Introduction
Cancer is the leading cause of morbidity and
mortality worldwide, among which lung cancer accounts for more
mortalities than other cancers (1).
The occurrence of lung cancer is associated with complicated
factors, including family history, infection (2), air pollution and tobacco use (3). Numerous genes are involved in the
occurrence of lung cancer. Among them, numerous members of heat
shock proteins (HSPs) are overexpressed in a wide range of human
cancers and are implicated in tumor cell proliferation,
differentiation and invasion, and also recognition by the immune
system (4).
The heat shock responses were identified by Ritossa
in 1962 in Drosophila in response to elevated temperatures
(5). The heat shock responses are
ubiquitous, existing in all organisms to protect cells against
harmful conditions, including heat shock, oxidative stress or
inflammation (6,7). The synthesis of HSPs is the typical
cellular response to stress. HSPs aid cells to facilitate refolding
or degradation of misfolded and aggregated proteins induced by
stress. HSPs are involved in numerous basic cell processes,
including cell proliferation, cell differentiation and apoptosis.
Cancer is characterized by an aberrant level of cell growth, cell
differentiation and apoptosis. It was previously found that altered
expression of HSPs has been reported in almost all classes of
tumors. Increased levels of HSP27, relative to its level in
non-transformed cells, have been detected in a number of cancers,
such as breast cancer, endometrial cancer and leukemia (6,8). Elevated
expression of members of the HSP70 family has also been reported in
high-grade malignant tumors (9).
HSP90 family members, including Hsp90α and Hsp90β, are
overexpressed in numerous types of cancers (10,11).
A stress-responsive promoter element can be found
upstream of the site of transcription initiation of HSP genes, and
it is termed the heat shock element (HSE). Heat shock factors
(HSFs) can bind to HSEs, and thus regulate the expression of HSPs
(12). In total 3 HSFs (HSF1, HSF2
and HSF4) have been characterized in human cells (13). Among them, HSF1 has been associated
with the occurrence of cancer (14).
HSF4 is also associated with cancer, and inactivation of HSF4
induces cellular senescence and suppresses tumorigenesis in
vivo (15). Thus, in the present
study, the expression level of HSF2 in lung cancer was investigated
and the cellular role of HSF2, including cell proliferation and
cell migration, was characterized.
Materials and methods
Ethics
The present study was approved by the Medical Ethics
Committee of Kunming University of Science and Technology (Kunming,
China). Human samples were used in accordance with the requirements
of Medical Ethics Committee of Kunming University of Science and
Technology, under the guidelines of the World Medical Assembly
(Declaration of Helsinki). Written informed consent was obtained
from the patients' families.
Lung tissue samples
Lung specimens (n=50) were obtained from the tumor
and an adjacent non-cancerous area ≥6 cm from the tumor tissues of
50 patients with lung cancer from Yunnan Province at the First
People's Hospital of Yunnan Province (Kunming, China) between April
2014 and January 2015, as previously described (16,17). The
non-neoplastic tissue was confirmed to lack tumor cell infiltration
using histological analysis. The tissues were immediately placed in
liquid nitrogen and stored at −80°C until use.
RNA extraction and polymerase chain
reaction (PCR)
RNA extraction and first-strand cDNA synthesis was
assessed as previously described (18). For quantitative PCR (qPCR) of HSF2,
the following primers were used: HSF2 forward,
5′-AAGTTCAGGCAGTGATGGCA-3′ and reverse,
5′-TGCACAGAACTAGTGAAAAGATCA-3′; glyceraldehyde 3-phospahte
dehydrogenase forward, 5′-GAAGGTCGGAGTCAACGGAT-3′ and reverse,
5′-GAGGGATCTCGCTCCTGGAAG-3′. qPCR was assessed using a continuous
fluorescence detector (Opticon Monitor; Bio-Rad Laboratories Inc.,
Hercules, CA, USA) and PCR was assessed using an SYBR Green qPCR
kit according to the manufacturer's protocol (Tiangen Bio, Inc.,
Beijing, China) with the following reaction conditions: Initial
denaturation at 95°C for 1 min followed by 40 cycles at 95°C for 15
sec, 60°C for 15 sec and 72°C for 20 sec. Fluorescence curve
analysis was assessed using Opticon Monitor software. The
identities of qPCR products were confirmed by DNA sequencing. The
relative gene expression in 2 different samples were compared using
the 2−ΔΔCq method (19).
Western blot analysis
Western blot analysis of the tissues and the cells
were performed as previously described (16,20,21).
Tissue and cell samples were homogenized in
radioimmunoprecipitation assay buffer containing a protease
inhibitor cocktail (Roche Applied Science, Penzburg, Germany).
Samples (containing 50 µg of protein) were loaded onto sodium
dodecyl sulfate-polyacrylamide electrophoresis gel, electrophoresed
and then electro-transferred onto a polyvinylidene fluoride
membrane. The membrane was subsequently blocked with 3% bovine
serum albumin and incubated with a rabbit anti-human HSF2
polyclonal antibody (dilution, 1:1,000; cat. no. SC-13056; Santa
Cruz Biotechnology, Inc., Dallas, TX, USA) and a horseradish
peroxidase-conjugated goat anti-rabbit IgG secondary antibody
(dilution, 1:5,000; cat. no. SC-2004; Santa Cruz Biotechnology,
Inc.). Protein bands were visualized using Super Signal reagents
(Thermo Fisher Scientific, Inc., Waltham, MA, USA). For detection
of HSP27, HSP47, HSP70 and HSP90, the same methods were used. The
antibodies were purchased from Santa Cruz Biotechnology, Inc and
listed as follows: Rabbit anti-human Hsp27 polyclonal antibody
(dilution, 1:1,000; cat. no. SC-9012); rabbit anti-human Hsp47
polyclonal antibody (dilution, 1:1,000; cat. no. SC-8352); rabbit
anti-human Hsp70 polyclonal antibody (dilution, 1:1,000; cat. no.
SC-25837); and rabbit anti-human Hsp90 polyclonal antibody
(dilution, 1:1,000; cat. no. SC-7947).
HSF2 overexpression plasmid
construction
The cDNA of HSF2 was amplified from normal lung
tissue using an RNA extraction kit (Tiangen Bio, Inc.) and the
first strand cDNA sythesis kit (Takara Biotechnology Co., Ltd.,
Dalian, China). The PCR primers used were as follows: Forward,
5′-CGCGTTCGGGTGTAGAATTT-3′; and reverse,
5′-CATCCCACCCCCGATCTTTC-3′. The cDNA with XhoI and
BamHI restriction sites and flag tag at N-terminal were
amplified from the HSF2 cDNA with the following primers: Forward,
5′-AGATCTCGAGCCTGCGCCGCGTTAACAATGA-3′; and reverse,
5′-AGATGGATCCTTACTTATCGTCGTCATCCTTGTAATCTTAGCTATCAATAAGTGGCAT-3′
(bold and underlined nucleotides are the restriction site of the
enzymes). The PCR products and the plasmid pIRES2-EGFP were
digested with XhoI and BamHI (Takara Biotechnology
Co., Ltd.) and ligated with T4 ligase (Takara Biotechnology Co.,
Ltd.) and transformed into Escherichia coli DH5a competent
cells (Tiangen Bio, Inc.). The positive clones were confirmed by
sequencing, then the plasmid was amplified and purified for cell
transfection.
Cell culture and transfection
The BEAS-2B and A549 cells were cultured in DMEM/F12
with 10% fetal bovine serum (FBS). The cells were grown in a
humidified atmosphere containing 5% CO2 at 37°C.
Transfection of cells was performed using Lipofectamine 2000
(Invitrogen; Thermo Fisher Scientific, Inc.) as previously
described (22).
Cell proliferation assay
The cells were seeded onto a 96-well plate (1,000
cells/well) 24 h following transfection. Fresh medium supplemented
with 10% FBS was added into the plates and changed every 24 h. The
number of viable cells was estimated by the methyl thiazolyl
tetrazolium (MTT) method every 24 h.
Cell migration assay
Cell migration was tested with a Boyden chamber
assay, which utilized 24-well plates (Corning Incorporated,
Corning, NY, USA) and Transwell plates (8 µm pore; EMD Millipore,
Billerica, MA, USA). The bottom side of the Transwell plates were
coated with collagen type 1 (10 µg/ml; Sigma-Aldrich; Merck
Millipore, Darmstadt, Germany). Subsequent to being starved for 12
h with starvation medium (DMEM/F12) containing 0.25% bovine serum
albumin, the cells were detached with cell dissociation buffer,
then 1×105 cells were planted into the top of each
chamber. Medium containing 1% FBS was added into the bottom of the
chamber for 16 h. Migrated cells were dyed with crystal violet.
Bound crystal violet was eluted with 1 ml 10% acetic acid and the
migration activity was expressed as the value monitored at 586 nm
of extraction.
Statistical analysis
Differences in the numerical data between the cancer
and adjacent normal tissues were evaluated using the paired
Wilcoxon test. The in vitro data were analyzed with
Student's t-test for variance. Experimental values were expressed
as the mean ± standard deviation. A value of P<0.05 was
considered to indicate a statistically significant difference.
Results
HSF2 is upregulated in tumor tissues
in lung cancer patients
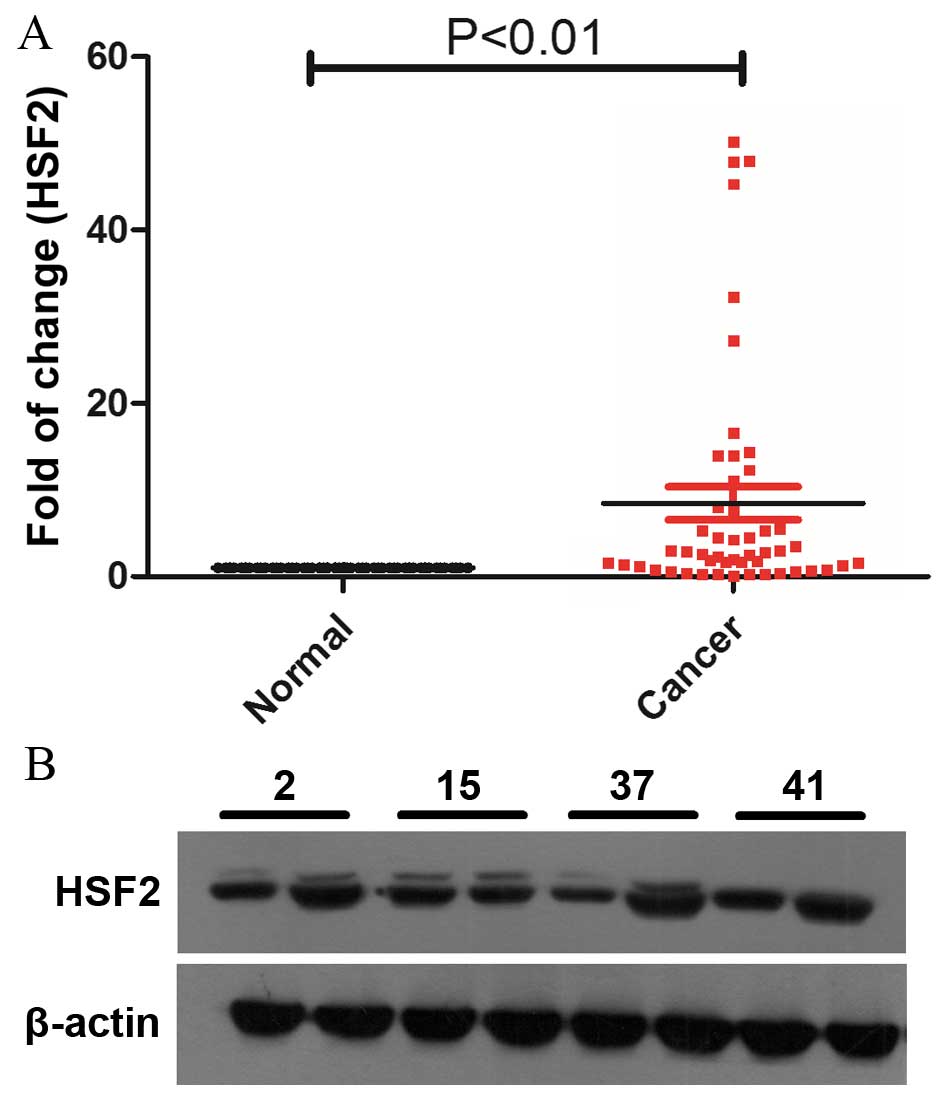
A total of 50 matched normal lung and lung cancer
tissues were analyzed with RT-qPCR to detect the expression level
of HSF2 (Fig. 1A). The results showed
that 76% (38/50) of the HSF2 expression levels in cancer tissues
were upregulated compared with the normal lung tissues (P=0.0003;
Fig. 1A). The protein levels of HSF2
were also detected by western blotting, and the results showed that
the protein levels of HSF2 were upregulated, which is consistent
with the results from RT-qPCR (Fig.
1B).
HSF2 overexpression enhances the cell
proliferation and cell migration in human lung epithelial cells and
lung cancer cells
Since HSF2 is upregulated in lung cancer, it is
reasonable to infer that it may affect the basic routine of lung
epithelial cells and cancer cells, including cell proliferation and
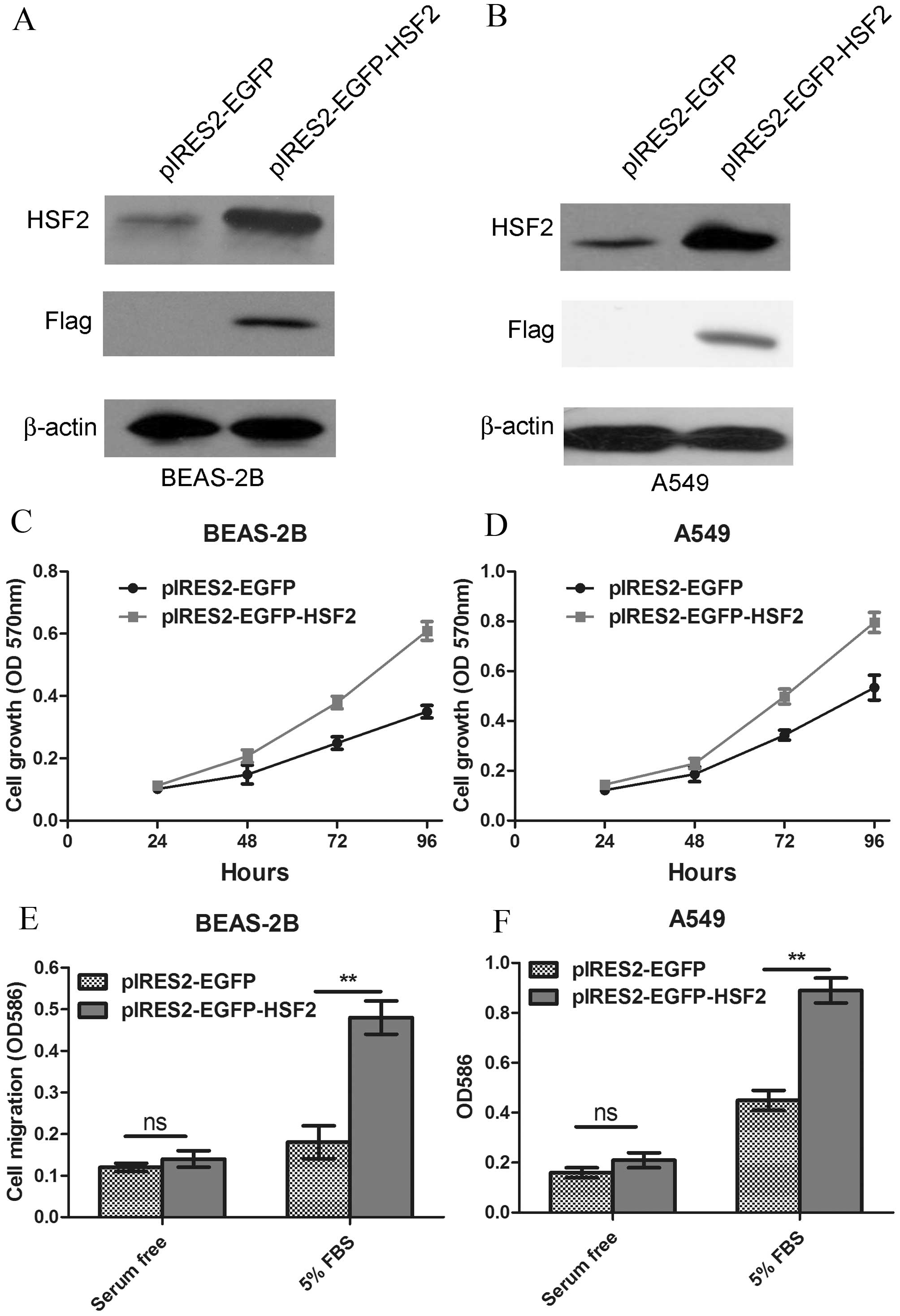
cell migration. Thus, a plasmid encoding HSF2 was constructed and
transfected into human lung epithelia cell line BEAS-2B (Fig. 2A). The growth of HSF2-overexpressing
BEAS-2B cells was monitored by MTT assay. The results showed that
the HSF2 overexpression markedly enhanced the proliferation ability
of BEAS-2B cells (P=0.0006; Fig. 2C).
The effect of HSF2 overexpression on the lung cancer A549 cell line
was also investigated (Fig. 2B), and
similar results were acquired (P=0.0021; Fig. 2D). Apart from cell proliferation,
enhanced cell migration was another characteristic of
tumorigenesis. Thus, cell migration was tested using a Boyden
chamber assay. The results showed that HSF2 overexpression
significantly promoted the cell migration of BEAS-2B cells and A549
cells (Fig. 2E and F).
HSF2 overexpression promotes the
expression of HSPs
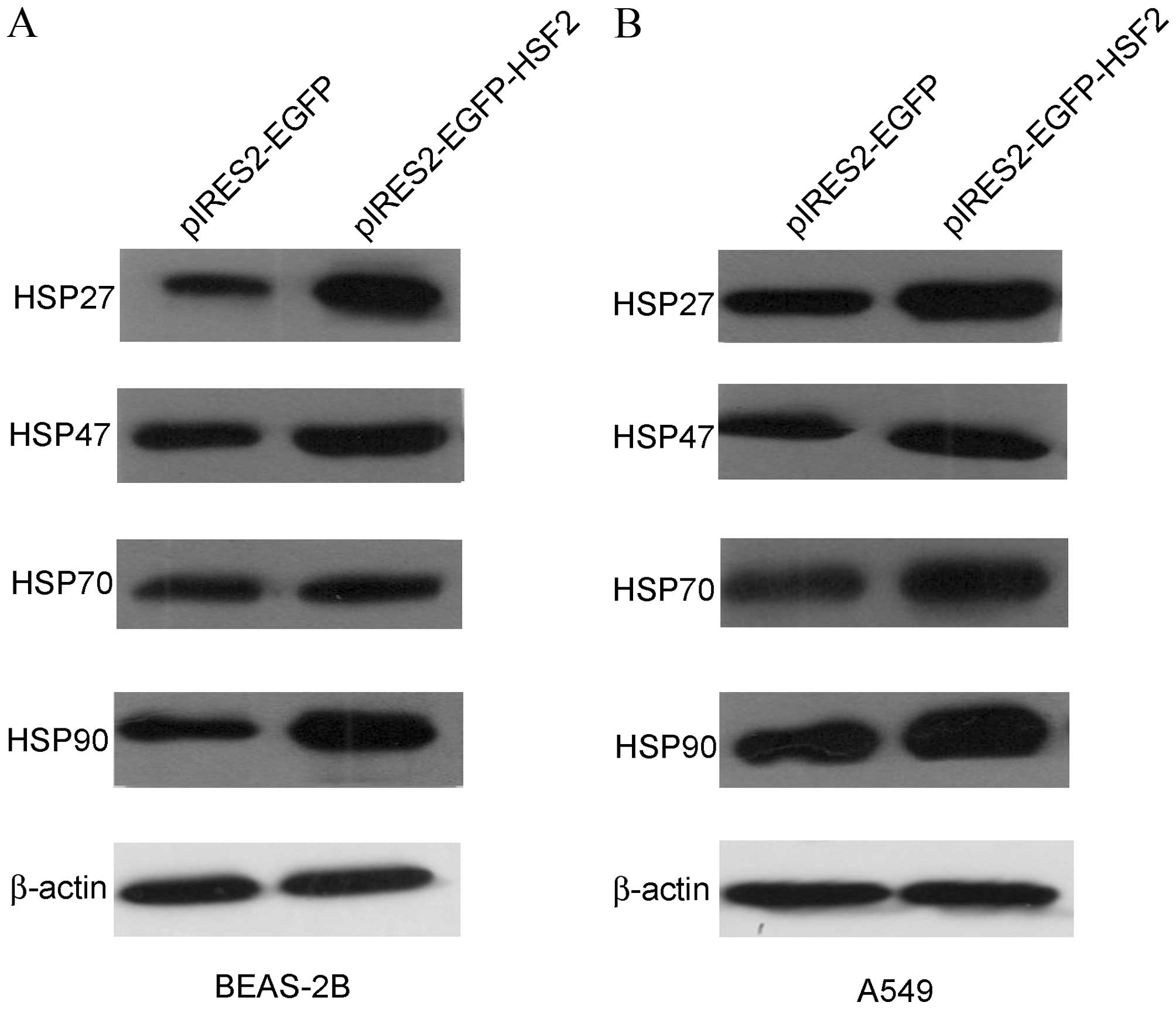
As the upstream regulator of HSP, HSF2 may regulate
the expression level of HSP27, HSP47, HSP70 and HSP90. The present
study additionally tested whether HSF2 overexpression can affect
the expression of these HSPs. Subsequent to overexpression of HSF2
in BEAS-2B cells, the expression level of HSPs was semi-quantified
by western blot analysis. All 4 HSPs, consisting of HSP27, HSP47,
HSP70 and HSP90, were upregulated following HSF2 overexpression.
Among them, HSP47 and HSP70 were only slightly upregulated, but the
expression level of HSP27 and HSP90 were increased (Fig. 3A). However, HSF2 overexpression
upregulated all 4 HSPs in A549 cells (Fig. 3B). These inconsistent results suggest
that the regulation mechanism through which HSF2 affects the
expression of HSPs may be different in normal epithelial cells and
cancer cells.
HSF2 promoted cell proliferation and
cell migration is dependent of its induced upregulation of HSP27
and HSP90
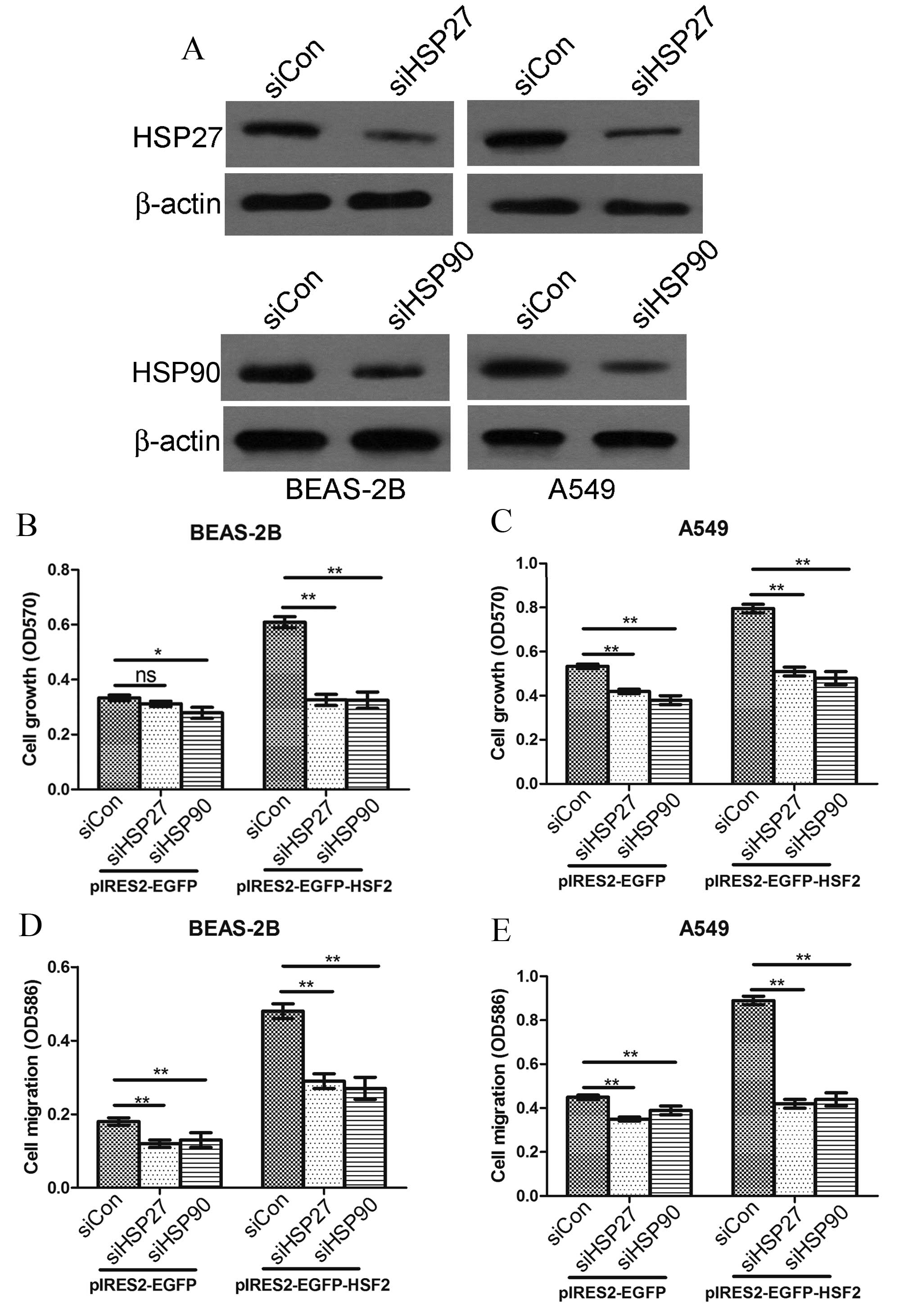
Since HSF2 overexpression can promote lung
epithelial cell and lung cancer cell proliferation and migration,
and at the same time upregulate HSP, particularly HSP27 and HSP90,
it is reasonable to infer that HSP27 and HSP90 is pivotal for HSF2
functioning in promoting cell proliferation and cell migration.
Thus, in the present study, HSP27 or HSP90 siRNA (or a combination
of the two) were co-transfected with HSF2-overexpressing plasmids
into BEAS-2B and A549 cells to assess the role of HSP27 and HSP90
in HSF2 function (Fig. 4A). The
results showed that HSP27 and HSP90 RNA interference significantly
attenuated HSF2 enhanced cell growth (Fig. 4B) and cell migration (Fig. 4D) on BEAS-2B cells. Similar results
were obtained in A549 cells (Fig. 4C and
E).
Discussion
Cancer is the leading cause of morbidity and
mortality worldwide, and lung cancer accounts for a large
proportion. Therefore, exploring the molecular and cellular
mechanisms of lung cancer is important. Early screening strategies
can be developed, and deciphering the mechanism underlying
tumorigenesis may facilitate the development of new strategies to
successfully treat cancer and cancer-associated diseases (23).
Increasing evidence suggests that changes in the
cellular microenvironment contribute to tumorigenesis (24,25). There
are numerous bodily responses to microenvironmental changes,
including inflammation. Among these responses, heat shock responses
are the most conserved among all types of life, from bacterial to
mammalian (26). Heat shock responses
are generally accepted to be associated with tumor growth and
metastasis (4). Numerous studies have
focused on the role of HSPs on tumor growth and metastasis
(27–29). As the upstream regulators of HSPs,
HSFs may be also involved in cancer. Among the 3 members of HSFs,
HSF1 has been well characterized for its association with cancer. A
clinical study showed that high levels of HSF1 are associated with
poor prognosis in breast cancer (30). It is also reported that HSF1 is
involved in tumor progression via regulation of hypoxia-inducible
factor 1 and RNA-binding protein HuR (31). However, the other two members, HSF2
and HSF4, have not been well characterized in cancer. Thus, in the
present study, the expression level of HSF2 in lung cancer tissues
was detected in 50 lung cancer patients. It was found that HSF2
level is upregulated in the majority of lung cancer tissues,
suggesting that HSF2 may be involved in lung cancer growth and/or
metastasis.
HSF1 is activated in response to the traditional
heat shock stimuli. However, it is generally accepted that HSF2 is
not activated in response to classical stress stimuli, but under
developmentally associated conditions (32,33).
Coincidentally, with the recent progress in developmental biology
and cancer biology, more and more similarities between early embryo
development and tumorigenesis have been found (34). Thus, the upregulation of HSF2 in
tumorigenesis complements current research. Therefore, in the next
step, the biological effect of HSF2 on basic cellular effects was
tested. The results showed that HSF2 promoted cellular
proliferation and cellular migration in vitro, which may
contribute to tumorigenesis in vivo.
The underlying mechanism of HSF2 promoting cell
proliferation and cell migration may be complicated. A previous
study showed that HSF2 binds to the HSP90, HSP27, HSP70 and c-Fos
and modulates their expression (35).
In the present study, it was assessed whether HSF2 affected the
expression of these HSPs, and found that HSF2 overexpression indeed
promoted the expression of HSP27, HSP47, HSP70 and HSP90,
particularly HSP27 and HSP90. This may partially explain why HSF2
promotes cell proliferation and cell migration. The knockdown of
HSP27 or HSP90 can significantly attenuate the HSF2 induced cell
proliferation and cell migration. However, additional research is
required to understand the role of HSF2 in cancer. The involvement
of certain signaling pathways, including the MAP kinase pathway,
which utilizes ERK1/2, P38 and JNK in HSF2-regulated cell
proliferation and cell motility, may be investigated in additional
studies. There are a limited number of studies about the HSF2
expression and human diseases. A previous study showed that HSF2 is
upregulated in inflammatory bowel diseases by enhancing the
production of inflammatory cytokines. Inflammation, particularly
chronic inflammation, is closely associated with the occurrence of
cancer. Whether it is the same or a similar condition in lung
cancer requires investigation. Additional detailed studies are
required to answer whether the upregulation of HSF is the cause or
the result of lung cancer, which is important for understanding the
association between HSF2 upregulation and lung cancer
occurrence.
In conclusion, to the best of our knowledge, the
present study showed for the first time that HSF2 is upregulated in
lung cancer and may be involved in tumorigenesis. HSF2 asserts its
biological function by upregulating the expression of HSP and
consequently affecting cell proliferation and cell migration.
Whether HSF2 acts alone or with other proteins may require
additional investigation. In addition, whether HSF2 is a good
diagnostic marker or drug target requires additional research.
Acknowledgements
This study was supported by Yunnan Department of
Natural Science and Kunming Medical University (grant no.,
2013FB197) and The First People's Hospital of Yunnan Province
Internal Institutions (grant no., 2016NS204).
References
|
1
|
Siegel R, Naishadham D and Jemal A: Cancer
statistics, 2013. CA Cancer J Clin. 63:11–30. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Zhan P, Suo LJ, Qian Q, Shen XK, Qiu LX,
Yu LK and Song Y: Chlamydia pneumoniae infection and lung cancer
risk: A meta-analysis. Eur J Cancer. 47:742–747. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Hecht SS: Tobacco smoke carcinogens and
lung cancer. J Natl Cancer Inst. 91:1194–1210. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Ciocca DR and Calderwood SK: Heat shock
proteins in cancer: Diagnostic, prognostic, predictive, and
treatment implications. Cell Stress Chaperones. 10:86–103. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Ritossa F: A new puffing pattern induced
by temperature shock and DNP in Drosophila. Experientia.
18:571–573. 1962. View Article : Google Scholar
|
|
6
|
Jolly C and Morimoto RI: Role of the heat
shock response and molecular chaperones in oncogenesis and cell
death. J Natl Cancer Inst. 92:1564–1572. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Morimoto RI: Cells in stress:
Transcriptional activation of heat shock genes. Science.
259:1409–1410. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Ciocca DR, Oesterreich S, Chamness GC,
McGuire WL and Fuqua SA: Biological and clinical implications of
heat shock protein 27,000 (Hsp27): A review. J Natl Cancer Inst.
85:1558–1570. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Ralhan R and Kaur J: Differential
expression of Mr 70,000 heat shock protein in normal, premalignant,
and malignant human uterine cervix. Clin Cancer Res. 1:1217–1222.
1995.PubMed/NCBI
|
|
10
|
Jameel A, Skilton RA, Campbell TA, Chander
SK, Coombes RC and Luqmani YA: Clinical and biological significance
of HSP89 alpha in human breast cancer. Int J Cancer. 50:409–415.
1992. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Yufu Y, Nishimura J and Nawata H: High
constitutive expression of heat shock protein 90 alpha in human
acute leukemia cells. Leuk Res. 16:597–605. 1992. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Wu C: Heat shock transcription factors:
Structure and regulation. Annu Rev Cell Dev Biol. 11:441–469. 1995.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Morimoto RI: Regulation of the heat shock
transcriptional response: Cross talk between a family of heat shock
factors, molecular chaperones and negative regulators. Genes Dev.
12:3788–3796. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Engerud H, Tangen IL, Berg A, Kusonmano K,
Halle MK, Oyan AM, Kalland KH, Stefansson I, Trovik J, Salvesen HB
and Krakstad C: High level of HSF1 associates with aggressive
endometrial carcinoma and suggests potential for HSP90 inhibitors.
Br J Cancer. 111:78–84. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Jin X, Eroglu B, Cho W, Yamaguchi Y,
Moskophidis D and Mivechi NF: Inactivation of heat shock factor
Hsf4 induces cellular senescence and suppresses tumorigenesis in
vivo. Mol Cancer Res. 10:523–534. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Jiang P, Yu GY and Zhang Y, Xiang Y, Hua
HR, Bian L, Wang CY, Lee WH and Zhang Y: Down-regulation of
protease-activated receptor 4 in lung adenocarcinoma is associated
with a more aggressive phenotype. Asian Pac J Cancer Prev.
14:3793–3798. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Yu G, Jiang P, Xiang Y and Zhang Y, Zhu Z,
Zhang C, Lee S, Lee W and Zhang Y: Increased expression of
protease-activated receptor 4 and trefoil factor 2 in human
colorectal cancer. PLoS One. 10:e01226782015. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Xiang Y, Gao Q, Su W, Zeng L, Wang J, Hu
Y, Nie W, Ma X and Zhang Y, Lee W and Zhang Y: Establishment,
characterization and immortalization of a fibroblast cell line from
the Chinese red belly toad Bombina maxima skin. Cytotechnology.
64:95–105. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) Method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Xiang Y, Wang X, Yan C, Gao Q, Li SA, Liu
J, Zhou K, Guo X, Lee W and Zhang Y: Adenosine-5′-triphosphate
(ATP) protects mice against bacterial infection by activation of
the NLRP3 inflammasome. PLoS One. 8:e637592013. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Wang YJ, Guo XL, Li SA, Zhao YQ, Liu ZC,
Lee WH, Xiang Y and Zhang Y: Prohibitin is involved in the
activated internalization and degradation of protease-activated
receptor 1. Biochim Biophys Acta. 1843:1393–1401. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Huang F, Lin C, Shi YH and Kuerban G:
MicroRNA-101 inhibits cell proliferation, invasion and promotes
apoptosis by regulating cyclooxygenase-2 in Hela cervical carcinoma
cells. Asian Pac J Cancer Prev. 14:5915–5920. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Lei YM, Zu YF, Wang J, Bai S, Shi YF, Shi
R, Duan J, Cui D, Chen J, Xiang Y and Dong J:
Interleukin-1β-mediated suppression of microRNA-101 and
upregulation of enhancer of zeste homolog 2 is involved in
particle-induced lung cancer. Med Oncol. 32:3872015. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Hu M, Yao J, Cai L, Bachman KE, van den
Brûle F, Velculescu V and Polyak K: Distinct epigenetic changes in
the stromal cells of breast cancers. Nat Genet. 37:899–905. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Gao F, Liang B, Reddy ST, Farias-Eisner R
and Su X: Role of inflammation-associated microenvironment in
tumorigenesis and metastasis. Curr Cancer Drug Targets. 14:30–45.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Fulda S, Gorman AM, Hori O and Samali A:
Cellular stress responses: Cell survival and cell death. Int J Cell
Biol. 2010:2140742010. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Tan JS, Ong Kc KC and Rhodes A: The role
of heat shock proteins and glucose regulated proteins in cancer.
Malays J Pathol. 38:75–82. 2016.PubMed/NCBI
|
|
28
|
Conroy SE and Latchman DS: Do heat shock
proteins have a role in breast cancer? Br J Cancer. 74:717–721.
1996. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Lianos GD, Alexiou GA, Mangano A, Mangano
A, Rausei S, Boni L, Dionigi G and Roukos DH: The role of heat
shock proteins in cancer. Cancer Lett. 360:114–118. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Santagata S, Hu R, Lin NU, Mendillo ML,
Collins LC, Hankinson SE, Schnitt SJ, Whitesell L, Tamimi RM,
Lindquist S and Ince TA: High levels of nuclear heat-shock factor 1
(HSF1) are associated with poor prognosis in breast cancer. Proc
Natl Acad Sci USA. 108:18378–18383. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Gabai VL, Meng L, Kim G, Mills TA,
Benjamin IJ and Sherman MY: Heat shock transcription factor Hsf1 is
involved in tumor progression via regulation of hypoxia-inducible
factor 1 and RNA-binding protein HuR. Mol Cell Biol. 32:929–940.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Loones MT, Rallu M, Mezger V and Morange
M: HSP gene expression and HSF2 in mouse development. Cell Mol Life
Sci. 53:179–190. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Pirkkala L, Nykänen P and Sistonen L:
Roles of the heat shock transcription factors in regulation of the
heat shock response and beyond. FASEB J. 15:1118–1131. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Ma Y, Zhang P, Wang F, Yang J, Yang Z and
Qin H: The relationship between early embryo development and
tumourigenesis. J Cell Mol Med. 14:2697–2701. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Wilkerson DC, Skaggs HS and Sarge KD: HSF2
binds to the Hsp90, Hsp27 and c-Fos promoters constitutively and
modulates their expression. Cell Stress Chaperones. 12:283–290.
2007. View Article : Google Scholar : PubMed/NCBI
|