Introduction
Osteosarcoma is a type of malignant bone tumor,
which is characterized by pain and bone destruction, and may
potentially lead to pathological bone fracture with the progression
of tumor growth. Patients with osteosarcoma often exhibit reduced
activity in daily life, as well as reduced quality of life. Tumor
growth stimulates osteoclast activity, resulting in bone resorption
and bone fracture (1). Therefore,
promoting osteogenic differentiation over osteoclast activity,
without increasing tumor cell proliferation, migration and
invasion, may be helpful for patients by reducing pain and
improving their quality of life. The SaOS2 human osteosarcoma cell
line resembles osteoblasts in their differentiative, proliferative
and mineralization ability. SaOS2 cells are able to differentiate
and form hydroxyapatite nodules under osteogenic conditions, which
are critical for bone formation. In addition, SaOS2 cells can be
induced to form mineral nodules, and express several markers of
bone differentiation, including osteocalcin (OCN), alkaline
phosphatase (ALP) and runt-related transcription factor 2 (Runx2),
in medium containing dexamethasone, β-glycerophosphate and ascorbic
acid. These previous findings suggested that SaOS2 may possess
potent osteogenic capacity (2,3).
Osteosarcoma is generally considered a disease
associated with differentiation, which is caused by genetic and
epigenetic disruptions in the terminal differentiation of
osteoblasts. In addition, >80% of cases of osteosarcoma are
poorly differentiated histopathologically; therefore, novel
therapeutic strategies based on the non-cytotoxic induction of cell
differentiation-responsive pathways may represent a significant
advance (4).
Interferon-induced transmembrane protein 5 (IFITM5)
encodes bone-restricted IFITM-like protein (BRIL), which is
involved in mineralization and is expressed in the skeleton. IFITM5
was initially observed at embryonic day 14.5, when undifferentiated
cells differentiate into osteoblasts and begin to form mineralized
structures (5,6). IFITM5 is highly expressed at the early
stage of mineralization and is considered to serve an essential
role in bone formation. Furthermore, IFITM proteins are involved in
early development, cell adhesion and cell growth regulation; and
therefore may be considered to act as tumor suppressors (7). The C>T transition at position-14 of
the 5′ untranslated region of IFITM5 has previously been identified
as the underlying cause of Type V osteogenesis imperfecta (OI)
(8,9).
It has been reported that Type V OI primary osteoblasts display
increased mineralization, despite decreased collagen type I, alpha
1 expression (10). However, to the
best of our knowledge, there are currently no reports on the
effects of IFITM5 on proliferation, migration and osteogenic
differentiation of human osteosarcoma cells. Therefore, there
remains a need to investigate the effects of IFITM5 and IFITM5
c.-14C>T mutation on cancer cells. The present study
investigated the effects of IFITM5 and IFITM5 c.-14C>T mutation
transfection on tumor proliferation, migration, invasion and
osteogenic differentiation in human osteosarcoma cells.
Materials and methods
Materials
Dexamethasone, ascorbate acid, Alizarin Red S, 10%
cetylpyridinium chloride and β-glycerophosphate sodium were
purchased from Sigma-Aldrich; Merck Millipore (Darmstadt, Germany).
McCoy's 5A medium and fetal bovine serum (FBS) were obtained from
Gibco; Thermo Fisher Scientific, Inc. (Waltham, MA, USA).
Penicillin and streptomycin (P/S) were purchased from Beyotime
Institute of Biotechnology (Haimen, China).
For transfection, X-tremeGENE™ HP DNA Transfection
Reagent was purchased from Roche Diagnostics (Basel, Switzerland).
Endo-Free Plasmid Mini kit II was purchased from Omega Bio-Tek,
Inc. (Norcross, GA, USA). Wild type IFITM5 (pcDNA4-IFITM5-E12-W)
and IFITM5 c-.14C>T mutation (pcDNA4-IFITM5-E12-MU) plasmids
were derived from pcDNA4 plasmids obtained from the Chinese Academy
of Sciences Cell Bank (Shanghai, China).
For proliferation and invasion assays,
3-dimethylthiazol-2,5-diphenyltetrazolium bromide (MTT), dimethyl
sulfoxide (DMSO) and Wright-Giemsa stain were obtained from Beijing
Solarbio Science & Technology Co., Ltd. (Beijing, China).
Hoechst kit was purchased from Beyotime Institute of Biotechnology.
MTT solution was dissolved in PBS to ensure the final concentration
reached 5 mg/ml.
For western blotting, bicinchoninic acid (BCA)
reagent kit and Tris-buffered saline-0.1% Tween 20 (TBST) were
purchased from Beyotime Institute of Biotechnology.
SDS-polyacrylamide gel electrophoresis gels were purchased from
Bio-Rad Laboratories, Inc. (Hercules, CA, USA). Polyvinylidene
difluoride (PVDF) membranes were purchased from EMD Millipore
(Billerica, MA, USA). Rabbit polyclonal antibodies against IFITM5
(SAB2105607) and GAPDH (SAB4300645) were obtained from
Sigma-Aldrich; Merck Millipore. Horseradish peroxidase
(HRP)-conjugated goat anti-rabbit immunoglobulin G (IgG; A0208) was
obtained from Beyotime Institute of Biotechnology.
For reverse transcription quantitative polymerase
chain reaction (RT-qPCR), TRIzol® reagent was purchased
from Invitrogen; Thermo Fisher Scientific, Inc. Random Primers,
dNTP mixture, 5X PrimeScript buffer, RNase inhibitor (40 U/µl) and
PrimeScript reverse transcriptase (200 U/µl) used in the reverse
transcription were obtained from Takara Bio, Inc. (Otsu, Japan).
SYBR Green used for qPCR was purchased from Roche Diagnostics.
Cell culture, osteogenic
differentiation and transfection
SaOS2 cells were obtained from the Chinese Academy
of Sciences Cell Bank. The cells were stored in liquid nitrogen and
were thawed in a 37°C water bath prior to culture. The cells were
cultured in McCoy's 5A medium supplemented with 10% FBS and 1% P/S.
The cultures were incubated at 37°C in a 95% humidified atmosphere
containing 5% CO2. Once the cells reached 90%
confluence, they were detached by mild treatment with trypsin.
SaOS2 cells were plated at a density of
2×104 cells/well in 96-well plates for 24 h prior to
transfection. Transfection was performed once the cells reached
80–90% confluence. The pcDNA4 plasmids were obtained from
Escherichia coli, according to the Endo-Free Plasmid Mini
kit II protocol. Construction of the plasmid IFITM5 3′untranslated
region (UTR) including the predicted binding site of miR-762 was
amplified by RT-PCR and inserted into multiple cloning sites of the
T-Vector pMD19 (pMD19-UTR) (Takara Bio, Inc.) using the SacI
and XbaI restriction sites. A site-directed gene mutagenesis
kit (Takara Bio, Inc.) was used to construct a mutant type of
miR-762-binding site vector (pMD19-mUTR) with 4 base mutations
within the seed region in accordance with the manufacturer's
protocol. SaOS2 cells were transfected with 0.01 µg/µl
pcDNA4-IFITM5-E12-W, which contains IFITM5, and
pcDNA4-IFITM5-E12-MU, which contains IFITM5 c.-14C>T mutation,
using X-tremeGENE™ HP DNA Transfection Reagent. The control (C)
group was treated with the same volume of X-tremeGENE™ HP DNA
Transfection Reagent. The cells were harvested for protein and mRNA
expression analyses at 24, 48 and 72 h post-transfection.
In addition, SaOS2 cells (8×104) were
cultured overnight in 24-well plates and were transfected with
pcDNA4-IFITM5-E12-W or pcDNA4-IFITM5-E12-MU using X-treme GENE™ HP
DNA Transfection Reagent for 3 days. The control group was just
treated with the transfection reagent for 3 days. Subsequently, the
cells were induced under osteogenic conditions (McCoy's 5A medium
supplemented with 10% FBS, 1% P/S, l0−8 M dexamethasone,
50 µg/ml ascorbate acid and 10 mmol/l β-glycerophosphate sodium).
After 3 days, the cells were harvested for protein and mRNA
analysis.
SaOS2 cell proliferation assay
To determine the effects of IFITM5 and IFITM5
c.-14C>T mutation overexpression on cell proliferation, an MTT
assay was carried out, according to the manufacturer's protocol.
The assay was conducted 24, 48 and 72 h post-transfection of cells
plated in a 96-well plate. Briefly, 20 µl MTT was added to each
well and incubated at 37°C for 4 h. After draining off the solution
in the well, 120 µl DMSO was added to each well and thoroughly
mixed. Subsequently, absorbance of the plate was read at 492 nm,
after complete elution, using a microtiter plate reader.
SaOS2 cell migration assay
SaOS2 cells were plated at a density of
3×105/well in 6-well plates and were cultured until
confluent. The confluent cells were scraped with a pipette tip and
cells scratched off were washed with PBS. The wells of each 6-well
plate were divided into three groups: W group, which was
transfected with pcDNA4-IFITM5-E12-W; MU group, which was
transfected with pcDNA4-IFITM5-E12-MU; and C group, which remained
untransfected. Cell migration into the wound surface was observed
at 0 and 48 h post-transfection by microscopy. Cells that migrated
into the wound surface were counted and the average number of
nuclei was determined by counting five random fields per well under
an optical microscope. Each experiment was performed in
duplicate.
SaOS2 cell apoptosis assay
SaOS2 cells were plated at a density of
3×105/well in 6-well plates containing cell slides, and
were cultured until they reached confluence. The W group was
transfected with pcDNA4-IFITM5-E12-W, the MU group was transfected
with pcDNA4-IFITM5-E12-MU, and the C group was not transfected. A
total of 48 h post-transfection, cells were fixed with 0.5 ml fix
solution (C0003-1; Beyotime Institute of Biotechnology) per well
for 10 min. After three washes, 0.5 ml Hoechst 33258 dye solution
was added to each well. A drop of anti-fade mounting medium was
added to the slides before they were covered. The average number of
nuclei was assessed by counting five random fields per well under
an optical microscope. Each experiment was performed in
duplicate.
SaOS2 cell Transwell migration
assay
Cells were seeded in the upper chamber of Transwell
plates and cultured at a density of 2×104/well. After,
24 h cells were transfected with pcDNA4-IFITM5-E12-W or
pcDNA4-IFITM5-E12-MU for 48 h at 37°C. The medium in the upper
chamber was then replaced with FBS-free medium, and medium
supplemented with 20% FBS was added to the lower chamber. After a
24 h incubation at 37°C, the upper sides of the filters were
carefully washed three times with PBS, and cells remaining on the
upper sides were removed with a cotton wool swab. The Transwell
filters were then fixed with 4% paraformaldehyde for 30 min, washed
three times with PBS, and stained with Wright-Giemsa stain for 1 h.
Cells that had migrated to the bottom side of the filter were
counted under an optical microscope, and the average number of
cells was determined by counting five random fields per filter.
Each experiment was conducted in duplicate.
Alizarin Red S staining
After 3 days of culturing in osteogenic medium,
group C, W and MU cells were washed three times with PBS and fixed
with 500 µl 4% paraformaldehyde for 20 min. The fixed cells were
then stained with 500 µl 0.1% Alizarin red S solution (pH 8.3). For
quantification, 10% cetylpyridinium chloride solution was added to
each well to elute the dye. The absorbance of the eluted solution
was measured at 560 nm using a microtiter plate reader after
complete elution.
RT-qPCR
Total RNA was isolated from the cells using
TRIzol® reagent and was reverse-transcribed into cDNA,
according to the manufacturer's protocol.
qPCR was performed using specific primers, the
SYBR® PrimeScript® RT-PCR kit, and with cDNA
as a template, using an Applied Biosystems 7500 Real-Time PCR
system (Applied Biosystems; Thermo Fisher Scientific, Inc.). The
qPCR reaction mixture consisted of 5 µl SYBR Green PCR Master mix
(Roche Diagnostics), 1 µl cDNA, 1 µl forward primer (Beijing
Genomics Institute, Shenzhen, China), 1 µl reverse primer (Beijing
Genomics Institute) and 2 µl RNase-free ddH2O (Tiangen
Biotech Co., Ltd., Beijing, China). The total PCR reaction volume
was 10 µl. cDNA was used to conduct gene-specific PCR for IFITM5,
Runx2, ALP and OCN. The primers used were as follows: IFITM5,
forward 5′-TTGATCTGGTCGGTGTTCAG-3′, reverse
5′-GTCAGTCATAGTCCGCGTCA-3′; Runx2, forward
5′-GCCGGGAATGATGAGAACTA-3′, reverse 5′-GGTGAAACTCTTGCCTCGTC-3′;
ALP, forward 5′-TGGCTCTGCCTTTATTCCCTAGT-3′, reverse
5′-AAATAAGGTGCTTTGGGAATCTGT-3′; OCN, forward
5′-GCCATCACCCTGTCTCCTAA-3′, reverse 5′-GCTGTGGAGAAGACACACGA-3′; and
GAPDH, forward 5′-CACCATCTTCCAGGAGC-3′ and reverse
5′-AGTGGACTCCACGACGTA-3′. The qPCR was carried out according to the
following cycling conditions: 45 cycles at 94°C for 2 min, 94°C for
30 sec, 60°C for 30 sec, 72°C for 40 sec and 72°C for 5 min. The
relative levels of target gene transcripts were normalized to the
control gene GAPDH by 2−ΔΔCq (11).
Western blot analysis
Cells were harvested for IFITM5 protein expression
analysis 72 h post-transfection with pcDNA4-IFITM5-E12-W or
pcDNA4-IFITM5-E12-MU. radioimmunoprecipitation lysis buffer
(Beyotime Institute of Biotechnology) was added to cells and lysed
for 30 min on ice. Protein was obtained from the cell lysates by
centrifugation at 16,363 × g for 15 min at 4°C, and protein
concentrations were determined using the BCA reagent. The samples
were heated at 95°C for 5 min, separated by 10% SDS-PAGE, and
transferred to methanol-activated PVDF membranes. After blocking
with 5% defatted milk in TBST for 2 h at room temperature, the
membranes were incubated with rabbit anti-IFITM5 and anti-GAPDH
polyclonal antibodies (diluted 1:1,000 in TSBT) at 4°C overnight,
followed by incubation with HRP-conjugated IgG secondary antibodies
(diluted 1:5,000 in TBST) at 4°C for 1 h. The protein bands were
observed by UMAX PowerLook 2100XL-USB (Umax Technologies, Dallas,
TX, USA) and quantitatively analyzed for pixel value by ImageJ
analysis system (version 1.48u; National Institutes of Health,
Bethesda, MD, USA). The relative level of target protein was
presented as the ratio of pixel value for the target protein to
pixel value for GAPDH.
Statistical analysis
Data are presented as the mean ± standard deviation.
One way analysis of variance (for initial multiple comparisons) and
post-hoc Least Significant Difference tests (comparison between two
groups) were conducted for data analysis. Statistical analyses were
performed using SPSS version 17.0 (SPSS, Inc., Chicago, IL, USA)
P<0.05 was considered to indicate a statistically significant
difference.
Results
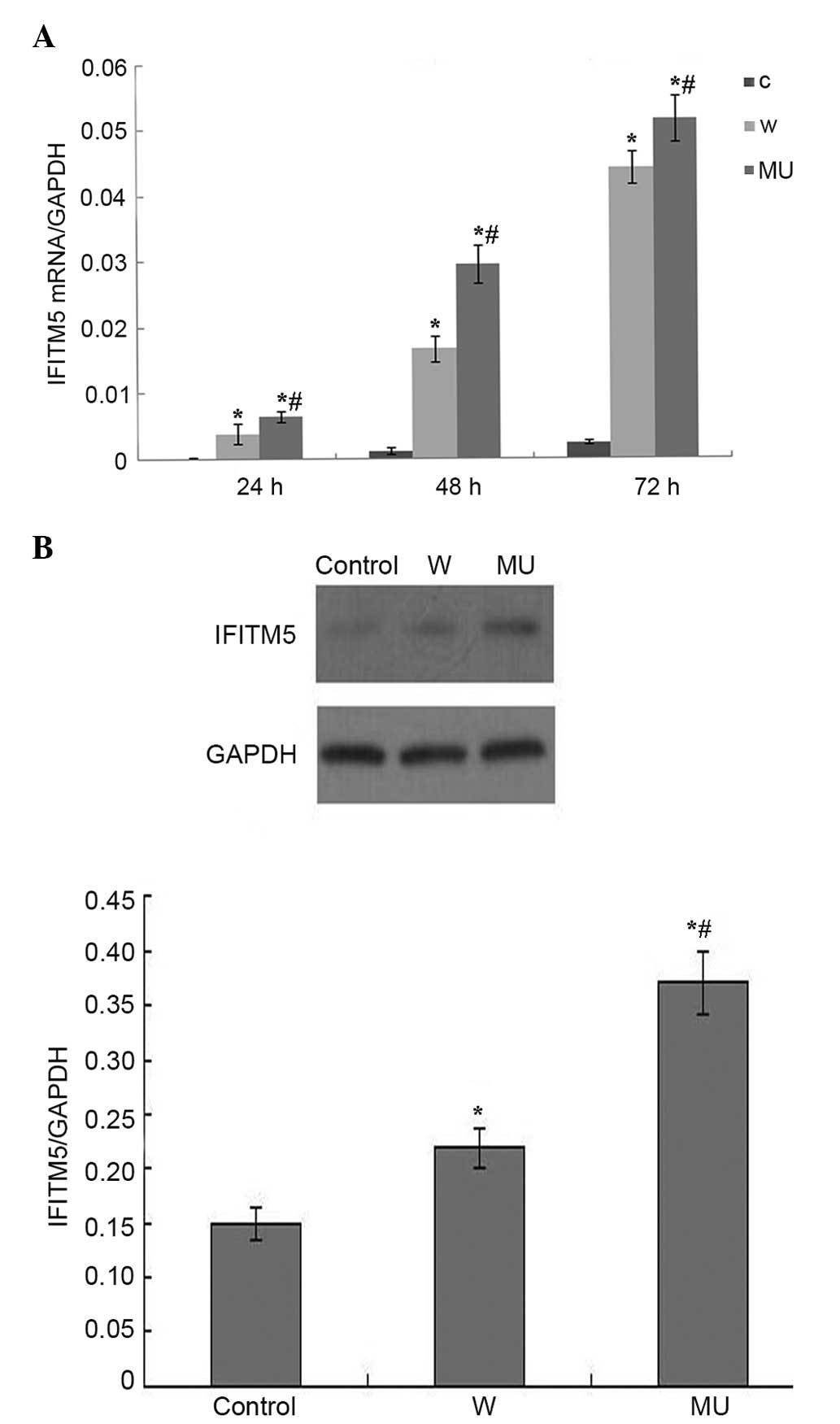
IFITM5 expression in vitro
RT-qPCR and western blotting indicated that IFITM5
was stably expressed in IFITM5 and IFITM5 c.-14C>T
mutation-transfected SaOS2 cells (Fig. 1A
and B). The mRNA expression levels of IFITM5 were increased in
a time-dependent manner. Protein expression was examined 72 h
post-transfection. The mRNA and protein expression levels were
markedly higher in W and MU groups compared with in C group. SaOS2
cells expressed the highest levels of IFITM5 72 h
post-transfection. In addition, there was a statistically
significant difference in IFITM5 expression between MU group and
the other two groups (P<0.05).
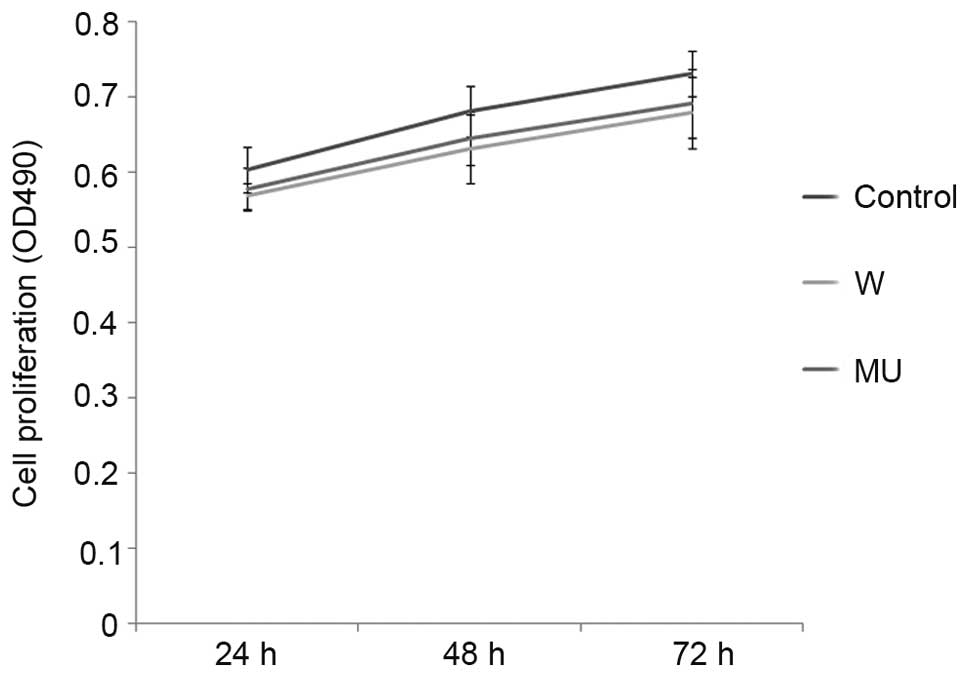
Effects of IFITM5 on cell
proliferation and apoptosis
Proliferative ability was measured by MTT assay 24,
48 and 72 h post-transfection in the three groups. Cell number
increased in a time-dependent manner, with little difference
between the W and MU groups, and the C group. There were no
significant differences between the groups (Fig. 2).
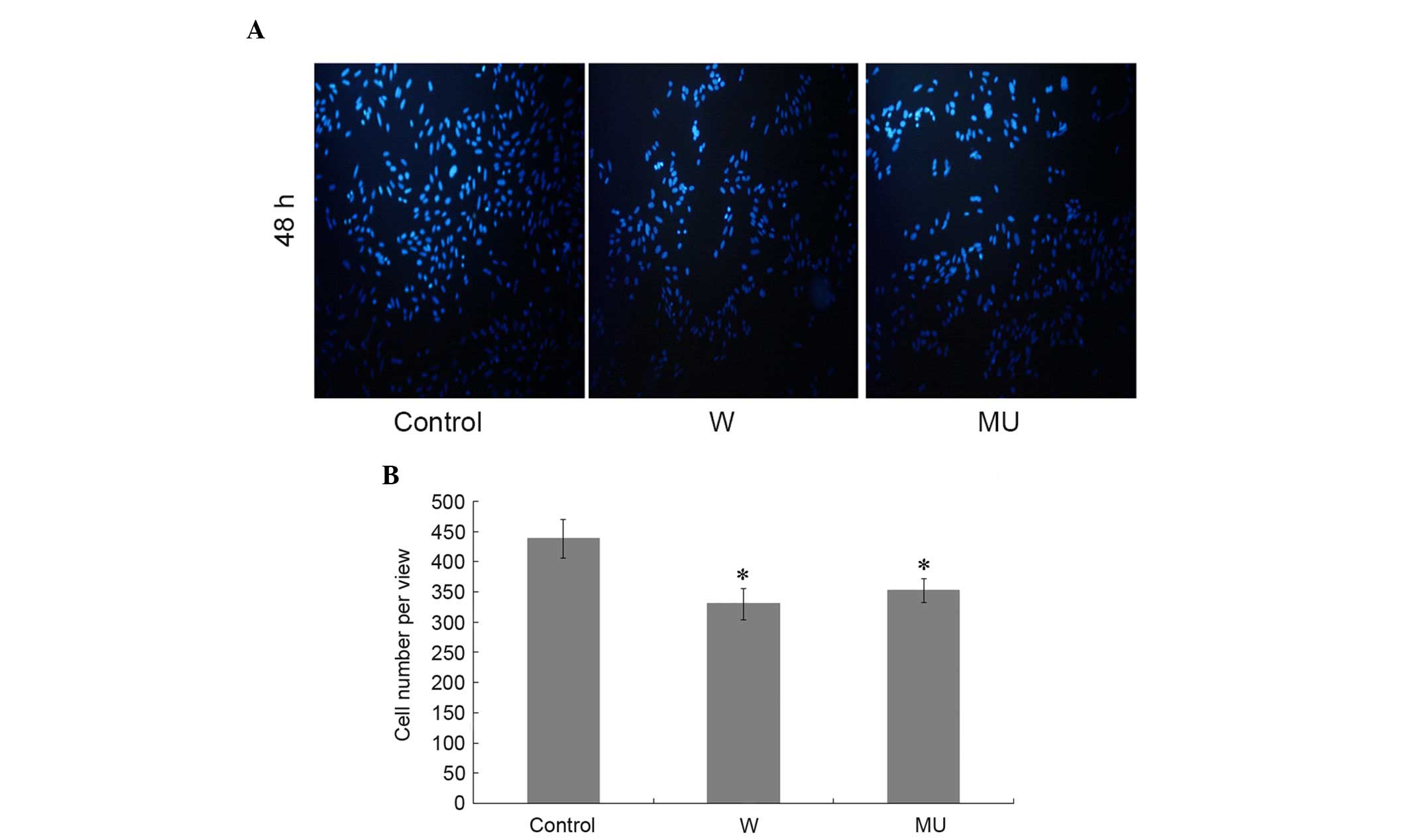
Hoechst dye solution was used to detect apoptosis in
the different groups 48 h post-transfection. The number of nuclei
was markedly decreased in W and MU groups compared with in C group.
There was no significant difference between groups W and MU
(Fig. 3A and B). These results
indicate that transfection with pcDNA4-IFITM5-E12-W or
pcDNA4-IFITM5-E12-MU enhance the apoptosis of SaOS2 cells. However,
the c.-14C>T mutation of IFITM5 appears to have no additional
effects on cell proliferation and apoptosis compared with IFITM5.
Furthermore, the treatments have no influence on the proliferation
of cells.
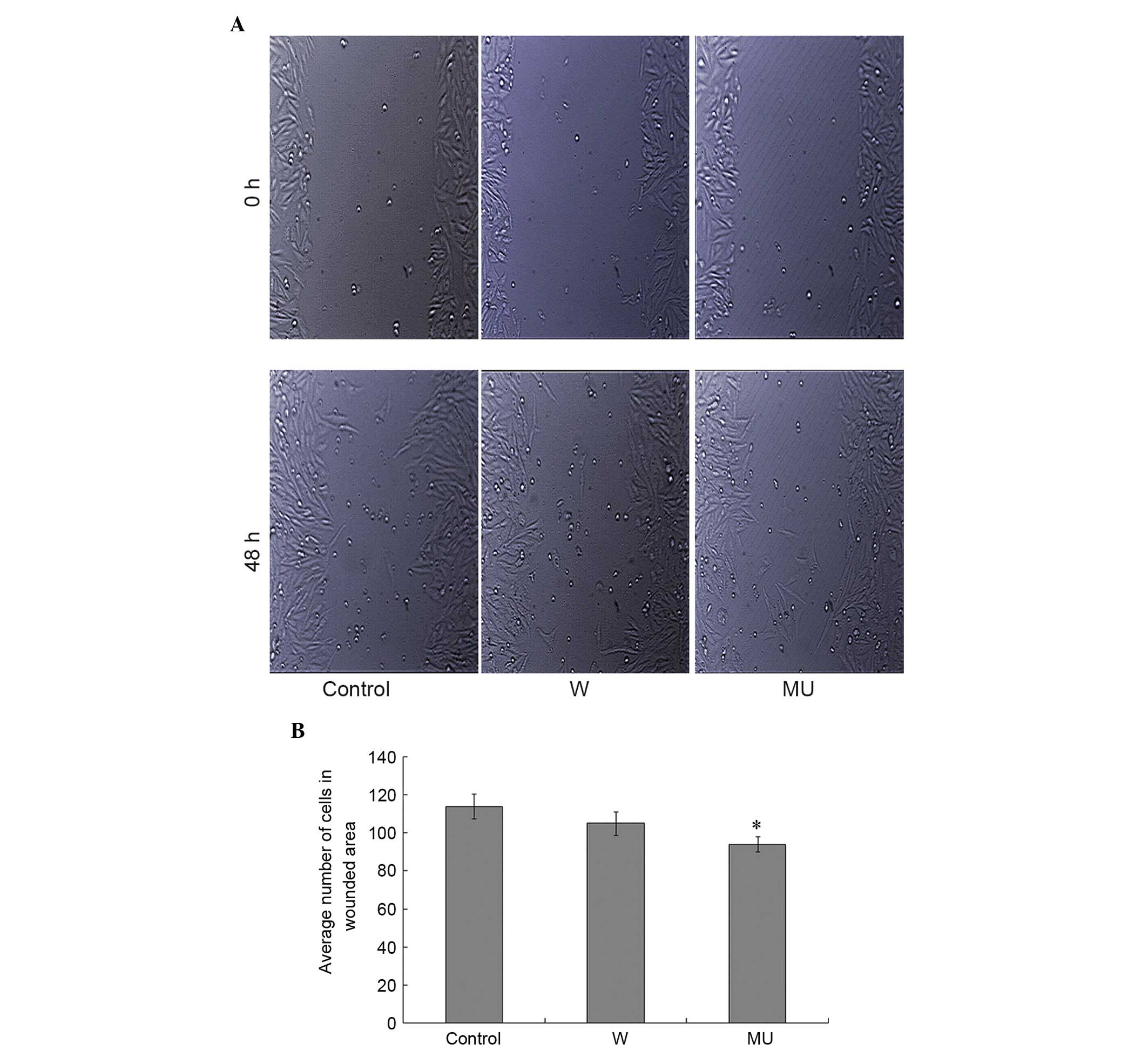
Effects of IFITM5 on cell
migration
In order to investigate the effects of
pcDNA4-IFITM5-E12-W and pcDNA4-IFITM5-E12-MU transfection on cell
migration, the number of cells that migrated into the wound surface
were counted 48 h post-transfection. The results indicated that
pcDNA4-IFITM5-E12-MU, rather than pcDNA4-IFITM5-E12-W,
significantly decreased SaOS2 cell migration compared with in C
group. There was no significant difference between W and MU groups
(Fig. 4A and B).
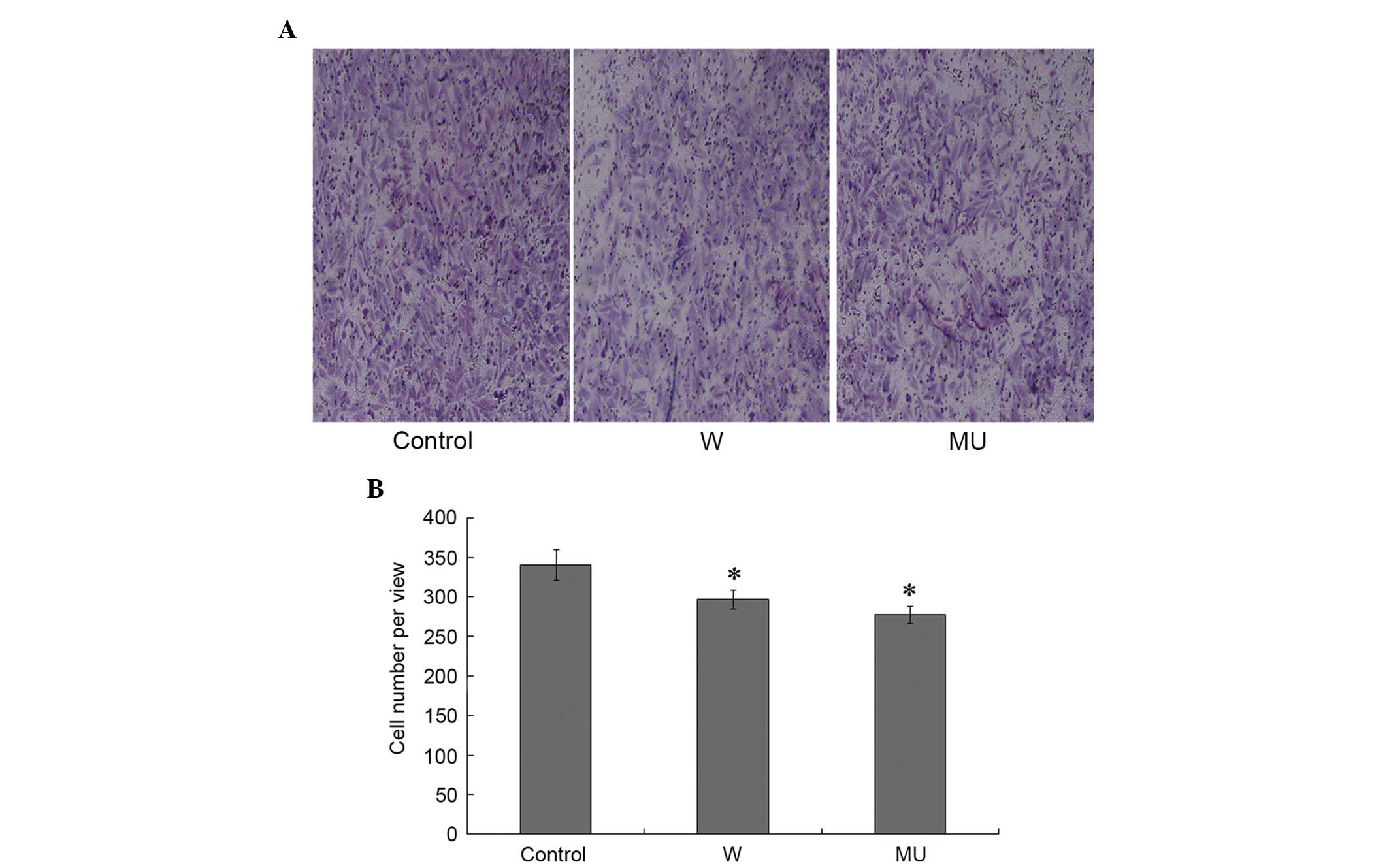
Effect of IFITM5 on Transwell
migration
Cell migration is a characteristic of malignant
tumor cells. The present study adopted a Transwell migration assay
to measure this ability. Cells that traversed the membrane were
counted under an optical microscope, in order to determine tumor
cell invasion. The number of migrated cells was significantly
decreased in W and MU groups compared with in C group. There were
no significant differences between W and MU groups (Fig. 5A and B). These results indicate that
IFITM5 and IFITM5 c.-14C>T mutation may suppress cell
migration.
IFITM5 and IFITM5 c.-14C>T mutation
stimulates osteogenic differentiation of SaOS2 cells
To determine whether IFITM5 and IFITM5 c.-14 C>T
mutation could affect mineralization in SaOS2 cells,
pcDNA4-IFITM5-E12-W and pcDNA4-IFITM5-E12-MU were transfected into
SaOS2 cells. Subsequently, cells in the three groups were induced
under osteogenic condition for 72 h. Alizarin Red S staining was
performed to detect calcification during differentiation (Fig. 6A and B). Calcium nodes in the cells
transfected with pcDNA4-IFITM5-E12-W and pcDNA4-IFITM5-E12-MU were
demonstrated increased staining compared with the control cells.
This suggested that IFITM5 and IFITM5 c.-14C>T mutation may
promote mineralization of SaOS2 cells. Previous studies have
reported that osteogenic differentiation is characterized by the
synthesis of ALP, OCN and Runx2 (12,13).
Therefore, the expression levels of ALP, Runx2 and OCN were
detected. Compared with the control cells, the expression levels of
ALP, Runx2 and OCN were increased following transfection with
pcDNA4-IFITM5-E12-W or pcDNA4-IFITM5-E12-MU. Compared with cells
transfected with pcDNA4-IFITM5-E12-W, the expression levels of ALP,
Runx2 and OCN were significantly increased in cells transfected
with pcDNA4-IFITM5-E12-MU (Fig.
6C-E). The results of the osteogenic gene marker detection were
concordant with the results of Alizarin Red S staining (Fig. 6A and B).
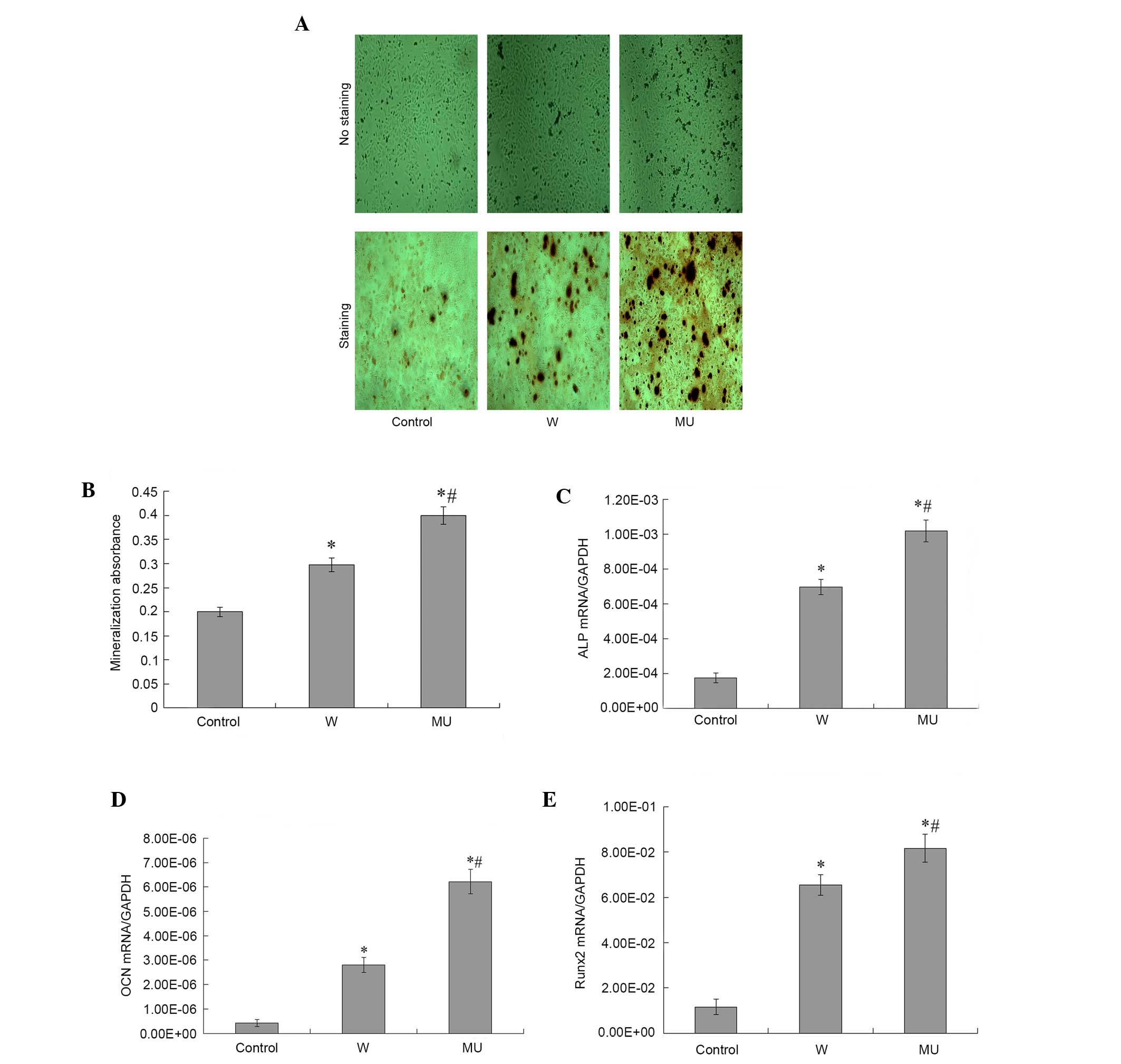 | Figure 6.Effects of IFITM5 on osteoblast
mineralization in SaOS2 cells. (A) Mineralized nodules were
measured by Alizarin Red S staining 72 h post-transfection with
pcDNA4-IFITM5-E12-MU or pcDNA4-IFITM5-E12-W. Magnification, ×100.
(B) For quantification, 10% cetylpyridinium chloride solution was
added to each well to elute Alizarin Red S. After complete elution,
the absorbance of the eluted solution was measured at 560 nm on a
microtiter plate reader. The absorbance was significantly decreased
in W and MU groups compared with in the control group. The
absorbance was markedly increased in MU group compared with W
group. (C-E) Effects of IFITM5 on the expression of osteogenic
markers. The expression of (C) ALP was compatible with the results
of Alizarin Red S staining. *P<0.05 vs. control group
(untransfected SaOS2 cells), #P<0.05 vs. W group. The
expression of (D) OCN and (E) Runx2 were compatible with the
results of Alizarin Red S staining. *P<0.05 vs. control group
(untransfected SaOS2 cells), #P<0.05 vs. W group
IFITM5, interferon-induced transmembrane protein 5; W,
pcDNA4-IFITM5-E12-W-transfected cells; MU,
pcDNA4-IFITM5-E12-MU-transfected cells; ALP, alkaline phosphatase,
OCN, osteocalcin; Runx2, runt-related transcription factor 2. |
Discussion
Osteosarcoma results in progressive pain and
pathological fracture or spinal cord compression, since a part of
intact bone is substituted with malignant cells with the growth of
metastatic bone tumor. During tumor growth, the balance of
osteoclast and osteoblast activity is disturbed. It is well-known
that stimulation of osteoclasts results in bone resorption, which
can be suppressed by the bone formation that results from
osteoblast differentiation. Bone resorption should be coupled to
bone formation in order to maintain skeletal homeostasis, and
imbalances in this process lead to various human diseases, such as
osteoporosis and osteopetrosis. Tumor growth stimulates activation
of osteoclasts, leading to bone resorption rather than bone
formation, thus resulting in bone destruction. Therefore, the
identification of a method that promotes SaOS2 cell mineralization,
without increasing tumor cell proliferation, migration and
invasion, is important.
IFITM5 encodes BRIL, which is involved in
mineralization and is expressed in the skeleton. Type V OI is
characterized by C>T transition at position-14 of the 5′
untranslated region of IFITM5. It has previously been reported that
Type V OI primary osteoblasts display increased mineralization
(10). The effects of
pcDNA4-IFITM5-E12-W and pcDNA4-IFITM5-E12-MU on mineralization in
SaOS2 cells were investigated in the present study. Furthermore,
the effects of pcDNA4-IFITM5-E12-W and pcDNA4-IFITM5-E12-MU were
examined on tumor cell proliferation, migration and invasion.
Osteosarcoma cells share several similar features to
undifferentiated osteoprogenitor cells, including a high
proliferative capacity and similar expression profiles of
osteogenic markers, such as Runx2, ALP and OCN (14,15).
Osteosarcoma cells can be induced to differentiate into mature
osteoblasts by certain compounds (16). Clinically, all-trans retinoic
acid-based differentiation therapy in acute promyelocytic leukemia
has achieved great success, and differentiation-based approaches
for the treatment of other malignant tumors has garnered attention
(17,18). Differentiation therapy may be
considered a promising alternative to conventional chemotherapy for
some malignancies (18). The aim of
this type of therapy is to activate endogenous differentiation
programs in cancer cells, resulting in cellular maturation of the
tumor and concurrent loss of the tumor phenotype (4). In the present study, osteoblast
differentiation of SaOS2 cells transfected with IFITM5 and
c.-14C>T mutation IFITM5 was induced by certain reagents. The
markers of osteoblast differentiation and mineralized bone nodules
were subsequently detected.
Compared with C group, W and MU groups exhibited
increased mRNA and protein expression levels of IFITM5. These
results indicate that IFITM5 was stably expressed in IFITM5- and
IFITM5 c.-14C>T mutation-transfected SaOS2 cells. Furthermore,
the mRNA and protein expression levels of IFITM5 were increased in
MU group compared with in W group. The stability of expression is
required for the subsequent steps.
The present study demonstrated that overexpression
of IFITM5 and IFITM5 c.-14C>T mutation had no effect on the
proliferation of SaOS2 cells; however, they did induce apoptosis.
Overexpression of IFITM5 and IFITM5 c.-14C>T mutation decreased
the migratory and invasive ability of tumor cells, and also induced
osteoblast differentiation of SaOS2 cells alongside increased bone
mineralization. These results may alleviate symptoms associated
with excessive bone resorption, and may promote tumor cell
differentiation and maturation. Previous studies have been
conducted regarding the therapeutic potential of treatments that
overcome differentiation defects associated with osteosarcoma and
prevent tumorigenesis (4,19). The results of the present study may
shed light on improving osteosarcoma treatment; however, the
mechanisms underlying IFITM5 and IFITM5 c.-14C>T mutation
overexpression-mediated osteoblast differentiation and
mineralization of SaOS2 cells remains unclear.
In conclusion, the present study examined the
effects of IFITM5 and IFITM5 c.-14C>T mutation overexpression on
SaOS2 osteosarcoma cells, with regards to tumor features, and
osteoblast differentiation and mineralization. IFITM5 is involved
in osteoblast differentiation and mineralization, and exerted
beneficial effects on tumor cell proliferation, apoptosis,
migration and invasion. These results may provide information
regarding the development of a novel treatment method that targets
IFITM5, and may provide a platform for future treatments of human
osteosarcoma. Future studies aim to develop a detailed
understanding of the role of IFITM5 in tumor biological
characteristics and osteogenic differentiation.
Acknowledgements
The present study was funded by the National
Science-Technology Support Plan (grant no. 2013BAI07B01) and the
Innovation Project of Shandong Academy of Medical Sciences.
References
|
1
|
Mundy GR: Metastasis to bone: Causes,
consequences and therapeutic opportunities. Nat Rev Cancer.
2:584–593. 2002. View
Article : Google Scholar : PubMed/NCBI
|
|
2
|
Wiens M, Wang X, Schlossmacher U,
Lieberwirth I, Glasser G, Ushijima H, Schröder HC and Müller WE:
Osteogenic potential of biosilica on human osteoblast-like (SaOS-2)
cells. Calcify Tissue Int. 87:513–524. 2010. View Article : Google Scholar
|
|
3
|
Thouverey C, Strzelecka-Kiliszek A,
Balcerzak M, Buchet R and Pikula S: Matrix vesicles originate from
apical membrane microvilli of mineralizing osteoblast-like Saos-2
cells. J Cell Biochem. 106:127–138. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Zhang N, Ying MD, Wu YP, Zhou ZH, Ye ZM,
Li H and Lin DS: Hyperoside, a flavonoid compound, inhibits
proliferation and stimulates osteogenic differentiation of human
osteosarcoma cells. PLoS One. 9:e989732014. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Hanagata N, Li X, Morita H, Takemura T, Li
J and Minowa T: Characterization of the osteoblast-specific
transmembrane protein IFITM5 and analysis of IFITM5-deficient mice.
J Bone Miner Metab. 29:279–290. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Moffatt P, Gaumond MH, Salois P, Sellin K,
Bessette MC, Godin E, de Oliveira PT, Atkins GJ, Nanci A and Thomas
G: Bril: A novel bone-specific modulator of mineralization. J Bone
Miner Res. 23:1497–1508. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Siegrist F, Ebeling M and Certa U: The
small interferon-induced transmembrane genes and proteins. J
Interferon Cytokine Res. 31:183–197. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Semler O, Garbes L, Keupp K, Swan D,
Zimmermann K, Becker J, Iden S, Wirth B, Eysel P, Koerber F, et al:
A mutation in the 50-UTR of IFITM5 creates an in-frame start codon
and causes autosomal-dominant osteogenesis imperfecta type V with
hyperplastic callus. Am J Hum Genet. 91:349–357. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Cho TJ, Lee KE, Lee SK, Song SJ, Kim KJ,
Jeon D, Lee G, Kim HN, Lee HR, Eom HH, et al: A single recurrent
mutation in the 5′-UTR of IFITM5 causes osteogenesis imperfecta
type V. Am J Hum Genet. 91:343–348. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Reich A, Bae AS, Barnes AM, Cabral WA,
Hinek A, Stimec J, Hill SC, Chitayat D and Marini JC: Type V OI
primary osteoblasts display increased mineralization despite
decreased COL1A1 expression. J Clin Endocrinol Metab.
100:E325–E332. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) Method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Noda M and Denhardt DT: Regulation of
osteopontin gene expression in osteoblasts. Ann N Y Acad Sci.
760:242–248. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Liu PP, Leung KS, Kumta SM, Lee KM and
Fung KP: Bone-specific alkaline phosphatase in plasma as tumour
marker for osteosarcoma. Oncology. 53:275–280. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Pensak MJ and Lieberman JR: Gene therapy
for bone regeneration. Curr Pharm Des. 19:3466–3473. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Glass DA II and Karsenty G: In vivo
analysis of Wnt signaling in bone. Endocrinology. 148:2630–2634.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Luo P, Yang X, Ying M, Chaudhry P, Wang A,
Shimada H, May WA, Adams GB, Mock D, Triche TJ, et al:
Retinoid-suppressed phosphorylation of RARalpha mediates the
differentiation pathway of osteosarcoma cells. Oncogene.
29:2772–2783. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Testi AM, Biondi A, Lo Coco F, Moleti ML,
Giona F, Vignetti M, Menna G, Locatelli F, Pession A, Barisone E,
et al: GIMEMA-AIEOPAIDA protocol for the treatment of newly
diagnosed acute promyelocytic leukemia (APL) in children. Blood.
106:447–453. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Pitha-Rowe I, Petty WJ, Kitareewan S and
Dmitrovsky E: Retinoid target genes in acute promyelocytic
leukemia. Leukemia. 17:1723–1730. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Gobin B, Moriceau G, Ory B, Charrier C,
Brion R, Blanchard F, Redini F and Heymann D: Imatinib mesylate
exerts anti-proliferative effects on osteosarcoma cells and
inhibits the tumour growth in immunocompetent murine models. PLoS
One. 9:e907952014. View Article : Google Scholar : PubMed/NCBI
|