Introduction
Esophageal cancer is one of the most common types of
malignant neoplasms and ranks eighth in incidence and sixth
(1) in mortality among cancers
worldwide. In China, it ranks fourth (2) in incidence among all cancers and its
major pathohistological subtype is esophageal squamous cell
carcinoma (ESCC). The majority of esophageal cancer patients
hospitalized are often diagnosed with advanced ESCC that
metastasizes to local lymph nodes (3,4). Thus, the
outcomes and prognosis of patients with ESCC are poor (5), despite the improvements in therapy
(6), with the 5-year survival rate
being no more than 15% (7).
Therefore, it is necessary to understand the mechanism of lymph
node metastasis of ESCC (8).
KIAA serial genes, also termed Cep126, encode
large proteins (9,10). The functions of numerous KIAA
genes remain unclear, although several studies (11–13) found
that certain KIAA genes were involved in certain diseases
using DNA microarray or proteomics-based approaches. KIAA1377 was
first reported to be localized to the midbody in the cytoplasm,
which may govern the process of cytokinesis through forming protein
complexes with other midbody proteins (14). Subsequently, Tipton et al
(15) found that KIAA1377 is a novel
centrosomal protein that also associates with microtubules and the
midbody. Another study reported KIAA1377 as a susceptible gene for
monomelic amyotrophy (16). There
have been no relevant studies on possible roles mediated by
KIAA1377 in the neoplasm.
In our previous study (Zheng et al,
unpublished data), using the comparative genomic hybridization
approach it was found that the copy numbers of the KIAA1377
gene were significantly amplified in ESCC tissues that metastasized
to local lymph nodes as compared to ESCC without metastasis (Zheng
et al, unpublished data), suggesting that KIAA1377 may play
a role in the lymph node metastasis of ESCC. In the present study,
the expression of KIAA1377 in ESCC clinical samples and cell lines
and its potential roles in metastasis were examined.
Materials and methods
Tissue microarray
The present study was approved by the Medical Ethics
Committee of the First Affiliated Hospital of Xinjiang Medical
University (Urumqi, China). The ESCC tissue microarray, consisting
of 86 cases of ESCC and 78 cases of paired adjacent normal control
tissues, was obtained from Shanghai Outdo Biotech Company
(Shanghai, China). The clinicopathological information accompanying
the tissue array, including tumor-node-metastasis grading and
staging, were also obtained. Staging and grading were assessed in
accordance with the World Health Organization 2014 version
classification and grading system (17). Prognosis and demographics were all
available.
Cell culture and transfection
The human ESCC ECa109, EC9706, TE-1, KYSE-30,
KYSE-150, KYSE-450 and KYSE-510 cell lines were grown in RPMI-1640
medium supplemented with 10% fetal bovine serum (FBS) and
penicillin/streptomycin in a 5% CO2 humidified incubator
at 37°C. For knockdown experiments, KIAA1377 short hairpin RNA
(shRNA) vector and shRNA scramble control were purchased from Santa
Cruz Biotechnology, Inc. (cat. no. sc-96891-SHH, Dallas, TX, USA).
ESCC cells were grown to 80–90% confluence in 6-well plates and
were transfected with 4.0 µg of shRNA-KIAA1377 vector or control
vector together with 10 µl Lipofectamine 2000 according to the
manufacturer's protocol (Invitrogen; Thermo Fisher Scientific,
Inc., Waltham, MA, USA). Cells were grown for two days, and media
was changed daily.
Immunohistochemistry (IHC)
Immunohistochemical stains were performed using
heat-induced epitope retrieval (18)
and avidin-biotin complex (19)
method. The rabbit anti-human KIAA1377 polyclonal antibody (clone,
N-16; catalogue no. sc-168347; Santa Cruz Biotechnology, Inc.) was
diluted at 1:200. The sections were evaluated by light microscopy,
and cellular localization of the protein and immunostaining level
in each section was assessed by 2 pathologists. The staining
patterns were scored as follows: negative, defined as <15%; weak
(≥15% but <30% of cells with positive staining), defined as +;
moderate (≥30% but <60%) of cells with positive staining),
defined as ++; and strong positive (≥60% of cells with positive
staining), defined as +++, according to the immunostaining
intensity area.
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR)
Total RNA was extracted from frozen tissue with
TRIzol reagent (Invitrogen; Thermo Fisher Scientific, Inc.) and
reverse transcribed to cDNA using the Prime Script RT-qPCR kit
(Takara Biotechnology Co., Ltd., Dalian, China). The RT-qPCR assay
was performed with the IQ5 system (Bio-Rad Laboratories, Inc.,
Hercules, CA, USA) using SYBR Premix Ex Taq (Takara Biotechnology
Co., Ltd.), according to the manufacturer's protocol. All reactions
were performed in triplicate. Subsequent to normalization with
β-actin controls, relative gene expression was determined using a
comparative standard curve. PCR was performed with the following
primer sets: KIAA1377 forward, 5′-TCCTAACACAGCCCTAAATGC-3′
and reverse, 5′-TCCAGATTGTAAAGCGTCCAG-3′; and β-actin forward,
5′-TGGCACCCAGCACAATGAA-3′ and reverse,
5′-CTAAGTCATAGTCCGCCTAGAAGCA-3′. The reaction mixtures for
KIAA1377 and β-actin were incubated at the following thermal
cycling conditions: 95°C for 3 min; 40 cycles at 95°C for 5 sec;
and 58°C for 30 sec. the final relative expression of KIAA1377 was
calculated after normalization to β-actin, as internal loading
control, using standard curve RT-qPCR approach (20).
Western blot analysis
In total, 72 h after transfection, TE-1 and EC-9706
cells were harvested in radioimmunoprecipitation assay lysis buffer
(BioTeke Corporation, Beijing, China) and 80 µg of cellular protein
was subjected to 8% sodium dodecyl sulfate-polyacrylamide gel
electrophoresis separation. Proteins were transferred to
polyvinylidene fluoride microporous membrane (EMD Millipore,
Billerica, MA, USA) and blots were probed with go at polyclonal
antibody against human KIAA1377 (catalogue no. sc-168347; dilution
at 1:800) and mouse monoclonal antibody against human
glyceraldehyde 3-phosphate dehydrogenase (GAPDH; catalogue no.
sc-25778; dilution at 1:500) were obtained from Santa Cruz
Biotechnology Inc. GAPDH was chosen as an internal control and the
blots were visualized with Western Breeze kit (catalogue no.,
WB7105; Invitrogen; Thermo Fisher Scientific, Inc.), according to
the manufacturer's protocol.
Methyl thiazolyl tetrazolium (MTT)
assay
MTT spectrophotometric dye assay (Sigma-Aldrich;
Merck Millipore, Darmstadt, Germany) was used to observe and
compare cell proliferation ability. TE-1 and EC-9706 cells were
plated in 96-well plates at a density of 5×103 cells per
well. Subsequent to transfection experiments, cell proliferation
was assessed. Cells were incubated for 4 h in 20 µl MTT at 37°C.
The color was developed by incubating the cells in 150 µl dimethyl
sulfoxide; the absorbance was detected at 490 nm wavelength. The
data were obtained from 3 independent experiments.
Cell migration and invasion assays in
vitro
Cell migration ability was calculated by the
wound-healing assay. TE-1 and EC-9706 cells were plated in 6-well
plates at a concentration of 4×105 cells/well and
allowed to form a confluent monolayer for 24 h. Subsequent to the
transfection experiment, the monolayer was scratched with a sterile
pipette tip (10 µl), washed with serum free medium to remove
floated and detached cells and images were captured (at 0 and 48 h)
by an inversion fluorescence microscope (Olympus Corporation,
Tokyo, Japan). Cell culture inserts (24-well, pore size 8 µm; BD
Biosciences, Franklin Lakes, NJ, USA) were seeded with
5×103 cells in 100 µl of medium with 0.1% FBS. Inserts
pre-coated with Matrigel (40 µl, 1 mg/ml; BD Biosciences) were used
for invasion assays. Medium with 10% FBS (400 µl) was added to the
lower chamber and served as a chemotactic agent. Noninvasive cells
were wiped from the upper side of the membrane and cells on the
lower side were fixed in cold methanol (−20°C) and air-dried. Cell
were stained with 0.1% crystal violet (dissolved in methanol) and
counted using the inverted microscope. Each individual experiment
had triplicate inserts, and 4 microscopic fields (magnification,
×100) were counted per insert.
Statistical analysis
Data were expressed as the mean ± standard deviation
and were analyzed by Student's t-test, one-way analysis of variance
and χ2 test, as appropriate, using SPSS for Windows
version 16.0 (SPSS, Inc., Chicago, IL, USA). Kaplan-Meier survival
curves were plotted and log rank test was performed. The
significance of various variables for survival was analyzed by Cox
proportional hazards model in a multivariate analysis. P<0.05
was considered to indicate a statistically significant
difference.
Results
KIAA1377 expression was significantly
upregulated in ESCC tissues and was associated with lymph node
metastasis and differentiation
To understand the clinicopathological significance
of KIAA1377 expression, a total of 78 distal normal tissues and 86
ESCC tissues were analyzed for KIAA1377 expression using IHC.
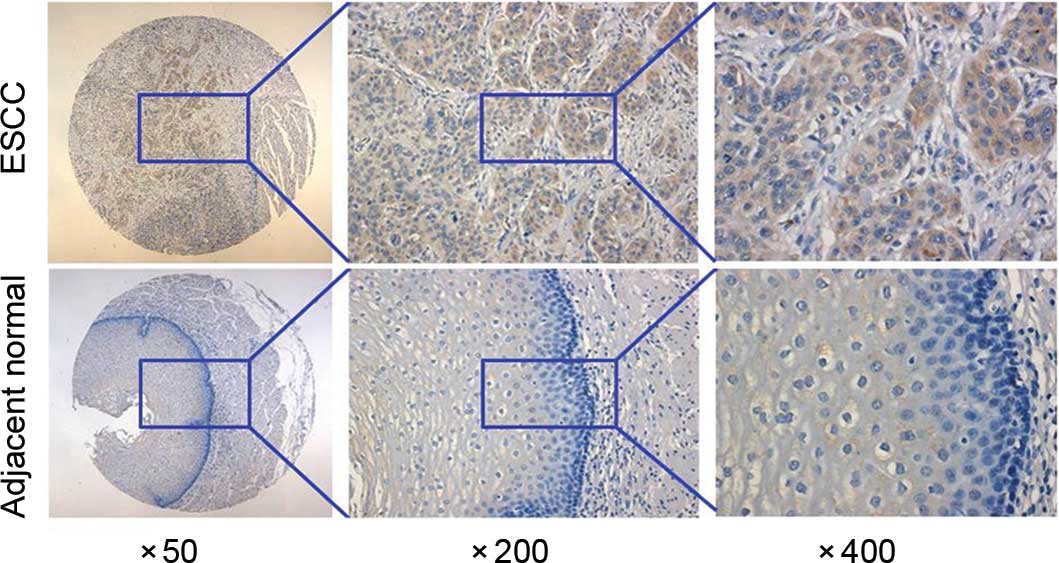
Expression of KIAA1377 was heterogeneous in ESCC tissues, ranging
from weak to moderate to strong positive staining. All paired
normal control tissues had positive expression of KIAA1377 compared
with ESCC tissues. It can be observed that KIAA1377 positive
staining localized at the cytoplasm region of tumor and normal
mucosal cells (Fig. 1).
Representative results of KIAA1377 expression are shown in Fig. 1. Clinicopathological analyses of
KIAA1377 expression are shown in Table
I. The KIAA1377 expression level is significantly different in
ESCC and normal tissues (P=0.001). There was a significant
difference of KIAA1377 expression levels when the tumor samples
were classified based on differentiation (P=0.030) and lymph node
metastases (P=0.043). No significant association was found between
KIAA1377 expression and other parameters (Table I).
 | Table I.Association between KIAA1377
expression and clinicopathological characteristics of ESCC. |
Table I.
Association between KIAA1377
expression and clinicopathological characteristics of ESCC.
|
|
| KIAA1377
expression |
|
|
|---|
|
|
|
|
|
|
|---|
| Characteristic | Total, n | High (++, +++) | Low (−, +) | χ2 | P-value |
|---|
| Tissue type |
|
|
| 41.539 | 0.001 |
| Adjacent
normal tissues | 78 | 1 | 77 |
|
|
|
ESCC | 86 | 38 | 48 |
|
|
| Gender |
|
|
| 1.286 | 0.257 |
|
Male | 64 | 26 | 38 |
|
|
|
Female | 22 | 12 | 10 |
|
|
| Age |
|
|
| 1.008 | 0.315 |
| >60
years | 57 | 23 | 34 |
|
|
| ≤60
years | 29 | 15 | 14 |
|
|
| Clinical stage |
|
|
| 4.807 | 0.090 |
| I | 6 | 2 | 4 |
|
|
| II | 43 | 24 | 19 |
|
|
|
III | 32 | 10 | 22 |
|
|
| N
classification |
|
|
| 4.965 | 0.043 |
|
N0 | 50 | 27 | 23 |
|
|
|
N1–3 | 34 | 10 | 24 |
|
|
| T
classification |
|
|
| 1.235 | 0.379 |
|
T1 | 5 | 1 | 4 |
|
|
|
T2–4 | 79 | 35 | 42 |
|
|
|
Differentiation |
|
|
| 6.999 | 0.030 |
|
Well | 8 | 4 | 4 |
|
|
|
Moderately | 60 | 31 | 29 |
|
|
|
Poorly | 18 | 3 | 15 |
|
|
| Tumor volume |
|
|
| 1.828 | 0.401 |
| <10
cm3 | 22 | 6 | 16 |
|
|
| 10–20
cm3 | 27 | 11 | 16 |
|
|
| >20
cm3 | 21 | 5 | 16 |
|
|
| Gross
classification |
|
|
| 1.855 | 0.603 |
|
Ulcerative type | 45 | 20 | 25 |
|
|
|
Fungating type | 6 | 2 | 4 |
|
|
|
Medullary type | 24 | 11 | 13 |
|
|
|
Protrude type | 2 | 0 | 2 |
|
|
To confirm whether or not there was an association
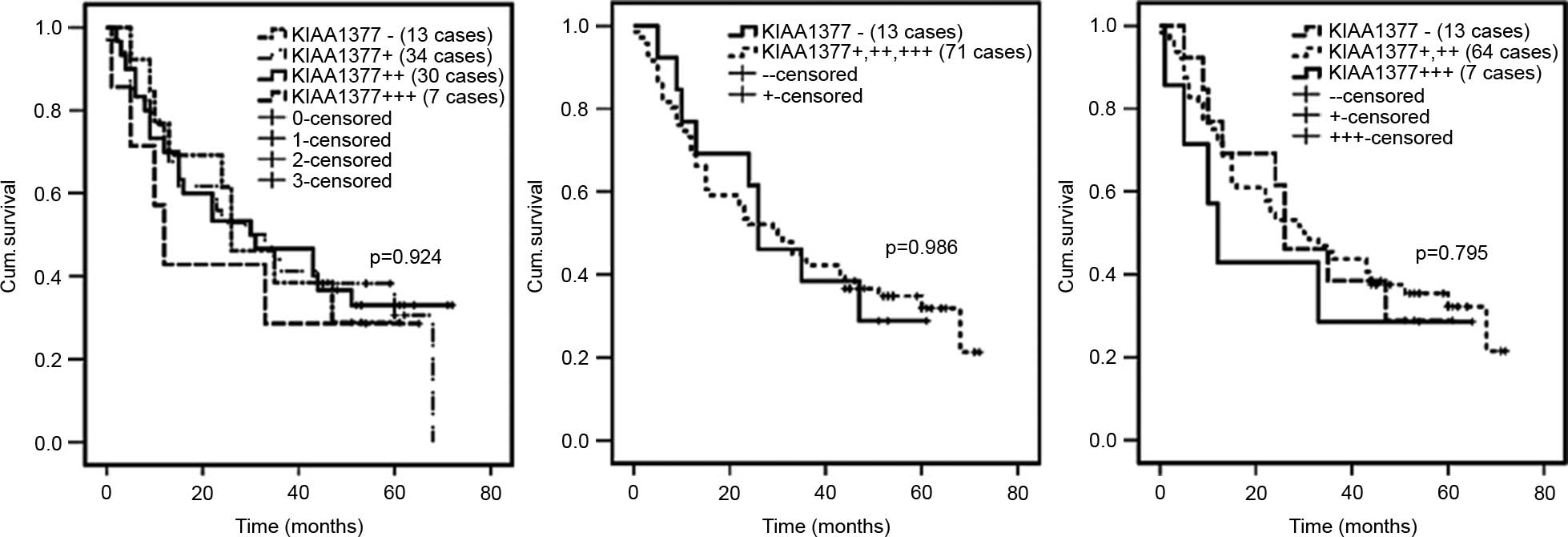
between KIAA1377 expression and prognosis, the Kaplan-Meier
survival curve for different expression statuses of KIAA1377 was
used. Based on 86 patients with ESCC, no significant difference in
survival was found comparing patients with weak, moderate or strong
KIAA1377 staining in tumors (P=0.924) (Fig. 2). Similarly, multivariate analysis
showed there were no significant differences in survival without
lymph node metastasis (N0) and with lymph node
metastasis (N1–3) or between stage I and II KIAA1377
immunostaining in tumors (data not shown).
Knockdown of KIAA1377 cannot
significantly affect the proliferation, migration and invasion of
ESCC cells
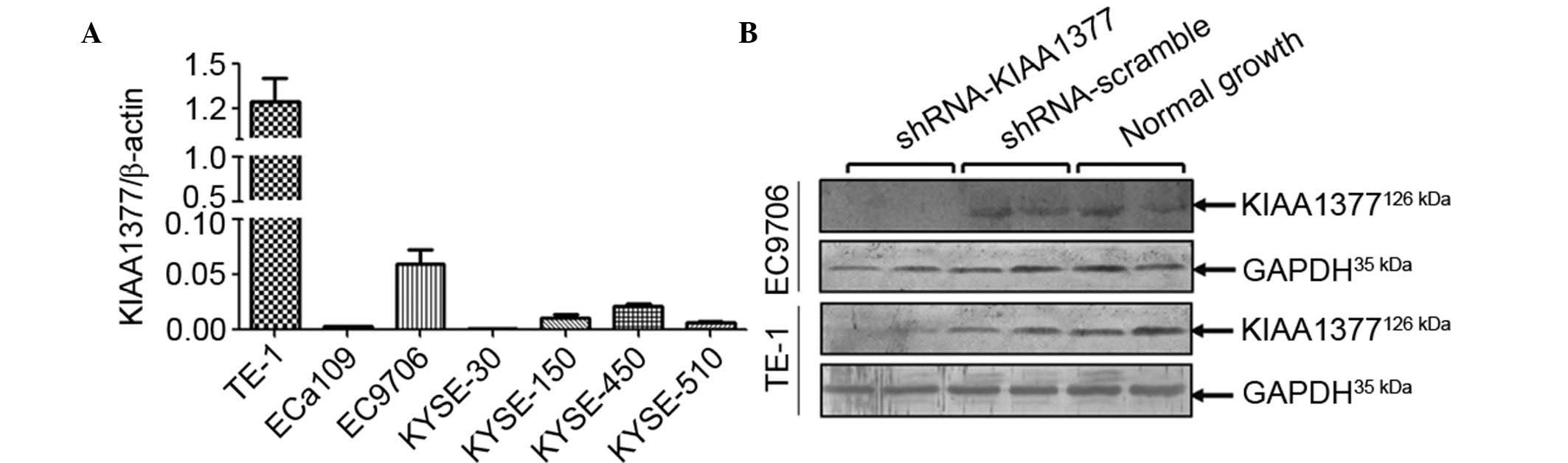
To determine the basal expression of KIAA1377 in a
panel of ESCC cell lines, RT-qPCR was performed on a panel of 7
different types of ESCC cell lines. It was found that in TE-1 and
EC9706, the 2 different ESCC cell lines, endogenous mRNA level of
KIAA1377 were higher than that in other cell lines (Fig. 3A). To understand the role of
KIAA1377 in the metastasis of ESCC, specific short hairpin
RNA (shRNA) interference vectors were transfected into TE-1 and
EC9706 cells. The knockdown efficiency was evaluated using western
blot analysis. It was shown that the shRNA vectors against
KIAA1377 may effectively knockdown the KIAA1377 protein
level (Fig. 3B). Based on which, the
proliferation, migration and invasion were assessed by MTT, wound
healing and Transwell assays, respectively. It was observed that
knockdown of KIAA1377 hardly affected the proliferation
(Fig. 4A and B), migration (Fig. 4C and D) and invasion (Fig. 4E and F) abilities subsequent to
statistical analysis.
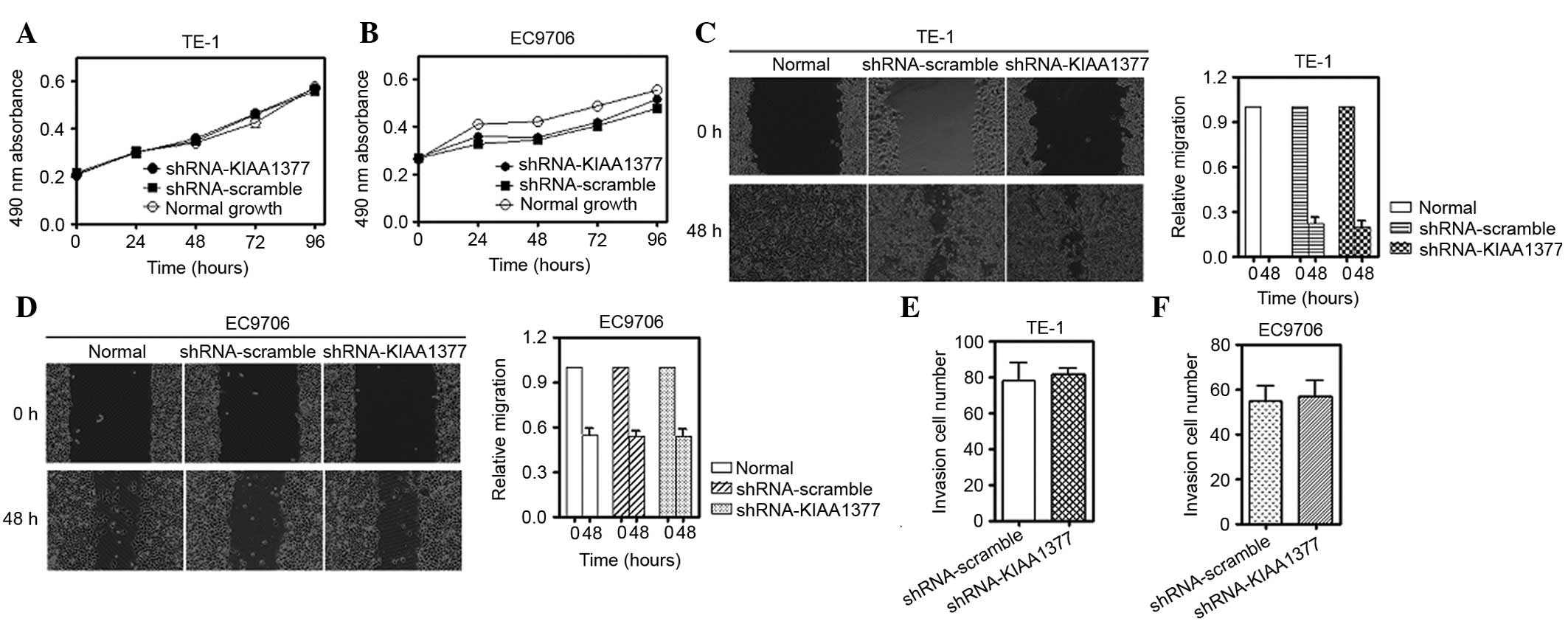 | Figure 4.KIAA1377 does not affect the
proliferation, migration and invasion ability of cells subsequent
to transient knockdown. (A) Methyl thiazolyl tetrazolium assay for
the ESCC TE-1 and EC9706 cell lines, (B) subsequent to knockdown of
KIAA1377 using transient transfection with a specific shRNA vector
for 0, 24, 48, 72 and 96 h. (C) Qualitative wound-healing assay
(left) and quantitative assay (right) for TE-1. (D) Similarly,
qualitative wound-healing assay (left) and quantitative assay
(right) for EC9706 subsequent to transient transfection for 48 h.
(E) Quantitative Transwell assay for TE-1 and EC9706, (F)
subsequent to knockdown for 48 h (P>0.05 compared with control
group). Images of migratory cells from the scratched boundary were
observed and acquired by light microscopy (magnification, 10×10).
Similar results were obtained in 3 independent experiments, and
representative images are shown. ESCC, esophageal squamous cell
carcinoma. |
Discussion
In the present study, it was found that
KIAA1377 expression was significantly associated with lymph
node metastasis and differentiation in ESCC clinical samples,
whereas to the best of our knowledge was not required in the
proliferation and mobility ex vivo in ESCC cell lines.
The molecular variations underlying the lymph node
metastasis of ESCC remain largely unclear. In our previous study
(Zheng et al, unpublished data), differentially expressed
genes were screened between ESCC tissues that metastasized to lymph
nodes and ESCC tissues without metastasis using comparative genomic
hybridization approach. It was found that the copy number of
several genes, including KIAA1377, was significantly
amplified in ESCC tissues with lymph node metastasis post-surgery
as compared with control. Due to the rarity of data of
KIAA1377 in cancers and its highest frequency of copy number
variation, KIAA1377 was targeted among the differential
genes screened out and hypothesized that amplification of
KIAA1377 may facilitate ESCC cells metastasis into lymph
nodes. Based on the hypothesis, it has firstly confirmed the
expression of KIAA1377 using IHC with ESCC tissue array. It was
identified that KIAA1377 expression was significantly associated
with lymph node metastasis and cell differentiation degree, which
was consistent with our previous finding on copy number variations.
However, no significant association was observed between KIAA1377
expression and prognosis, as well as other clinicopathological
parameters.
To additionally verify the supposed role of
KIAA1377 in facilitating the lymph node metastasis in ESCC,
KIAA1377 was knocked down in TE-1 cells, which demonstrated
the higher basal expression level among the 7 different ESCC cell
lines available. It was shown that KIAA1377 has little
effect on the proliferation, migration and invasion subsequent to
transient knockdown, suggesting that KIAA1377 maybe
dispensable in the proliferation and mobility of ESCC cell lines
ex vivo.
With regard to the role of KIAA1377 in lymph
node metastasis, there appears to be somewhat controversial and
inconsistent findings in vivo and ex vivo. However,
in consideration of studies (14–16)
available regarding the way KIAA1377 works in physiological or
pathophysiological setting, KIAA1377 may co-ordinate with other
unknown proteins (14,21) located on the midbody or in the
cytoplasm through protein-protein interactions (15,21,22). Thus
it would make sense that only knockdown of KIAA1377 may
cause few phenotypic defects as other compensation mechanisms maybe
provided by other interacting proteins (23,24).
Furthermore, Bonavita et al (25) found that depletion of Cep-126, also
known as KIAA1377, was required for the formation of primary
cilium in hTERT-RPE-1 and IMCD3 cells. Based on this study, it is
hypothesized that KIAA1377 may be associated with
micro-metastasis in ESCC cells. Therefore, it may be due to the
detection limit in the experimental setting that no significant
phenotypic variation was observed ex vivo subsequent to
knockdown of KIAA1377 in ESCC cells.
Despite the limited cases studied and lack of
mechanistic information provided by in vitro KIAA1377
experiments, to the best of our knowledge, this is the first study
that shows KIAA1377 is significantly associated with lymph
node metastasis and differentiation in ESCC, but is dispensable in
the motility of ESCC cell lines ex vivo. Overall,
KIAA1377 may play a role in the lymph node micrometastasis
of ESCC.
Acknowledgements
The present study was supported by National Science
Foundation of China (grant no. 81360357). The authors thank Dr
Antonino Colanzi [Institute of Protein Biochemistry (CNR), Naples,
Italy] and Associate Professor Songtao Liu (Department of
Biological Sciences, University of Toledo, OH, USA) for
constructive proofreading and comments.
References
|
1
|
Torre LA, Bray F, Siegel RL, Ferlay J,
Lortet-Tieulent J and Jemal A: Global cancer statistics, 2012. CA
Cancer J Clin. 65:87–108. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Chen W, Zheng R, Baade PD, Zhang S, Zeng
H, Bray F, Jemal A, Yu XQ and He J: Cancer statistics in China,
2015. CA Cancer J Clin. 66:115–132. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Pech O: Nodes or no nodes? The lymph node
metastasis risk of T1 esophageal cancer revisited. J Natl Cancer
Inst. 106:dju1742014. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Vidovic V, Nikolic I, Vukojević J,
Samardzija G, Kukic B, Bogdanović B and Petrović N: Unusual
metastasis of esophageal cancer. Vojnosanit Pregl. 71:975–977.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Lin D and Leichman L: The current status
of neoadjuvant therapy for esophageal cancer. Semin Thorac
Cardiovasc Surg. 26:102–109. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Gaur P, Kim MP and Dunkin BJ: Esophageal
cancer: Recent advances in screening, targeted therapy, and
management. J Carcinog. 13:112014. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Ford HE: Gefitinib for oesophageal cancer:
A cog in need of a wheel? Lancet Oncol. 15:790–791. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
He Z, Wu S, Li Q, Lin Q and Xu J: Use of
the metastatic lymph node ratio to evaluate the prognosis of
esophageal cancer patients with node metastasis following radical
esophagectomy. PloS One. 8:e734462013. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Koga H, Shimada K, Hara Y, Nagano M, Kohga
H, Yokoyama R, Kimura Y, Yuasa S, Magae J, Inamoto S, et al: A
comprehensive approach for establishment of the platform to analyze
functions of KIAA proteins: Generation and evaluation of anti-mKIAA
antibodies. Proteomics. 4:1412–1416. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Nakajima D, Okazaki N, Yamakawa H, Kikuno
R, Ohara O and Nagase T: Construction of expression-ready cDNA
clones for KIAA genes: Manual curation of 330 KIAA cDNA clones. DNA
Res. 9:99–106. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Mete O, Lopes MB and Asa SL: Spindle cell
oncocytomas and granular cell tumors of the pituitary are variants
of pituicytoma. Am J Surg Pathol. 37:1694–1699. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Zhang KQ, Salzman SA, Reding DJ, Suarez
BK, Catalona WJ and Burmester JK: Genetics of prostate cancer. Clin
Med Res. 1:21–28. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Sugita Y, Nakamura Y, Yamamoto M,
Ogasawara S, Ohshima K and Shigemori M: Expression of KIAA 0864
protein in neuroepithelial tumors: An analysis based on the
presence of monoclonal antibody HFB-16. J Neurooncol. 89:151–158.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Chen TC, Lee SA, Hong TM, Shih JY, Lai JM,
Chiou HY, Yang SC, Chan CH, Kao CY, Yang PC and Huang CY: From
midbody protein-protein interaction network construction to novel
regulators in cytokinesis. J Proteome Res. 8:4943–4953. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Tipton AR, Wang K, Oladimeji P, Sufi S, Gu
Z and Liu ST: Identification of novel mitosis regulators through
data mining with human centromere/kinetochore proteins as group
queries. BMC Cell Biol. 13:152012. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Lim YM, Koh I, Park YM, Kim JJ, Kim DS,
Kim HJ, Baik KH, Choi HY, Yang GS, Also-Rallo E, et al: Exome
sequencing identifies KIAA1377 and C5orf42 as susceptibility genes
for monomelic amyotrophy. Neuromuscul Disord. 22:394–400. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
McGuire S: World Cancer Report 2014.
Geneva, Switzerland: World Health Organization, International
Agency for Research on Cancer, WHO Press, 2015. Adv Nutr.
7:418–419. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Shi SR, Shi Y and Taylor CR: Antigen
retrieval immunohistochemistry: Review and future prospects in
research and diagnosis over two decades. J Histochem Cytochem.
59:13–32. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Cardiff RD, Miller CH and Munn RJ: Manual
immunohistochemistry staining of mouse tissues using the
avidin-biotin complex (ABC) technique. Cold Spring Harb Protoc.
2014:659–662. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Percopo CM, Dyer KD, Karpe KA, Domachowske
JB and Rosenberg HF: Eosinophils and respiratory virus infection: A
dual-standard curve qRT-PCR-based method for determining virus
recovery from mouse lung tissue. Methods Mol Biol. 1178:257–266.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Murakami M, Shimada K, Kawai M and Koga H:
InCeP: Intracellular pathway based on mKIAA protein-protein
interactions. DNA Res. 12:379–387. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Hermjakob H, Montecchi-Palazzi L, Bader G,
Wojcik J, Salwinski L, Ceol A, Moore S, Orchard S, Sarkans U, von
Mering C, et al: The HUPO PSI's molecular interaction format-a
community standard for the representation of protein interaction
data. Nat Biotechnol. 22:177–183. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Hall EA, Keighren M, Ford MJ, Davey T,
Jarman AP, Smith LB, Jackson IJ and Mill P: Acute versus chronic
loss of mammalian Azi1/Cep131 results in distinct ciliary
phenotypes. PLoS Genet. 9:e10039282013. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Stelzl U, Worm U, Lalowski M, Haenig C,
Brembeck FH, Goehler H, Stroedicke M, Zenkner M, Schoenherr A,
Koeppen S, et al: A human protein-protein interaction network: A
resource for annotating the proteome. Cell. 122:957–968. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Bonavita R, Walas D, Brown AK, Luini A,
Stephens DJ and Colanzi A: Cep126 is required for pericentriolar
satellite localisation to the centrosome and for primary cilium
formation. Biol Cell. 106:254–267. 2014. View Article : Google Scholar : PubMed/NCBI
|