 |
| hsa-let-7e |
1.32×10−2 |
1.54×10−2 |
3.73×10−2 | −1.17 |
| hsa-miR-126 |
4.78×10−2 |
2.75×10−2 |
4.28×10−2 | −1.20 |
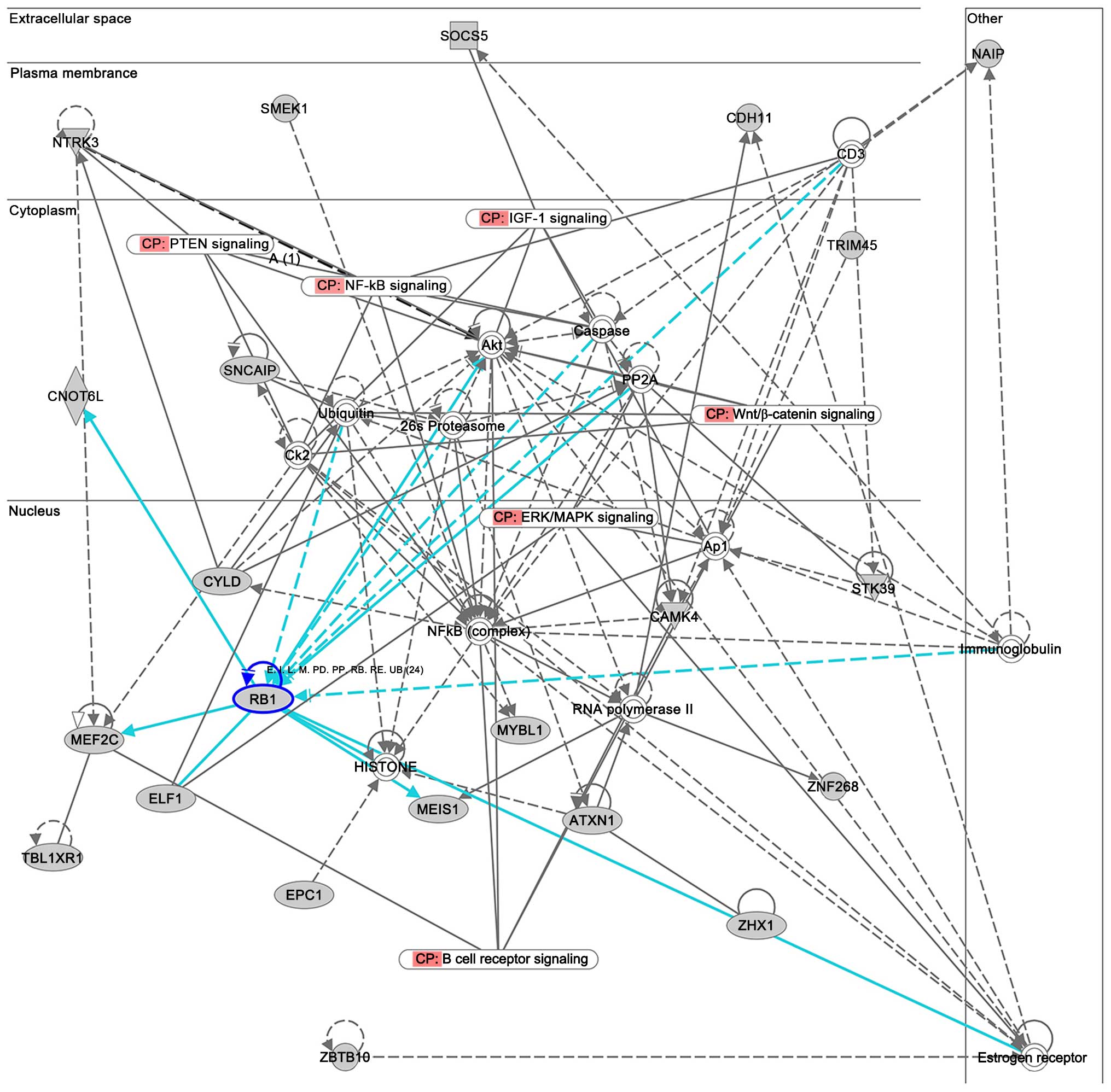
Target genes prediction
Since miRNAs serve their functions by targeting
mRNAs, the predicted target genes of the differentially expressed
miRNAs were retrieved. The most significant deregulated miRNA,
miR-1297, attracted our attention because one of its target genes
supported by multiple evidences is the tumor-suppressor gene
RB1 (Fig. 2), which is
involved in the regulation of the cell cycle and in human cancer
pathways (hsa04110 and hsa05200 Kyoto Encyclopedia of Genes and
Genomes pathways) (15–17).
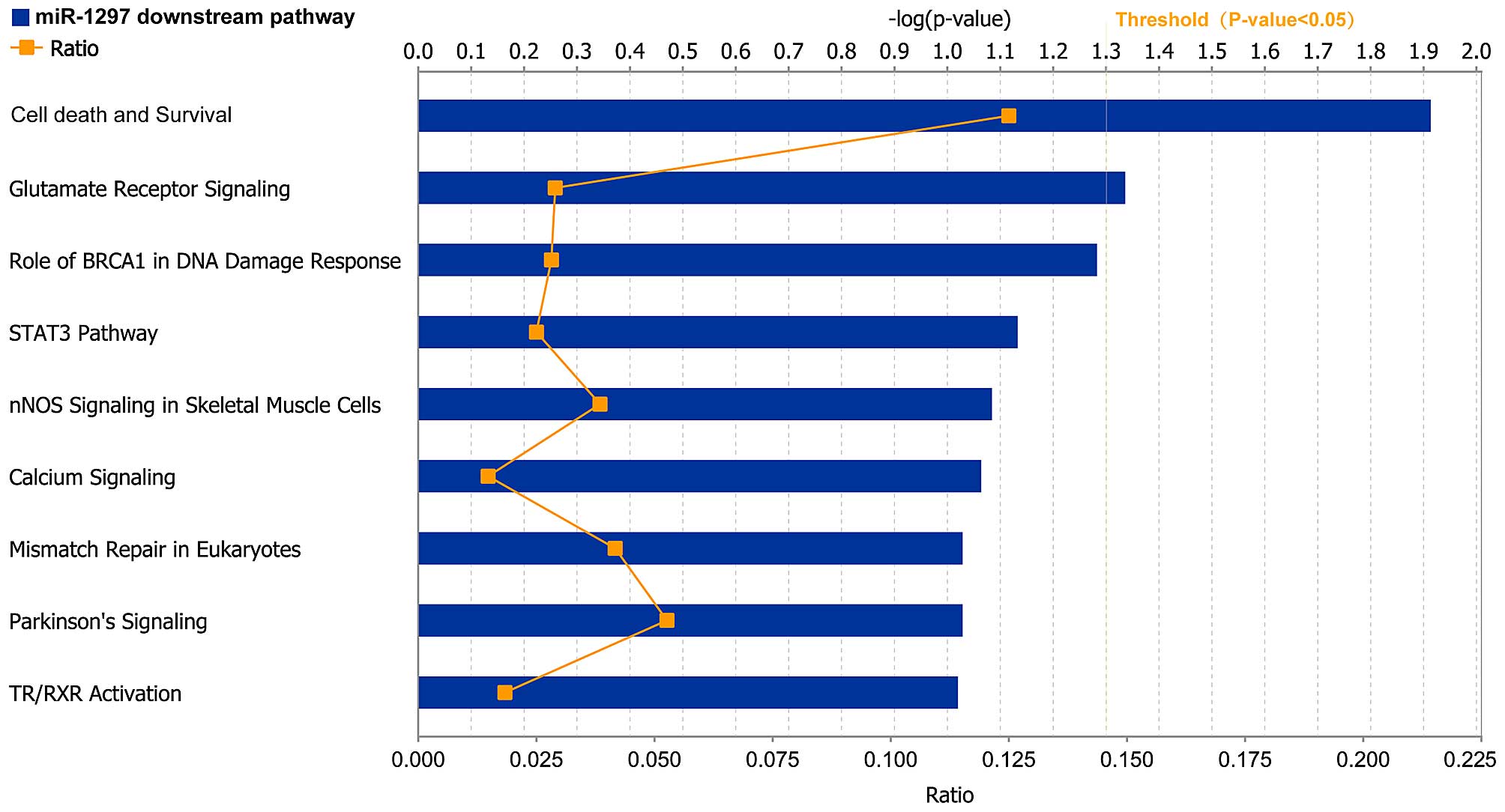
IPA
The predicted target genes of miR-1297 were
collected and imported into the IPA system to investigate their
biological functions in liver cancer. Table II contains the top five most
significant networks identified by IPA. Among these networks, cell
death and survival was the most frequent function, with a
significant score of 43 (Table II).
IPA also indicated that miR-1297 target genes were involved in
various biological functions, including cell cycle and cellular
development (Table III). Cell death
and survival as well as glutamate receptor signaling were the most
significant pathways enriched in target genes of miR-1297 (Fig. 3).
 | Table II.Top networks associated with
microRNA-1297 target genes. |
Table II.
Top networks associated with
microRNA-1297 target genes.
| Identity | Associated network
functions | Score |
|---|
| 1 | Cell death and
survival, behavior, nervous system development and function | 43 |
| 2 | Carbohydrate
metabolism, small molecule biochemistry, skeletal and muscular
disorders | 32 |
| 3 | Cell cycle, cell
morphology, cellular function and maintenance | 30 |
| 4 | Cancer,
gastrointestinal disease, auditory disease | 27 |
| 5 | Cell cycle, cell
death and survival, tumor morphology | 24 |
 | Table III.Diseases and functions associated with
microRNA-1297 target genes. |
Table III.
Diseases and functions associated with
microRNA-1297 target genes.
| A, Diseases and
disorders |
|---|
|
|---|
| Name | P-value, range | Molecules, n |
|---|
| Neurological
disease |
3.66×10−4-3.62×10−2 | 18 |
| Organismal injury and
abnormalities |
3.66×10−4-4.79×10−2 | 13 |
| Cancer |
6.07×10−4-4.88×10−2 | 74 |
| Gastrointestinal
disease |
6.07×10−4-2.93×10−2 | 44 |
| Respiratory
disease |
5.38×10−3-3.34×10−2 | 11 |
|
| B, Molecular and
cellular functions |
|
| Name | P-value, range | Molecules, n |
|
| Cell cycle |
4.70×10−5-4.79×10−2 | 14 |
| Cellular
development |
1.37×10−3-4.79×10−2 | 17 |
| RNA
post-transcriptional modification |
4.21×10−3-3.62×10−2 | 5 |
| Molecular
transport |
5.91×10−3-4.46×10−2 | 8 |
| Carbohydrate
metabolism |
6.12×10−3-2.43×10−2 | 1 |
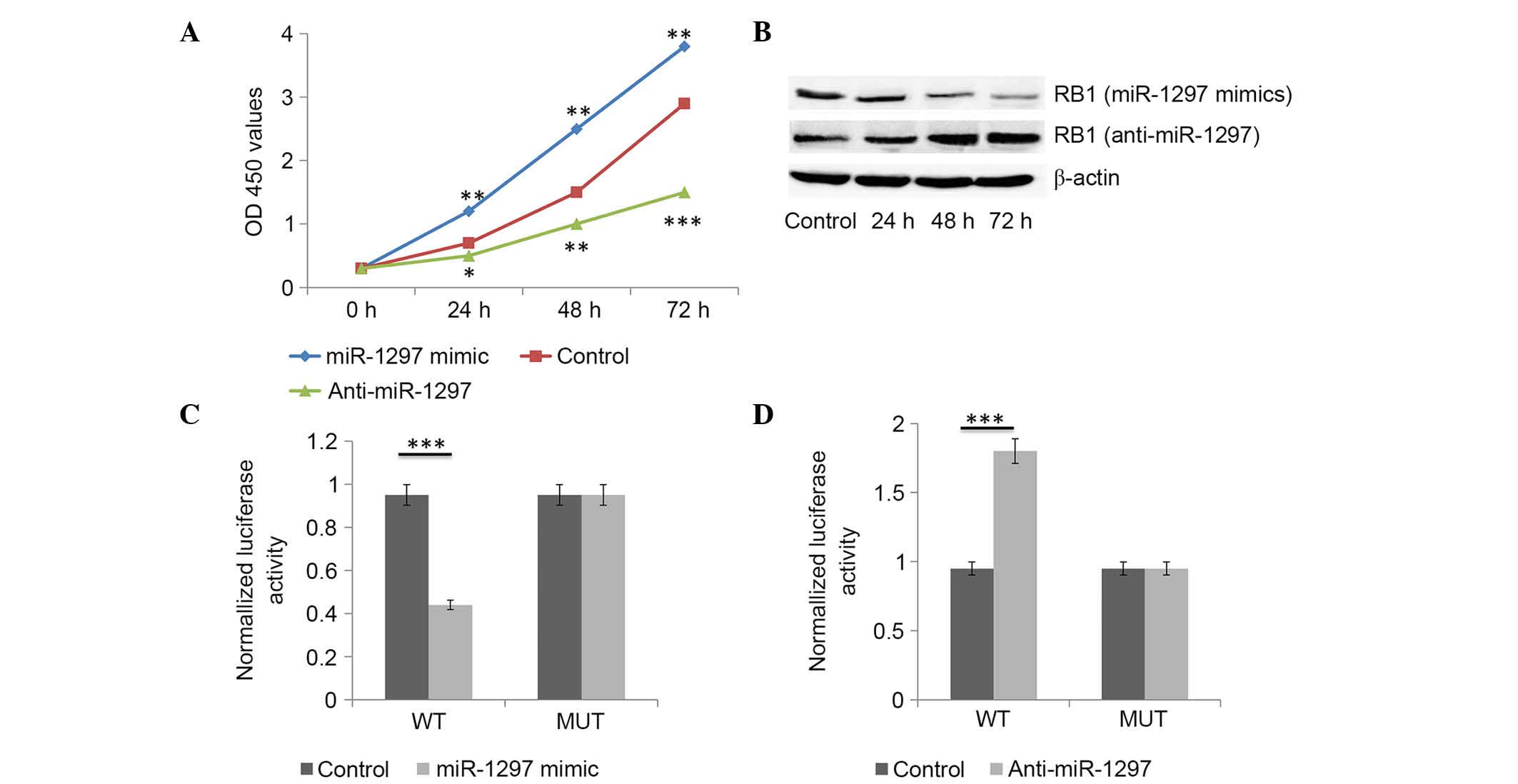
miR-1297 promotes liver cancer cell
proliferation
The potential impact of miR-1297 on liver cancer
cell proliferation was assessed in the HepG2 cell line. HepG2 cells
were transfected with miR-1297 mimics or miR-1297 inhibitor, or
with the inactive control cel-mir-67. CCK-8 proliferation assay
indicated that cell proliferation was significantly promoted in
miR-1297-mimics-transfected HepG2 cells compared with that in
inactive control cel-mir-67-transfected cells (Fig. 4A). Conversely, miR-1297 inhibitor
significantly inhibited the proliferation of HepG2 cells (Fig. 4A).
miR-1297 targets and negatively
regulates RB1 in liver cancer cells
miR-1297 mimics significantly reduced the protein
levels of RB1 in liver cancer cells (Fig.
4B). Conversely, miR-1297 inhibitor significantly increased the
protein levels of RB1 in liver cancer cells (Fig. 4B). As predicted by bioinformatics
analysis, there was complementarity between hsa-miR-1297 and the
3′UTR of RB1. The effect of miR-1297 on the translation of RB1 mRNA
into protein was then determined using a luciferase reporter assay.
miR-1297 mimics significantly reduced the luciferase activity of
the reporter gene with the wild-type construct but not with the
mutant RB1 3′UTR construct (Fig. 4C).
The inhibitor of miR-1297 significantly enhanced the luciferase
activity of the reporter gene with the wild-type construct but not
with the mutant RB1 3′UTR construct (Fig.
4D). These evidences indicate that miR-1297 directly binds to
the 3′UTR region of RB1. In general, miR-1297 targets and
negatively regulates RB1 in liver cancer cells.
Discussion
In the present study, using three independent tests
(Fisher's exact test, t-test and Wilcoxon test), five
differentially expressed miRNAs were identified, which may play
crucial roles in the carcinogenesis of liver cancer (Table I). Upon retrieving and analyzing the
target genes of these five miRNAs, the most significantly
deregulated miRNA, miR-1297, attracted our attention. One of its
target genes with various supporting evidences is the
tumor-suppressor gene RB1. The RB1 gene encodes a
negative regulator of the cell cycle, and was known to be a tumor
suppressor of multiple types of cancer (15–17),
including liver cancer (18). In
addition, RB1 is involved in the human cancer pathway (http://www.kegg.jp/kegg/pathway.html,
hsa04110 and hsa05200). This leads to the hypothesis that miR-1297
may be important in liver cancer.
Therefore, IPA was conducted to analyze the
biological function of the target genes of miR-1297. IPA is based
on the Ingenuity Knowledge Base, which derives known biological
functions and interactions of genes from published studies. IPA
allows the identification of biological networks, functions and
pathways that are associated with the target genes of miR-1297. The
results indicated that cell death and survival was the
highest-rated miR-1297 downstream biological network, with a
significance score of 43. The cell cycle was the most enriched
cellular function of miR-1297 target genes, as shown in Table II. These results revealed that
miR-1297 may participate in cancer through regulating cell death or
the cell cycle. Generally, miR-1297 may be important in the
carcinogenesis of liver cancer through regulation of its target
genes, particularly the tumor-suppressor gene RB1. To date,
no other studies have reported an association between miR-1297 and
liver cancer. However, miR-1297 has been reported to regulate the
carcinogenesis of colorectal cancer (19), lung adenocarcinoma (20) and laryngeal squamous cell carcinoma
(21). In addition, its predicted
target gene, RB1, is a negative regulator of the cell cycle and a
tumor-suppressor gene (22).
Therefore, the roles of miR-1297 in liver cancer were validated by
wet experiments.
CCK-8 proliferation assay indicated that cell
proliferation was promoted by miR-1297 in the HepG2 cell line,
while miR-1297 inhibitor could significantly inhibit the
proliferation of this cell line. Western blotting revealed that
miR-1297 suppressed the expression of RB1 at the protein level in
HepG2 cells. Furthermore, luciferase assays confirmed that miR-1297
directly bound to the 3′UTR of RB1 and suppressed its expression.
In conclusion, these results indicated that miR-1297 promotes cell
proliferation in liver cancer by negatively regulating the cell
cycle-inhibitory gene RB1. Therefore, miR-1297 may be a
potential therapeutic target for liver cancer in the future.
References
|
1
|
Ghouri YA, Mian I and Blechacz B: Cancer
review: Cholangiocarcinoma. J Carcinog. 14:12015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Tejeda-Maldonado J, Garcia-Juarez I,
Aguirre-Valadez J, González-Aguirre A, Vilatobá-Chapa M,
Armengol-Alonso A, Escobar-Penagos F, Torre A, Sánchez-Ávila JF and
Carrillo-Pérez DL: Diagnosis and treatment of hepatocellular
carcinoma: An update. World J Hepatol. 7:362–376. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Kinoshita A, Onoda H, Fushiya N, Koike K,
Nishino H and Tajiri H: Staging systems for hepatocellular
carcinoma: Current status and future perspectives. World J Hepatol.
7:406–424. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Kimhofer T, Fye H, Taylor-Robinson S,
Thursz M and Holmes E: Proteomic and metabonomic biomarkers for
hepatocellular carcinoma: A comprehensive review. Br J Cancer.
112:1141–1156. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Lee RC, Feinbaum RL and Ambros V: The C.
elegans heterochronic gene lin-4 encodes small RNAs with antisense
complementarity to lin-14. Cell. 75:843–854. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Malumbres M and Barbacid M: RAS oncogenes:
The first 30 years. Nat Rev Cancer. 3:459–465. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Garzon R, Calin GA and Croce CM: MicroRNAs
in cancer. Annu Rev Med. 60:167–179. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Nicolaidou V and Koufaris C: MicroRNA
responses to environmental liver carcinogens: Biological and
clinical significance. Clin Chim Acta. 445:25–33. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Hung CH, Chiu YC, Chen CH and Hu TH:
MicroRNAs in hepatocellular carcinoma: Carcinogenesis, progression
and therapeutic target. Biomed Res Int. 2014:4864072014. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Paraskevopoulou MD, Georgakilas G,
Kostoulas N, Reczko M, Maragkakis M, Dalamagas TM and Hatzigeorgiou
AG: DIANA-LncBase: Experimentally verified and computationally
predicted microRNA targets on long non-coding RNAs. Nucleic Acids
Res. 41:D239–D245. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
John B, Enright AJ, Aravin A, Tuschl T,
Sander C and Marks DS: Human MicroRNA targets. PLoS Biol.
2:e3632004. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Lewis BP, Burge CB and Bartel DP:
Conserved seed pairing, often flanked by adenosines, indicates that
thousands of human genes are microRNA targets. Cell. 120:15–20.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Irizarry RA, Hobbs B, Collin F,
Beazer-Barclay YD, Antonellis KJ, Scherf U and Speed TP:
Exploration, normalization and summaries of high density
oligonucleotide array probe level data. Biostatistics. 4:249–264.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Vergoulis T, Vlachos IS, Alexiou P,
Georgakilas G, Maragkakis M, Reczko M, Gerangelos S, Koziris N,
Dalamagas T and Hatzigeorgiou AG: TarBase 6.0: Capturing the
exponential growth of miRNA targets with experimental support.
Nucleic Acids Res. 40:D222–D229. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Gordon CA, Gulzar ZG and Brooks JD: NUSAP1
expression is upregulated by loss of RB1 in prostate cancer cells.
Prostate. 75:517–526. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Sabir M, Baig RM, Ali K, Mahjabeen I,
Saeed M and Kayani MA: Retinoblastoma (RB1) pocket domain mutations
and promoter hyper-methylation in head and neck cancer. Cell Oncol
(Dordr). 37:203–213. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Kansara M and Thomas DM: RB1-mediated
cell-autonomous and host-dependent oncosuppressor mechanisms in
radiation-induced osteosarcoma. Oncoimmunology. 3:e275692014.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Anwar SL, Krech T, Hasemeier B, Schipper
E, Schweitzer N, Vogel A, Kreipe H and Lehmann U: Deregulation of
RB1 expression by loss of imprinting in human hepatocellular
carcinoma. J Pathol. 233:392–401. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Chen P, Wang BL, Pan BS and Guo W:
MiR-1297 regulates the growth, migration and invasion of colorectal
cancer cells by targeting cyclo-oxygenase-2. Asian Pac J Cancer
Prev. 15:9185–9190. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Zhang C, Chi YL, Wang PY, Wang YQ, Zhang
YX, Deng J, Lv CJ and Xie SY: miR-511 and miR-1297 inhibit human
lung adenocarcinoma cell proliferation by targeting oncogene TRIB2.
PloS one. 7:e460902012. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Li X, Wang HL, Peng X, Zhou HF and Wang X:
miR-1297 mediates PTEN expression and contributes to cell
progression in LSCC. Biochem Biophys Res Commun. 427:254–260. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Di Fiore R, D'Anneo A, Tesoriere G and
Vento R: RB1 in cancer: Different mechanisms of RB1 inactivation
and alterations of pRb pathway in tumorigenesis. J Cell Physiol.
228:1676–1687. 2013. View Article : Google Scholar : PubMed/NCBI
|